Abstract
The Saccharomyces cerevisiae genome contains about 35 copies of dispersed retrotransposons called Ty1 elements. Ty1 elements target regions upstream of tRNA genes and other Pol III-transcribed genes when retrotransposing to new sites. We used deep sequencing of Ty1-flanking sequence amplicons to characterize Ty1 integration. Surprisingly, some insertions were found in mitochondrial DNA sequences, presumably reflecting insertion into mitochondrial DNA segments that had migrated to the nucleus. The overwhelming majority of insertions were associated with the 5′ regions of Pol III transcribed genes; alignment of Ty1 insertion sites revealed a strong sequence motif centered on but extending beyond the target site duplication. A strong sequence-independent preference for nucleosomal integration sites was observed, in distinction to the preferences of the Hermes DNA transposon engineered to jump in yeast and the Tf1 retrotransposon of Schizosaccharomyces pombe, both of which prefer nucleosome free regions. Remarkably, an exquisitely specific relationship between Ty1 integration and nucleosomal position was revealed by alignment of hotspot Ty1 insertion position regions to peak nucleosome positions, geographically implicating nucleosomal DNA segments at specific positions on the nucleosome lateral surface as targets, near the “bottom” of the nucleosome. The specificity is observed in the three tRNA 5′-proximal nucleosomes, with insertion frequency dropping off sharply 5′ of the tRNA gene. The sites are disposed asymmetrically on the nucleosome relative to its dyad axis, ruling out several simple molecular models for Ty1 targeting, and instead suggesting association with a dynamic or directional process such as nucleosome remodeling associated with these regions.
The Ty1 element is the most abundant retrotransposon in Saccharomyces cerevisiae. It targets very specific regions of the host genome upon integration, termed “integration windows,” lying 5′ of Pol III-transcribed genes, a set of genes principally consisting of the 275 nuclear tRNA genes. Prior studies have shown that these “windows” are open to Ty1 insertions only if the adjacent Pol-III transcribed gene is capable of being transcribed. These windows lie upstream of the site bound by transcription factor TFIIIB and in a genomic compartment that is mostly free of genes, also known as a “safe harbor” (Ji et al. 1993; Devine and Boeke 1996; Bolton and Boeke 2003; Bachman et al. 2004, 2005). As for target sequence requirements for Ty1 elements, an “anti-consensus” sequence consisting of the five base pairs comprising the target site duplication (TSD) was reported by Ji et al. as the target sequence requirement for Ty1 elements (Ji et al. 1993). However, a wide variety of sequences, including bacterial DNA, could serve as a target in vitro (Devine and Boeke 1996), showing there was no strict sequence requirement. Integration into DNA in vitro lacked the specificity for Pol III-transcribed genes observed in vivo (Ji et al. 1993; Devine and Boeke 1994; Rinckel and Garfinkel 1996), indicating that targeting requires some feature of functioning cellular DNA, presumably including both Pol III transcription factor binding and dynamic activities of upstream positioned nucleosomes. Deep sequencing has revolutionized the analysis of transposable element targeting due to its depth of coverage (Wang et al. 2007; Gangadharan et al. 2010; Guo and Levin 2010; Roth et al. 2011). Here, we examine the pattern of Ty1 insertions in considerable depth based on analysis of millions of independent Ty1 insertion events recovered via high-throughput sequencing of a tagged Ty1 element and its flanking yeast genomic sequences.
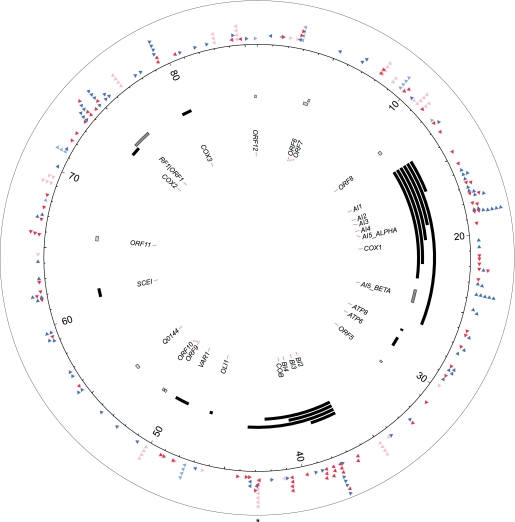
We find that Ty1 targets all types of Pol III genes and that the nuclear tRNA gene upstream regions are hit at a very high frequency; 93.6% of all of the recovered insertions occurred within 2 kb of the 275 nuclear tRNA genes, and 272 of these tRNA genes have an insertion within 2 kb upstream of the transcription start site. The 5S gene and most of the few other known Pol III genes are also preferentially hit in their 5′ flanking regions. In contrast, and in all cases, insertions were very rarely observed in the coding region of the Pol III genes. Unlike DNA transposon Hermes and Schizosaccharomyces pombe retrotransposon Tf1, both of which target nucleosome-free regions (Gangadharan et al. 2010; Guo and Levin 2010), Ty1 prefers targets occupied by nucleosomes. Detailed mapping of the sites of insertion relative to tRNA start sites revealed an exquisitely specific pattern of Ty1 insertions relative to the global pattern of nucleosome phasing upstream of tRNA genes. In this pattern, pairs of peaks separated by 70 bp rather precisely mirror the positions of peak nucleosome occupancy. The pattern is somewhat asymmetric with regard to Ty1 insertion in that the peak is slightly “right-shifted” and is observed at the three 5′-tRNA proximal nucleosomes, with the largest pair of peaks on the tRNA proximal nucleosome. This unusual specificity implicates DNA sequences defined by their specific geographical location on nucleosome lateral surfaces as the preferred sites of Ty1 integration. We suggest that these sites might be defined by a nucleosome remodeling process.
