Abstract
Evolutionary rate covariation (ERC) is a phylogenetic signature that reflects the covariation of a pair of proteins over evolutionary time. ERC is typically elevated between interacting proteins and so is a promising signature to characterize molecular and functional interactions across the genome. ERC is often assumed to result from compensatory changes at interaction interfaces (i.e., intermolecular coevolution); however, its origin is still unclear and is likely to be complex. Here, we determine the biological factors responsible for ERC in a proteome-wide data set of 4459 proteins in 18 budding yeast species. We show that direct physical interaction is not required to produce ERC, because we observe strong correlations between noninteracting but cofunctional enzymes. We also demonstrate that ERC is uniformly distributed along the protein primary sequence, suggesting that intermolecular coevolution is not generally responsible for ERC between physically interacting proteins. Using multivariate analysis, we show that a pair of proteins is likely to exhibit ERC if they share a biological function or if their expression levels coevolve between species. Thus, ERC indicates shared function and coexpression of protein pairs and not necessarily coevolution between sites, as has been assumed in previous studies. This full interpretation of ERC now provides us with a powerful tool to assign uncharacterized proteins to functional groups and to determine the interconnectedness between entire genetic pathways.
A protein's amino acid sequence does not evolve at a constant rate over time, as shown by the rate variation between different evolutionary lineages (Li et al. 1987). Although each individual protein has a unique pattern of rate variation, the rates of physically interacting proteins have been observed to covary over a phylogenetic tree, as has been observed among prokaryotes, abalone, yeast, and Drosophila species (Pazos and Valencia 2001; Hakes et al. 2007; Clark et al. 2009; Clark and Aquadro 2010). This signature, which we term evolutionary rate covariation (ERC), is detected by comparing a protein's individual branch rates to the corresponding branch rates of another protein (Fig. 1). As such, the rates of two proteins will covary if they have experienced similar acceleration and deceleration of their evolutionary rate over various branches of a phylogenetic tree. Note that a pair of proteins could evolve at very different average rates and still exhibit ERC. Similarly, consistently fast evolving proteins or slow evolving proteins would show correlated average rates but would not necessarily show ERC.
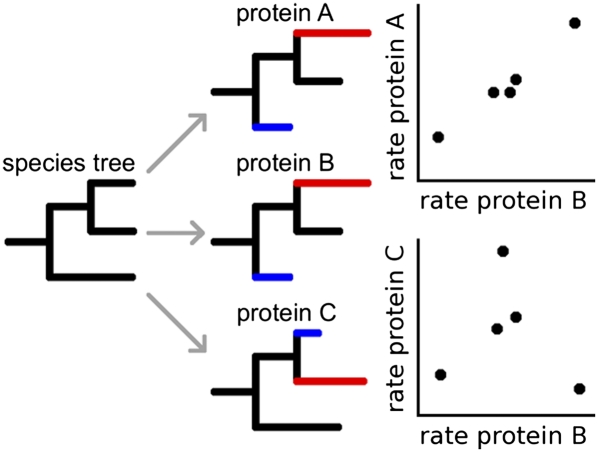
Figure 1.
Parallel change in evolutionary rate leads to rate covariation. Most proteins encoded in a genome evolve over the same species tree and so have evolved for the same amount of chronological time over each branch. Yet individual proteins experience varying rates of sequence evolution over those same branches. Hypothetical protein “A” experienced rapid evolution in one species lineage (red branch) and an exceptionally slow rate of evolution in another (blue branch). Another protein “B” experienced very similar rate variation during the evolution of these species, so that its branch rates are positively correlated with the rates of protein A (upper plot). Their evolutionary rate covariation suggests a relationship between A and B. Another protein, “C,” also experienced acceleration and deceleration, but its evolutionary pattern did not result in ERC with protein B (lower plot). Note that the values in these plots are rates of sequence evolution normalized to the expected rate given the species tree.
Many studies have aimed to improve the detection of ERC because it is thought to provide a means to infer new physical interactions (Fraser et al. 2004; Pazos et al. 2005; Sato et al. 2005). However, our incomplete understanding of the causes of ERC makes it difficult to confidently make such biological inferences. Early studies assumed that ERC resulted solely from intermolecular coevolution that occurs at the physical interface between interacting proteins (Goh et al. 2000; Pazos and Valencia 2001;). There is evidence that some residues across interaction interfaces change in a statistically correlated manner to maintain binding complementarity (Moyle et al. 1994; Travers and Fares 2007; Madaoui and Guerois 2008; Kann et al. 2009). However, it is unclear if intermolecular coevolution at the limited number of interface residues is sufficient to create the observed signatures of ERC that typically involve the entire protein sequence (Lovell and Robertson 2010). Furthermore, Hakes et al. (2007) found that ERC is not stronger for the actual interface residues compared to all surface residues, although Kann et al. (2009) found evidence to the contrary. However, we and others have observed that physically distant members of several protein complexes correlate despite their lack of direct physical interaction (Juan et al. 2008; Clark and Aquadro 2010).
Moving away from the simple intermolecular coevolution model, there is increasing emphasis in the field on additional forces that could affect evolutionary rate over the entire protein (Lovell and Robertson 2010). One potential force is the dispensability of a pathway. For example, as a species occupies a new environment, a particular metabolic pathway could become more important and, hence, become more constrained, while other pathways could be allowed to drift and diverge due to lack of use. Another potential force is a protein's expression pattern. Expression level is highly correlated with the rate of amino acid evolution (Duret and Mouchiroud 2000; Pal et al. 2001; Drummond et al. 2006), so that a change in expression for all the proteins in a pathway could affect their evolutionary rates in a correlated way.
We considered these and other potential driving forces of ERC on a proteome-wide scale using the well-annotated yeast (Saccharomyces cerevisiae) protein interactome and the full genome sequences of 18 budding yeast species. We demonstrate that physically and genetically interacting proteins exhibit ERC on a proteome-wide scale. We also reveal that noninteracting but functionally related proteins show significant ERC. Finally, we quantify the forces contributing to ERC, implicating cofunctionality and change in expression level as two major, independent contributors. Thus, ERC across the genome most reliably reflects shared biological function and does not necessarily imply direct physical interaction. In fact, we propose that physical interaction and coevolution, although they may be important for some ERC signals, are minor contributors to genome-wide patterns of ERC. Our findings widen the breadth of biological insight that can be gained from ERC to include entire pathways and functional groups.
Results
Calculating evolutionary rate covariation proteome-wide
We compiled 4459 orthologous protein groups from 18 budding yeast species (family Saccharomycetaceae) and estimated each protein's phylogenetic branch lengths (sequence divergence) over the 18-species tree (Fitzpatrick et al. 2006). Raw branch lengths were then transformed into their relative deviation from that expected in an average proteome-wide tree (Fig. 1). This transformation greatly improves power to discern functionally related from unrelated protein pairs (Sato et al. 2005). ERC was then calculated between all protein pairs (∼8.4 million pairs) as the correlation coefficient (r) of their transformed branch rates, so that possible ERC values range from −1 (negative correlation) to 1 (positive correlation).
Functionally related proteins exhibit evolutionary rate covariation
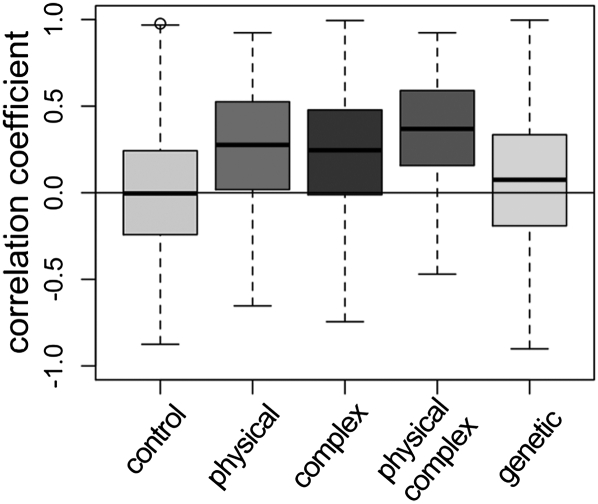
We first assessed the proteome-wide relationship between ERC and functionally related proteins. We designated a control set of protein pairs between which there were no annotated functional relationships or interactions; the control set was, on average, not correlated (median r = −0.004). Furthermore, the control set is very similar to the proteome-wide set of pairwise comparisons because the vast majority of all possible pairs are not functionally related. Physically interacting protein pairs, as discovered by yeast two-hybrid and coimmunoprecipitation, for example, were generally positively correlated (median r = 0.275, Fig. 2), and the difference between these and the control set was highly significant (Wilcoxon rank sum test, P < 2.2 × 10−16). Protein pairs in the same complex were also generally correlated (median r = 0.245, P < 2.2 × 10−16), and the intersection of cocomplexed and physically interacting pairs was even more pronounced (median r = 0.369, P < 2.2 × 10−16). Many individual protein complexes demonstrated highly correlated evolution, such as the CCR4-NOT transcriptional regulation complex. The mean RVC value between the nine members of the CCR4-NOT complex was >0.6, and this high degree of correlation was not observed in one million random sets of nine genes (P < 1 × 10−6). Using this permutation test, 62% of all annotated complexes had a significantly elevated mean ERC (P < 0.05) (Supplemental Table S1). We also examined genes showing epistatic genetic interactions, such as those detected in high-throughput synthetic genetic arrays (Tong et al. 2001), and found that protein pairs in genetic interactions were significantly correlated, although not to the same degree as physically interacting proteins (median r = 0.075, P < 2.2 × 10−16). Because our method includes some genes without all 18 species, we were concerned that these missing sequences could create some type of bias. However, we found the same patterns of ERC presented above when we restricted analysis to only genes whose orthologs were found in all 18 species (data not shown).