Results
Design of library
In order to collect very large numbers of independent Ty1 insertions, we utilized a Gal-Ty1 element bearing a tiny sequence tag placed so that after retrotransposition, it ends up very close to one end of the element (Lauermann et al. 1997). The donor element is like the original pGTy1-H3 plasmid described (Boeke et al. 1985), except that it carries a 25-bp synthetic DNA tag, referred to as ssb, inserted 24 bp from the 5′ end of the 3′ LTR, in its U3 region (Fig. 1A), exploited to specifically sequence newly transposed Ty1 elements without generating a background from the ∼35 endogenous Ty1 copies (Kim et al. 1998; Wheelan et al. 2006). Upon retrotransposition, this element duplicates the U3 region, resulting in progeny insertions in which the ssb-marked U3 element is at the 5′ end of the new insertion (Fig. 1B). A strategy directly adapted from earlier methods (Gangadharan et al. 2010) was used to design a custom primer that produced sequences consisting of the first 8 bp of the left LTR of the Ty1 element, followed by 30 bp of flanking sequences (Fig. 1C). Based on analysis of a Southern blotting experiment performed to measure the number of copies of the ssb tag in randomly selected colonies from the pool of cells analyzed (Fig. 1D), we conclude that the conditions used gave rise to ∼5 insertions per cell. In order to minimize the likelihood of “jackpot” events (early transposition events or PCR amplification events that might dominate the population), we pooled eight independent cultures prior to making DNA, then 12 sets of pooled cultures were separately PCR-amplified, and the amplicons were pooled prior to making the sequencing library (see Methods). The summary statistics for the sequencing are given in Table 1, and an extended set of mapping statistics is given in Supplemental Table S1.
Figure 1.
Experimental strategy for recovering genome-wide insertion data. (A) Donor plasmid structure showing ssb tag in 3′ LTR. (B) The 25-bp tag is transferred to the 5′ end during retrotransposition, placing the tag within 24 bp of the 5′ end of the element. (C) A custom sequencing primer was designed to produce sequences consisting of 8 bp of Ty1 terminus and 30 bp of flanking sequences. (D) Genomic Southern blot to estimate copy number of new insertions. The first 12 lanes represent random colonies isolated from 12 independent transposition libraries. (p) Diluted donor plasmid lane.
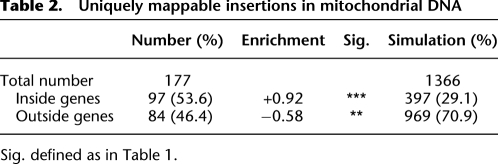
Table 1.
Ty1 targeting statistics
Analysis of target sequences
Ty1 duplicates five base pairs upon integration, defining the Ty1 target site duplication (Farabaugh and Fink 1980). Previous studies have noted that the base composition of the TSD is nonrandom, and can be summarized as an “anti-consensus sequence” of 5′ V-W-W-W-B 3′, where V is A, C, or G (“not T”), W is A or T, and B is T, C, or G (“not A”). It is important to note that none of these positions are invariant and thus represent statistical enrichments of specific nucleotides above or below what is expected from the overall genome-wide base composition. We examined the sequence properties of the nonredundant set of target sites identified by two methods. The first method, a standard sequence logo analysis, makes these patterns very clear (Fig. 2A). The second method was to align all of the pre-integration sequences, centered on the Ty1 insertion site and in the same orientation as the Ty1 insertion. By examining the counts of each base at each position, the anti-consensus can once again be seen clearly as a series of peaks and valleys symmetric about the middle base of the TSD, which we label position zero, with the TSD running from−-2 to +2 (Fig. 2B). We noted that the base pairs immediately surrounding the consensus also significantly deviate from randomness, allowing us to extend the symmetric anti-consensus sequence to read 5′ [T>A]3-V-W-W-W-B-[A>T]3 3′.
Figure 2.
Departures from randomness in Ty1 target sequences. (A) Sequence logo showing the TSD and bases immediately flanking it. (B) Plot of deviations from randomness of individual bases in the 100-bp window surrounding the TSD. For comparison, gray bars depict the span of the first and third quartile of composition of the yeast genome.
Virtually all tRNA gene upstream regions serve as targets
Our data firmly recapitulate the well-known fact that Ty1's most frequent type of target consists of nuclear tRNA genes (Table 1), the most abundant Pol III-transcribed gene class in the genome. As can be seen by plotting the positions of insertions as a function of chromosome length, the peaks of insertions that attain statistical significance (red peaks) are virtually all associated with tRNA genes (Fig. 3, red and blue triangles) and are associated with all but three tRNA genes. The expected number of insertions per kb along each chromosome is defined by a horizontal dashed blue line, (derived from simulations; see Methods). Even the small number of peaks associated with tDNAs that do fall below the random expectation cutoff show clearly visible peaks within the background. Earlier work suggests that certain tRNA genes are less good targets than others (Bachman et al. 2004). Zoomed-in views of these peaks, produced by the “CTY” viewer (Methods), show typical structures observed, consisting of Ty1 insertions clustered on the 5′ side of the tRNA target, with few to no insertions inside or 3′ from the gene. The insertions are symmetrically disposed in both Ty1 orientations relative to the target gene and span an integration window from about −70 to as far as −2000. A more complex pattern (bottom inset) results when there are two nearby tRNAs, and the tRNA-associated insertion peaks overlap. Finally, another pattern, in which one peak adjacent to a tRNA is separated by 6 kb from a second, non-tRNA associated peak, was shown to be an artifact of differences between reference annotations and the distribution of Ty1s in the host strain used in these experiments (see Supplement, “Analysis of double peaks”; Supplemental Fig. S1). Inset 2 shows the typical peak structure for a single DNA, obtained by analyzing the identical region shown in Inset 1 but mapped onto a “Ty-less” genome sequence derived from the reference. More extensive analysis of the tRNA targets in detail is described in the last section of the Results. First, we review other types of target and nontarget genomic compartments identified.