Figure 2.
ERC is elevated between functionally related proteins. Here, we contrast ERC between protein pairs that: (left to right) have no annotated relationship (control), physically interact, are in the same complex, physically interact and are in the same complex, and genetically interact. All classes are significantly different from the control class (Wilcoxon rank sum test, P < 2.2 × 10−16). The box limits are the upper and lower quartiles of each distribution, while the bold line represents the median. Whiskers extend to the most extreme data point outside the box that is no more than 1.5 times the interquartile range.
Evolutionary rate covariation is uniformly distributed over the protein primary sequence
If intermolecular coevolution contributes greatly to ERC, we would expect the correlation to be stronger between physically interacting subregions. We tested this hypothesis by dividing all sequence alignments at the midpoint into two subalignments. The rationale is that the subalignment containing the interaction domain would be more strongly correlated with its interacting partner, whereas in noninteracting protein pairs, there would be no reason to expect a consistent increase in correlation after subdivision.
The subdivided analysis did not increase statistical power to distinguish physically interacting proteins from control proteins compared to the original, full-protein analysis (power = 0.72 and 0.75 for subdivided and full data sets, respectively). To assure that shorter alignments do not result in a general decrease in power, we also tested a staggered subdivision based on odd- and even-numbered alignment columns. Its statistical power was equal to that of the full-protein analysis (power = 0.75). These results are consistent with a uniform distribution of ERC along the protein primary sequence and do not support the hypothesis that coevolution of interaction interfaces is responsible for genome-wide ERC in yeast. In addition, these findings are in agreement with those of Hakes et al. (2007) who reported that ERC was not localized to interaction interfaces.
Direct physical interaction is not required for evolutionary rate covariation
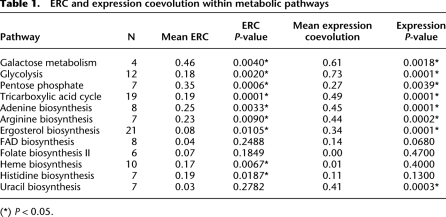
We hypothesized that ERC best reflects fluctuations in external evolutionary pressures that act on functionally related proteins that do not necessarily physically interact. To test this, we examined proteins involved in metabolic pathways that are unlikely to physically interact. We sampled 12 diverse pathways that metabolize carbohydrates, amino acids, cofactors, or lipids and included both anabolic (biosynthetic) and catabolic (degradation) pathways. The few cases of known direct physical interaction in these pathways were removed from analysis. If variation in shared evolutionary pressures contributes to ERC, we would predict that proteins in these pathways would be positively correlated. Indeed, each of the 12 pathways had positive median ERC values, and nine out of 12 (75%) had distributions that were significantly elevated at P < 0.05 (Table 1). We also found that the intensity of ERC between these noninteracting but cofunctional metabolic enzymes is not different from physically interacting protein complexes. The distribution of median ERC values in the 12 metabolic pathways is not significantly different from that of the 244 protein complexes analyzed above (Kolmogorov-Smirnov test, P = 0.47; metabolic median r = 0.19; complexes median r = 0.20) (Table 1; Supplemental Table S1).
Table 1.