Figure 3.
Map of Ty1 insertions. (Horizontal dashed line) Boundary of statistical significance. (Red lines) Statistically significant deviations from randomness that align to tRNA upstream regions. (Blue and red triangles) tRNA genes. (Pale blue arrows) Pairs of peaks in which one is tRNA gene-associated and one ~6 kb away is not; the arrow is associated with the latter. Note that peaks for chromosome M (mtDNA) are all blue, indicating they do not reach statistical significance for enrichment. A Ty1 element is found in each case lying between the two peaks in the reference sequence. (Gray arrow) Insertions on Chrs IV and XV where there are no known Pol III transcripts. (Upper inset) Chr XVI Double Ty1 peak associated with one tRNA gene. (Middle inset) Same region after pre-existing reference Ty1 sequence was removed; note merging of peaks. See Supplemental Figure S1 for explanation of Chr XVI double peaks. (Insets) Small red tick marks are individual insertion positions. (Lower inset) Chr XIV, typical single tRNA peaks associated with two nearby tRNA genes in high resolution. Note how integration windows map to the 5′ side of tRNAs in all cases.
Insertions in mitochondrial or mitochondrially derived DNA
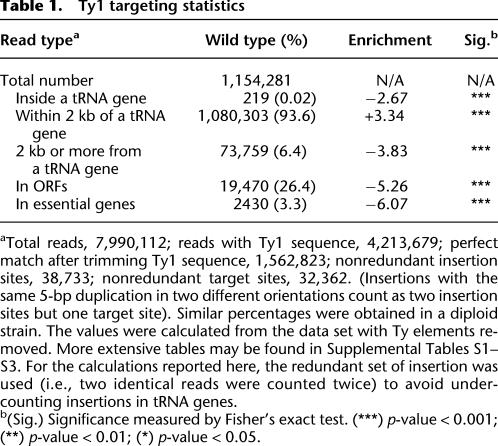
To our surprise, in each of the two libraries analyzed, we observed a significant number of reads corresponding to exact matches to mitochondrial DNA (mtDNA). Since these sequences were joined to a perfect match to the 5′ end of the Ty1 element and align to a wide variety of distinct and unique mtDNA locations, it is difficult to escape the conclusion that the Ty1 insertions into mtDNA are real. The number of insertions into mtDNA does not exceed the expectation for random integration into single copy DNA, and, of course, mtDNA is present in multiple copies per cell, i.e., retrotransposition into these sequences does not appear to be favored (Fig. 4). We believe that the most plausible explanation for this observation is that these events actually occurred in the nucleus (or perhaps even the cytoplasm), as will be discussed later. Ty1 insertions into mtDNA preferentially target regions of mtDNA that are transcriptionally active in mitochondria (Fig. 4; Table 2).
Figure 4.
Ty1 insertions in mitochondrially derived sequences. (Inner circle) mtDNA gene names; (rectangles) corresponding ORFs; (gray rectangles) dubious ORFs. Triangles in outer circle represent insertions; direction of triangle shows transcriptional orientation of Ty1.
Table 2.
Uniquely mappable insertions in mitochondrial DNA
Other Pol III-transcribed genes
The yeast ribosomal RNA (rRNA) is encoded by a 9.1-kb-long unit that is tandemly arrayed 100–200 times at a single locus on chromosome XII referred to as RDN1 (Petes 1979; Rustchenko et al. 1993). Within each repeat unit there are two distinct transcription units. The 35S pre-rRNA transcript is a product of RNA Pol I and is subsequently processed to form the mature 25S, 18S, and 5.8S rRNAs. The 5S gene is transcribed by RNA Pol III and is divergent from the 35S gene (Fig. 5). Because of the presence of 100–200 identical copies of yeast rDNA, all of which can serve as insertion targets, we can treat these sequences as a consensus to get an overview of Ty1 insertion specificity in that region. There is a large peak of Ty1 insertions upstream of the 5S genes. As is the case with the other Pol III transcribed genes, only the 5′ region is targeted, whereas the coding region is not, and the 3′ region is targeted but to a much lesser extent. The remainder of the rDNA is not hit at higher than expected frequencies (from simulation).
Figure 5.
Insertions in rDNA. Insertions are highly enriched in the rDNA on the 5′ side of the 5S gene and slightly enriched on the 3′ side but are otherwise randomly distributed within the rDNA region. (TSS) Pol I transcription start site.
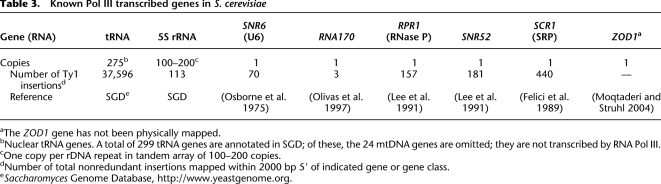
Six single-copy non-tRNA genes are reportedly transcribed by RNA Pol III (Table 3 and references therein). Interestingly, four of these genes have Ty1 insertion “windows” at their 5′ ends, very similar to those upstream of tRNA genes. However, two of the genes, RNA170 and ZOD1, lacked significant numbers of Ty1 insertions upstream, suggesting that these two genes somehow differ from all other Pol III genes. The RNA170 gene is unusual in that its “A box” and “B box” promoter elements are separated by nearly 100 bp, a much greater distance than is seen in tRNA genes. However, in SNR6, the distance between the boxes is even greater, and yet it is a good target for Ty1 (Devine and Boeke 1996), suggesting that this feature alone cannot account for the low frequency.
Table 3.
Known Pol III transcribed genes in S. cerevisiae
Protein coding genes
We examined which ORFs were most frequently hit by Ty1. As noted above, mitochondrially encoded genes were hit surprisingly often. When the nuclear (Pol II) genes with the largest number of insertions are examined, the vast majority of these are ORFs that lie relatively close to a Pol III gene, and when the distributions in those genes are examined, they are always biased toward the end of the gene closest to the nearest Pol III-transcribed gene. After filtering out tRNA-adjacent genes, the remaining ORFs are typically represented by zero, one, or a few insertions. Looking at the insertions in Pol II genes >5 kb away from a Pol III-transcribed gene, we do not find clusters of insertions, but we do see that the insertion sites cluster at the 5′ end of the gene, both at the −1 nucleosome upstream of the transcription start site and at the +1 nucleosome downstream from it, with a big trough at the nucleosome-free region (Supplemental Fig. S2). These results are consistent with earlier studies reporting insertions clustering at gene 5′ ends.
Genome-wide comparison to DNA transposon Hermes insertion sites
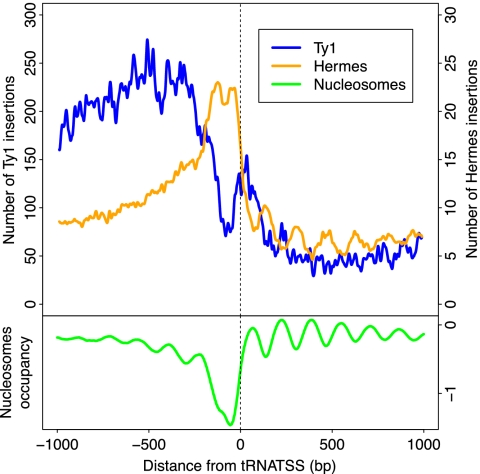
Previous studies of insertions of the Hermes DNA transposon have shown that this element prefers nucleosome-free regions as targets (Gangadharan et al. 2010). A comparison of Hermes and Ty1 integration patterns genome-wide is instructive. Whereas Hermes actively avoids nucleosome regions, Ty1 insertions are strongly anti-correlated with Hermes insertions (p < 0.001 by absolute minimum distance test) (SJ Wheelan, unpubl.) and correlated with nucleosomes (p < 0.001 by Jaccard test) (SJ Wheelan, unpubl.) (Fig. 6). We conclude that Ty1 elements target nucleosomes.
Figure 6.
Genome-wide comparison of Hermes and Ty1 insertion patterns, and nucleosome positions. The insertions are mapped relative to start or stop site of nearest pol II gene. (Blue line) Hermes insertions from Gangadharan et al. (2010); (yellow line) Ty1 insertions (this work); (green line) nucleosome positioning data (Lee et al. 2007) averaged across all tRNA genes.
tRNA gene hotspots
We examined the relative targetability of each tRNA isoacceptor type to be targeted. We expressed this as insertions/kb/tRNA copy across a window of 2 kb and found that these varied by only approximately twofold. All but three tRNA genes have insertions within 2 kb upstream; one of these three has an insertion at −2012, and the other two have nearby downstream insertions. It is difficult to map the landscape of insertions around these three genes, as they all lie within extremely repetitive regions, suggesting these are not outlier tRNA genes but simply ones for which our method is not effective at mapping insertions. When we examine the peak heights for different tRNA gene members within a single isoacceptor family we observe significant variability within each family, consistent with earlier findings (Bachman et al. 2004).
By mapping Ty1 insertions upstream of every tRNA gene, we observe distinct subsets of tRNA genes with different insertion patterns, which can be clustered into a number of groups (Fig. 7; Supplemental Fig. S3). While these subsets did not obviously correlate with isoacceptor type, they correlated both with the density of surrounding tRNA genes (shown in the figure) and with Pol III occupancy of the various tRNA genes (p < 0.002 by t-test, using Pol III binding data from Roberts et al. [2003]) (Moqtaderi and Struhl 2004). We presume that some specific aspects of the tRNA flanking sequences may share a structural or functional feature(s) yet to be uncovered that helps define the tRNA gene groups. This clustering revealed a clear periodic nucleosome-sized pattern in the insertions, extending at least 3–4 nucleosomes 5′ of the tRNA gene and, in some cases, considerably further.
Figure 7.
Ty1 insertion pattern clustergram. tRNA genes were clustered by integration site pattern similarity on 10-bp windows. Note nucleosome periodicity vertical streaks. Isoacceptors are coded on the left side with 42 distinct colors representing 42 isoacceptor species. On the right side of the map is tRNA density in the region flanking each tRNA shown. This density correlates with Ty1 insertion propensity in the 100-bp section 5′ of each tRNA but not with the overall Ty1 insertion patterns. We do not give a key to the exact identity of the tRNA behind each color code, as we are concerned only with whether or not the colors are clustered.
For every tRNA gene, the peaks are very heavily skewed toward the 5′ end and correspond to a window extending from approximately −60 to −700 (but occasionally as far as −2000) relative to the tRNA 5′ end. When plotted as a “butterfly” histogram, with insertions in the two orientations plotted above and below the x-axis, and aligned relative to the nearest tRNA 5′ end, the insertions form a roughly wedge-shaped distribution, with the largest number of insertions nearest the tRNA gene (Fig. 8A). The insertions in the two orientations insert with equal frequency at each hotspot, as can readily be seen in the butterfly histogram. The 3′ boundary of the integration window roughly corresponds to the boundary between the DNA region bound by the TFIIIB transcription factor and the first nucleosome positioned upstream of it. Interestingly, like the region bound by TFIIIB, the tRNA coding region itself is only very slightly enriched as a target; it is also non-nucleosomal, presumably because it is constitutively engaged with TFIIIC (Fig. 8A; Table 1). Insertions in the tRNA 3′ region are more highly enriched but not nearly as much as the 5′ region is, and nucleosome occupancy is once again observed, although not as strikingly as at the 5′ end.