ERC and expression coevolution within metabolic pathways
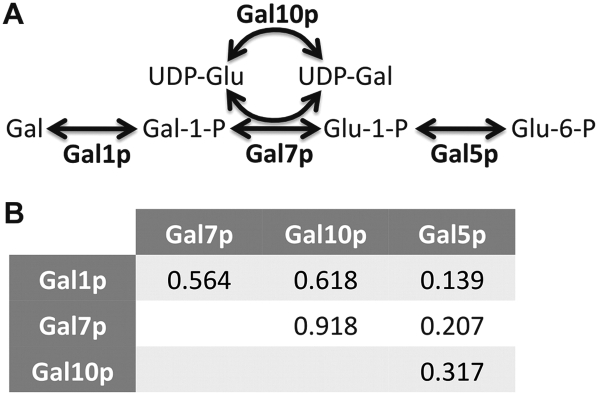
The well-characterized galactose pathway provides a useful illustration of ERC in a metabolic pathway. There are four metabolic steps carried out by the galactose pathway proteins Gal1p, Gal7p, Gal10p, and Gal5p (Fig. 3A). All of the correlation coefficients between these enzymes are positive (Fig. 3B), and the pathway as a whole shows significantly elevated ERC, despite there being no known physical interactions between them (P = 0.004). The most notable correlation is between Gal7p and Gal10p (r = 0.918), which is the highest ERC value observed for both proteins out of the entire analyzed proteome (>4000 proteins). Interestingly, the GAL7, GAL1, and GAL10 genes are colinear on the second chromosome of Saccharomyces cerevisiae. This raises the possibility that chromosomal vicinity could be involved in ERC. However, the other 11 metabolic pathways analyzed do not contain neighboring genes, except for heme biosynthesis in which HEM12 and HEM13 are separated by two genes. Expression of GAL7, GAL1, and GAL10 is regulated by a single group of genes (GAL4, GAL80, and GAL3), suggesting that gene expression pattern could contribute to ERC (De Robichon-Szulmajster 1958; Platt and Reece 1998).
Figure 3.
Galactose enzymes exhibit ERC. The classic galactose metabolic pathway (A) converts galactose (Gal) into glucose 6-phosphate (Glu-6-P) via four enzymes: Gal1p, Gal7p, Gal10p, and Gal5p. A pairwise comparison table (B) shows the strength of ERC (correlation coefficient) between each protein pair. RVC between Gal1p, Gal7p, and Gal10p is notably elevated, while that with Gal5p is less elevated but also positive. ERC between Gal7p and Gal10p is the highest value proteome-wide for both proteins.
Thus, our analysis of proteins in metabolic pathways demonstrates that direct physical interaction is not required for ERC in yeast. Rather, more general forces such as shared evolutionary pressures and coevolution of expression level, which we explore in the next section, could be enough to create the observed correlations throughout the proteome.
No simple relationship between coevolution of expression level and rate covariation
Since complexes and pathways perform better with balanced abundances of their members (Papp et al. 2003; Veitia et al. 2008), there may be strong selection for their expression levels to coevolve over phylogenetic lineages (Lemos et al. 2004). Such coevolution of expression could result in parallel changes in amino acid substitution rate and thus create ERC, because expression level and evolutionary rate are correlated (Drummond et al. 2006). Evidence for coevolution of expression level was previously reported using a set of four yeast species and employing codon bias as a proxy for gene expression level (Fraser et al. 2004). This proxy is justified because codon bias is strongly correlated with expression level in yeast (Bennetzen and Hall 1982). In fact, codon bias is perhaps better than directly measuring mRNA expression levels under an arbitrary laboratory condition that is not likely to reflect a species' current or past natural environment.
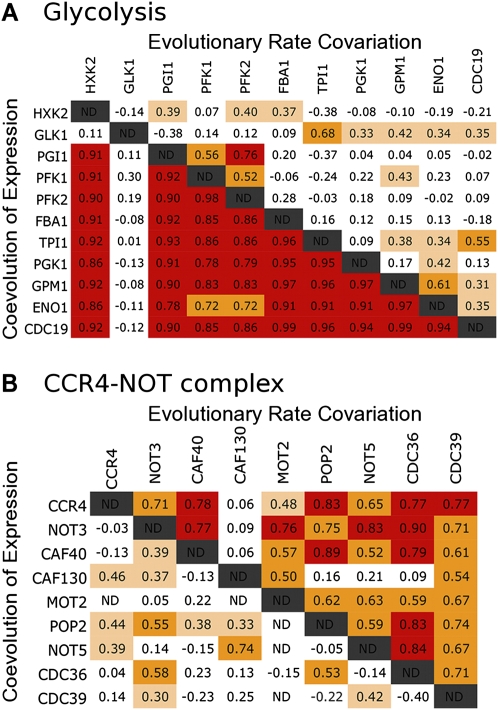
We, too, found that species-specific expression level was significantly correlated between physically interacting proteins compared to the control set (Wilcoxon rank sum test, P < 2.2 × 10−16). However, coevolution of expression level was not as statistically powerful as ERC (power = 0.62 and 0.74 for expression and ERC, respectively). Yet, there are certain sets of cofunctional genes that demonstrated very strong coevolution of expression level, suggesting that there is a great deal of functional information to glean from these correlations (Fig. 4, values below diagonal). For example, the mean correlation for expression coevolution between glycolysis enzymes was 0.73, compared to the genome-wide background mean of −0.01 (permutation test, P < 0.0001) (Table 1).