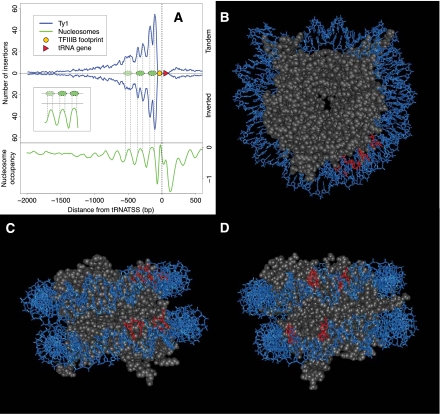
Figure 8.
Global distribution pattern of Ty1 insertions upstream of tRNA genes relative to nucleosome position. (A) The number of Ty1 insertions is plotted relative to all 275 nuclear tRNA genes. Blue curve above midline is Ty1 insertions inserted in the same transcriptional orientation as tRNAs (Tandem); line below is elements inserted in inverted orientation. (Triangle) tRNA gene; (oval) position of TFIIIB DNA footprint. (Green line) Nucleosome positioning data (Lee et al. 2007) averaged across all tRNA genes. (Inset) Relationship between nucleosome peaks and peaks of new insertions. (B) Modeling inferred site of dinucleotides targeted by Ty1 integration (red) on front view of yeast nucleosome (PDB 1ID3). Position zero on the nucleosome is oriented at “12 o'clock” in this view. The sites were inferred from an inter-peak spacing of 73 between the midpoints of the TSDs; this implicates TSDs spanning base pairs asymmetrically disposed relative to the nucleosome dyad axis (aligned to the vertical in this diagram; see Methods). In this view, only the first half of nucleosomal DNA is shown for clarity. (C) Bottom view. In this view, the two asymmetrically disposed dinucleotides targeted by Ty1 integration can be seen. The two phosphodiester bonds predicted to be cut by Ty1 integrase are directly across the major groove from each other. (D) Nucleosome lateral surface rotated ~30° clockwise relative to C for an “integrase-eye view.”
Specificity for nucleosomal DNA target segments in Ty1 hotspots
Insertion positions for all Ty1s upstream of tRNA genes were plotted as a high-resolution histogram relative to tRNA 5′ ends (i.e., oriented such that the tRNA is transcribed from left to right) (Fig. 8A). The butterfly histogram shows that within tRNA-adjacent hotspots, there is substantial substructure within the hotspot, with nucleosome-size periodicity, and the histogram also shows striking orientational symmetry, with equal height peaks for the two Ty1 orientations. Remarkably, the periodic pattern consists of two sharp peaks associated with each tRNA-adjacent nucleosome. The distance between each pair of peaks, ~70 bp, is remarkably consistent for the three tRNA-adjacent nucleosomes. Each pair of peaks is separated from the next by a center-to-center distance of 170 bp, aligning with the center-to-center distance of the tRNA-adjacent nucleosomes.
The nucleosome has strong rotational symmetry around the dyad axis, which consists of an imaginary vertical line drawn through the nucleosome, as visualized in Figure 8B. Surprisingly, alignment of the insertion distribution with nucleosome positioning data (green line) shows that the peak of nucleosome occupancy in each case does not lie precisely midway between the two Ty1 insertion peaks (inset of Fig. 8A). The Ty1 integration peak pairs are “right-shifted” from the nucleosome dyad axis of symmetry The pair of peaks closest to the tRNA contains the largest fraction of insertions, and each subsequent pair of peaks is smaller as one moves away to the left of the tRNA target gene.
The wedge shape and orientation independence of the high-resolution Ty1 insertion histogram is similar to previously described patterns of insertion at specific tRNA target genes (Devine and Boeke 1996; Bachman et al. 2004, 2005). However, although somewhat periodic patterns of insertion have previously been seen within some tRNA hotspots, the double peak pattern was only revealed by high-resolution deep sequencing analysis and has not been observed previously. The double peaks specifically implicate two highly specific regions of nucleosomal DNA as the highly preferred target for insertion (Fig. 8B–D). The region of the nucleosome in question can be identified by traversing along nucleosomal DNA 44 and 34 bp in each direction, starting at the dyad axis of the nucleosome (position zero), to define the presumed midpoint of the TSD (see Methods; Supplemental Fig. S4). We modeled the phosphodiester bonds inferred to be trans-esterified by Ty1 integrase by identifying the inferred 5′ ends of the TSD and coloring the adjacent nucleotides (Fig. 8C,D). This region lies in the “bottom half” of the nucleosome.
Discussion
Apparent insertions into mitochondrial DNA
There were a surprising number of insertions into mtDNA, and these appear to target transcribed regions (53.6% inserted into transcribed regions, an enrichment over the expected 29.1%; p-value < 0.001). The question is where in the cell the actual transposition event occurred and what state the mtDNA was in when it encountered the retrotransposition intermediate. The yeast nuclear genome is known to contain fragments of the mitochondrial genome, and it accumulates more fragments over time via double-strand break repair mechanisms (Thorsness and Fox 1990) If insertions occurred into mitochondrial-derived fragments present in the reference yeast nuclear genome sequence, they would not have been found in our search because the insertion sites would not have been uniquely mappable (as the sites are found both in the nuclear and mitochondrial genomes). We cannot definitively rule out the possibility that these molecules represent artefactual junctions made during library construction; however, the fact that regions transcribed in mitochondria argue rather strongly against this possibility. Thus, these insertions must have occurred either within the mitochondrion, which seems rather unlikely a priori as it is extremely difficult to introduce DNA into mitochondria, or they occurred into fragments of mtDNA from shattered mitochondrial DNAs either in the cytoplasm or in the nucleus (Ricchetti et al. 2004; Richly and Leister 2004; Lenglez et al. 2010). The apparent preference for transcribed regions could reflect insertion into in vivo cDNA previously formed in mitochondria, which can accumulate to significant levels, as well as into fragments of mtDNA itself.
Insertion sites deviate from randomness
The integration patterns of Ty1 as defined by deviations from randomness at specific distances from the target site duplication are characterized by close-range and long-range effects. This probably reflects steric constraints on integrase Ty1 DNA complexes that may ultimately be revealed by co-crystal structures of Ty1 integrase and a preferred target site. Such deviations from randomness are now well known for virtually every transposable element examined in detail (Waddell and Craig 1989; Bender and Kleckner 1992; Liao et al. 2000; Barr et al. 2006; Lewinski et al. 2006; Wang et al. 2007; Gangadharan et al. 2010; Guo and Levin 2010). Long-range deviations from randomness extending outward from the TSD (not shown) may reflect phased nucleosomes and suggest that tRNA genes occupy particular chromatin structures.
Ty1 targets nucleosomes in Pol III upstream regions
RNA polymerase III transcribes all tRNA genes as well as a few non-tRNA genes encoding structural RNAs in S. cerevisiae. The deep sequencing data obtained here validate numerous studies done on individual genes (Devine and Boeke 1996; Bachman et al. 2004) and a chromosome (Ji et al. 1993) that suggest Pol III gene upstream regions are the preferred targets of yeast retrotransposon Ty1. Previous studies examined a number of tRNA genes as well as the U6 gene, and most of the genes studied were found to be readily detectable as targets using a variety of assays; for example Ji et al. (1993) showed that all tRNA genes on chromosome III were targeted. Here, we expanded this type of analysis to the entire genome and provided evidence that every nuclear tRNA gene is, indeed, a target.
Some tRNA genes, however, appear to be significantly worse targets than others, a conclusion previously reached by Bachman et al. (2004). This is consistent with studies of Pol III transcription factor binding, which is associated with all tRNA genes (p < 0.002 by t-test, using Pol III binding data from Roberts et al. [2003]) (Moqtaderi and Struhl 2004). Pol III transcription factor binding strength is included in Figure 7 (left column) and correlates with the frequency of Ty1 insertion in the 100 bp just upstream of the tRNA gene. More surprisingly, two non-tRNA genes transcribed by RNA polymerase III, RNA170 and ZOD1, did not show any evidence of serving as targets. However, Pol III chromatin immunoprecipitation showed that these genes were very poor at recruiting Pol III and its associated factors or had unusually skewed amounts of the protein complexes normally associated with Pol III itself—TFIIIB and TFIIIC. RNA170 is a member of a set of eight ETC genes that efficiently bind TFIIIC but not TFIIB or Pol III itself (Moqtaderi and Struhl 2004). However, a transcript has clearly been detected from the region, suggesting that this transcript is either made at an extremely low rate or is not made by a standard Pol III transcription mechanism.
Clustering of the tRNA targets and alignments of all insertions relative to the tRNA start sites showed very clearly a nucleosome-spacing of the hotspots, which were highly correlated with the positions of nucleosomes in the region. Anti-correlation with Hermes integration sites further corroborated this trend. Insect DNA transposon Hermes, the insertion of which was analyzed in yeast, avoids nucleosomes, as does the Tf1 element in S. pombe (Guo and Levin 2010). However, recent studies on gammaretrovirus MLV as well as HIV show that these viruses also preferentially insert into nucleosomal DNA. However, the patterns of retroviral insertions into nucleosomes is distinct from that of Ty1, hitting a wide variety of outward-facing major groove nucleosomal DNA sites distributed throughout the nucleosome (Wang et al. 2007b; Roth et al. 2011).
Specificity for nucleosomal position in Ty1 hotspots
The hotspots for Ty1 integration, namely the collection of 275 tRNA upstream regions, are a diverse group of sequences but share arrays of well-phased nucleosomes. These are presumably phased in response to the tight binding of the transcription factors TFIIIC and TFIIIB to the body of the tRNA gene and the ∼70 bp upstream of it, respectively. Mutations in the gene encoding TFIIIB component Bdp1 lead to altered nucleosome phasing upstream of tRNA genes, consistent with the idea that the TFIIIB factor directly affects nucleosome positioning upstream of the tRNA gene (Bachman et al. 2005).
Although a consensus position on the nucleosome can be identified from these studies, the analyses of the patterns of insertion at individual tRNA genes show that there is significant variation at individual target genes. Clustering of the integration pattern shows these variations are not determined by tRNA isoacceptor type, and perhaps only slightly (in the tRNA-proximal nucleosome) by Pol III occupancy, but instead appear to be dominated by other genomic features, such as the presence of one or more additional Pol III gene targets nearby.
Next, using the distance between the sharp peaks in the tRNA proximal nucleosome as a guide (Fig. 8A) and knowing the relationship between the target site duplication and the phosphodiester bonds actually cut, we modeled the positions on the nucleosome recognized by the Ty1 integration machinery. Remarkably, this identified a pair of asymmetrically disposed phosphodiester bonds, consistent with attack on a nucleosomal DNA substrate at very specific preferred nucleosomal positions, near the “bottom” of the nucleosome (Fig. 8B,C). One remarkable aspect of the mapping relative to the structure is that the two “hot” regions lie adjacent to each other on the nucleosome lateral surface near “4 o'clock”; however, the symmetrically disposed counterpart of this structure (at “8 o'clock”) is not associated with high target activity. This fact, together with the finding of the two peak sites of insertion activity, tends to rule out certain models for how the nucleosomal regions are targeted. Had the sites been symmetrically disposed, several models would make sense; e.g., interactions, either direct or indirect, with nearby nucleosome surfaces might have led to the targeting. In this case, we would expect to see four hotspot peaks rather than two associated with each nucleosome, as the nucleosome is inherently rotationally symmetrical. The main conclusions of this paper agree well with the data reported by Baller et al. (2012).
It is clear that the insertions are targeted to a specific surface of the nucleosome. This nucleosome domain may be much more accessible in the highly dynamic chromatin located upstream of tRNA genes, which is among the most dynamic in the genome (Dion et al. 2007). These facts suggest integration may be associated with a dynamical process, such as chromatin remodeling. Asymmetry in remodeling makes sense, as remodeling is expected to be directional with respect to the tRNA target gene.
Methods
Strains, plasmids, and media
The strain used for the sequencing experiment was BY348 (MATα his3Δ200 ura3-167 GAL+) containing plasmid pJEF2365. pJEF2365 is a derivative of pVIT41 (Lauermann et al. 1997) with a higher performing 2μ origin fragment derived from pJEF1562 (Monokian et al. 1994). The Ty1 element in this plasmid is tagged with a sequence called ssb (Fig. 1A). The media used were YPD, SC–Ura containing either 2% glucose (glu), 2% galactose, (gal), or 1% raffinose (raf), and SC+ 5-FOA (fluoro-orotic acid). Media were prepared as described (Rose et al. 1990; Smith and Boeke 1997).
Sequencing library construction
Twelve individual colonies of strain BY348 were grown on SC–Ura glucose plates grown at 30°C. These were used as the starting material for 96 independent galactose inductions to minimize jackpot effects. The colonies were picked into 5 mL SC–Ura 1% Raf and grown at 30°C overnight. For each of the 12 cultures, corresponding to one column of a 96-deep-well plate, the following were added to each well: a sterile 3-mm glass bead, 200 μL of the above inoculum, and 1 mL SC–Ura gal. The plate was covered with Air Pore tape and grown at 22°C at 240 rpm for 48 h in a refrigerated air shaker. A total of 1150 μL were removed from each well and replaced with 1150 μL of YPD and grown at 30°C overnight with shaking to allow plasmid loss. A total of 1150 μL were removed from each well and replaced with 1150 μL of SC + 1 mg/mL 5-FOA medium and grown overnight with shaking to select for plasmid-free cells. The eight cultures in each column were pooled to make a DNA preparation as described (Boeke et al. 1985). The DNA was then extracted three times with phenol/chloroform/isoamyl alcohol, and ethanol precipitated.
Custom Illumina adapters were ordered as duplexes from IDT (top strand oligonucleotide, 5′-pTAGTCCCTTAAGCGGAG-NH2; bottom strand oligonucleotide, 5′-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC). Two μG gDNA was digested to completion with 60 U MseI and 120 U BglII in a 100-μL reaction overnight and purified on a QiaQuick column (Qiagen). MseI-compatible adaptors were ligated overnight at 16°C and heat-inactivated at 70° for 15 min.
The purpose of the BglII was to cut molecules derived from the 3′ LTR which would only give rise to internal Ty1 sequence. Each DNA was used to template 48 PCR reactions with a Clontech Advantage PCR kit using the following primers: 5′-aatgatacggcgaccaccgagatctTAGAGGATCCCGGGAGCTC and 5′-caagcagaagacggcatacgagctcttccgatctGTAATACGACTCACTATAGGGCTC. After a touchdown PCR was run (94°C, 1 min; [94°C, 2 sec, 75°C 1 min, 3 cycles], [94°C, 2 sec, 72°C 1 min, 4 cycles], [94°C, 2 sec, 68°C, 15 sec, 72C, 1 min, 7 cycles], [94°C, 2 sec, 62°C, 15 sec, 72C, 1 min, 30 cycles]), the 200–800-bp- long products were pooled and gel purified on a 1% agarose TTE gel, carefully avoiding the primer bands. Twenty μL of 14 ng/μL DNA was sequenced on an Illumina GAII sequencer at the University of California Riverside sequencing facility using custom primer JB13646 (GAGGATCCCGGGAGCTCTGATAGTTGATTTC).
Estimating Ty1 retrotransposition frequency in the population
A Southern blot was used to profile the number of ssb-containing bands in a sample of 12 randomly chosen colonies from the FOA-resistant (donor plasmid free) population. Genomic DNA was digested with AseI, run on an agarose gel, blotted, and then the ssb segment was radiolabeled and used as probe, ensuring that only new insertions would be identified. Seventy bands were counted in the 12 strains, using a conservative counting method in which darker bands were counted as two bands and lighter bands were counted as one band. In earlier work, we evaluated the efficiency with which new insertions inherited one or two tagged LTRs (Lauermann et al. 1997). Because only a fraction of the progeny Ty1 insertions carries two ssb-tagged LTRs, and some carry only one, each band corresponds to 0.65 Ty1 element ssb-tagged in the 5′ LTR. Thus, we estimate the average copy number of 5′-LTR marked elements to be ∼5 per cell in this population. As expected, many elements are tagged in the 3′ LTR; these uninformative reads were filtered out.
Filtering of the sequences
We considered only those Illumina reads that started with the Ty1 LTR, allowing one “N” mismatch. After trimming sequence not derived from the yeast genome, the reads were aligned to the yeast genome using Bowtie (Langmead et al. 2009), using a seed of 20 bp. Only uniquely and perfectly mapping reads were retained; reads mapping to identical genomic positions were counted only once to avoid biases due to PCR or other amplification artifacts.
CTY viewer
While chromosomal distribution histograms give a good “big picture” of the insertions, it is necessary to zoom in to look at effects on individual genes. A web viewer, “CTY,” was written to enable users to visualize the distribution of Ty1 insertions associated with yeast genome features, including tRNA genes and ORFs. The viewer is available at http://sjwheelan.som.jhmi.edu/data.html.
Data sets
Gene and tRNA coordinates are from Saccharomyces Genome Database (http://www.yeastgenome.org/). Nucleosome occupancy data are from Lee et al. (2007). The nucleosome mapping data used here (Lee et al. 2007) is from a 4-bp resolution tiling array. As we are combining hundreds of thousands of insertion events to achieve these results, our effective resolution for insertion mapping is 1 bp.
Clusters
Clusters of insertions are genomic regions where the density of insertions is higher than expected by chance (defined by the third quartile of peak height from 100 randomizations).
Simulation
We created a simulated data set of Ty1 insertions to account for biases introduced by limitations of the experimental procedure; virtual fragmentation followed by random sampling of insertion sites generated a set of sites the same size as the experimentally recovered set of insertions.
All analysis was done using the Python programming language (http://www.python.org/) and the R statistical package (R Development Core Team 2010).
Modeling integration site on the nucleosome
Several measurements were made on the inter-peak distance on both the raw data and the smoothed data (Supplemental Fig. S3). Because the integration data were “tightest” for the first nucleosome, both in terms of numbers of insertions associated and because this nucleosome appears to be the most well-positioned, we focused on mapping insertions relative to it. The pair of peaks corresponding to the nucleosome closest to the tRNA was, on average, 73 bp (even when independently calculated based on three separately clustering groups of tRNA genes for which first nucleosome spacing relative to the tRNA varied). The two peaks flanked the position of peak nucleosome occupancy (assumed to reflect the position of nucleosome position zero, or “12 o'clock” in Fig. 8B) asymmetrically, with the peaks of Ty1 integration displaced ~5 bp off-center relative to the position of peak nucleosome occupancy, toward the direction of the target tRNA, i.e., “right-shifted.” Averaging the data from the three hierarchically clustered groups of tRNAs, the middle base pairs of the 5-bp target site duplication were predicted to be 44 and 34 bp from the phosphodiester bond at position zero. This placed phosphodiester bonds predicted to be attacked by Ty1 integrase (assuming a 5′ overhang in the transposition intermediate) at dinucleotides 41/42 on the top strand and the complement of dinucleotides 46/47 on the bottom strand; the second site was similarly mapped to positions 112/113 (top strand) and 117/118 (complement of the bottom strand). These dinucleotides were mapped onto the yeast nucleosome crystal structure.
Data access
The data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE33986.
Acknowledgments
We thank Joshua Baller and Daniel Voytas for communicating results on matters of symmetry prior to publication and Nancy Craig for helpful discussions. This work was supported in part by NIH grant GM36481 to J.D.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.129460.111.
References
- Bachman N, Eby Y, Boeke JD 2004. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res 14: 1232–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev 19: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF 2012. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res (this issue). doi: 10.1101/gr.129585.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD 2006. HIV integration site selection: Targeting in macrophages and the effects of different routes of viral entry. Mol Ther 14: 218–225 [DOI] [PubMed] [Google Scholar]
- Bender J, Kleckner N 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci 89: 7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500 [DOI] [PubMed] [Google Scholar]
- Bolton EC, Boeke JD 2003. Transcriptional interactions between yeast tRNA genes, flanking genes, and Ty elements: A genomic point of view. Genome Res 13: 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine SE, Boeke JD 1994. Efficient integration of artificial transposons into plasmid targets in vitro: A useful tool for DNA mapping, sequencing, and genetic analysis. Nucleic Acids Res 22: 3765–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine SE, Boeke JD 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 10: 620–633 [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Fink GR 1980. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature 286: 352–356 [DOI] [PubMed] [Google Scholar]
- Felici F, Cesareni G, Hughes JM 1989. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol Cell Biol 9: 3260–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ, Craig NL 2010. Inaugural Article: DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc Natl Acad Sci 107: 21966–21972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Levin HL 2010. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res 20: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018 [DOI] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8: 464–478 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauermann V, Hermankova M, Boeke JD 1997. Increased length of long terminal repeats inhibits Ty1 transposition and leads to the formation of tandem multimers. Genetics 145: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Evans CF, Engelke DR 1991. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci 88: 6986–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Lenglez S, Hermand D, Decottignies A 2010. Genome-wide mapping of nuclear mitochondrial DNA sequences links DNA replication origins to chromosomal double-strand break formation in Schizosaccharomyces pombe. Genome Res 20: 1250–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. 2006. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog 2: e60 doi: 10.1371/journal.ppat.0020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao GC, Rehm EJ, Rubin GM 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci 97: 3347–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monokian GM, Braiterman LT, Boeke JD 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: Mutations, transposition, and dominance. Gene 139: 9–18 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K 2004. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol 24: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas WM, Muhlrad D, Parker R 1997. Analysis of the yeast genome: Identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res 25: 4619–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne JC Jr, Palumbo G, Brewer HB Jr, Edelhoch H 1975. The self-association of the reduced ApoA-II apoprotein from the human high density lipoprotein complex. Biochemistry 14: 3741–3746 [DOI] [PubMed] [Google Scholar]
- Petes TD 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci 76: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2010 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B 2004. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol 2: E273 doi: 10.1371/journal.pbio.0020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D 2004. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Rinckel LA, Garfinkel DJ 1996. Influences of histone stoichiometry on the target site preference of retrotransposons Ty1 and Ty2 in Saccharomyces cerevisiae. Genetics 142: 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci 100: 14695–14700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P 1990. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Roth SL, Malani N, Bushman FD 2011. Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol 85: 7393–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F 1993. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol 175: 7189–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11: 241–254 [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Fox TD 1990. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature 346: 376–379 [DOI] [PubMed] [Google Scholar]
- Waddell CS, Craig NL 1989. Tn7 transposition: Recognition of the attTn7 target sequence. Proc Natl Acad Sci 86: 3958–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD 2007. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17: 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD 2006. Transposon insertion site profiling chip (TIP-chip). Proc Natl Acad Sci 103: 17632–17637 [DOI] [PMC free article] [PubMed] [Google Scholar]