Figure 4.
ERC and coevolution of expression are not coincidental. Pairwise correlation matrices show values of ERC (above diagonal) and coevolution of expression level (below diagonal) between glycolysis (A) and CCR4-NOT complex (B) proteins. Both ERC and expression coevolution are significantly elevated for both sets of proteins (P < 0.01). However, ERC is much stronger than expression coevolution in the CCR4-NOT complex, while it is the opposite case between glycolysis proteins. (Red) Values greater than 0.75; (orange) values between 0.5 and 0.75; (beige) values between 0.3 and 0.5.
Our analysis suggests that expression coevolution and ERC are not entirely derivative of each other and contain independent information. In the full 18-species data set, there was only a weak correlation between them (r = 0.05), and there are multiple biological pathways in which ERC is stronger than expression coevolution and vice versa. For example, in glycolysis, expression coevolution (mean r = 0.73) was much stronger than ERC (mean r = 0.18), while in the CCR4-NOT complex, expression coevolution was substantially weaker (mean r = 0.18) than ERC (mean r = 0.60) (Fig. 4). In addition, the 12 metabolic pathways examined above demonstrate that ERC and expression coevolution are not always coincidental or of equal intensity (Table 1). If coevolution of expression level contributes to genome-wide ERC in yeast, the question is to what degree. In the next section, we quantify this and other potential biological contributors to ERC using multivariate analysis.
Cofunctionality and coevolution of expression both contribute to rate covariation
We compiled a set of six variables representing physical interaction, shared function, and coevolution of expression in order to dissect their relative contributions to genome-wide ERC. Physical interaction (variable 1) was scored using annotated physical interactions and protein complexes (see Methods). Coevolution of expression level (variable 2) was approximated by the covariation of codon bias between species, as described in the previous section. Shared function was represented by four variables: genetic interaction (variable 3) and the semantic similarity of Gene Ontology categories: biological process (variable 4), molecular function (variable 5), and cellular component (variable 6) (Ashburner et al. 2000). We found most of these predictor variables to be significantly correlated with ERC, but importantly, they also correlated with each other (Supplemental Tables S1, S2). Correlated predictors make it difficult to disentangle their potential influence on ERC. Thus, we separated the six predictor variables into six principal components, each independently explaining a portion of predictor variance. We then regressed each principal component against ERC and calculated the percent of variation in ERC that each principal component explained (i.e., principal components regression) (Mandel 1982). Note that principal components are ordered by the amount of explained predictor variance, not by explained ERC variance.
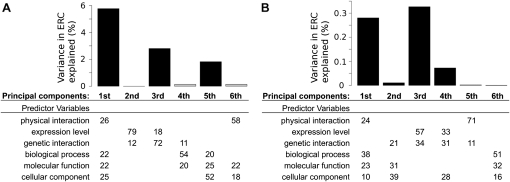
We first analyzed 59 proteins from two distinct processes, the nuclear pore complex and DNA mismatch repair, so that there would be similar numbers of related and unrelated protein pairs. Together, the six principal components explained 10.7% of the total variance in ERC (Fig. 5A). The first principal component (Fig. 5A, first column) was significantly associated with ERC (P = 1.1 × 10−9) and alone explained 5.8% of ERC variance (bar plot). The first principal component was composed of four predictor variables in roughly equal percentages (numerical values in first column): physical interaction and shared biological process, molecular function, and cellular component. Physical interaction and functional similarity were not separated in this principal component since physical interaction and function are often intertwined. The combination of these variables could be described as the general cofunctionality of a protein pair. The third principal component of predictor variables was composed of genetic interaction and coevolution of expression level and explained 2.8% of ERC variance (P = 2.5 × 10−5). The third principal component largely separated genetic interaction from other predictor variables, suggesting that it independently influences ERC.
Figure 5.
Multivariate analysis reveals biological variables associated with ERC. Two principal components regressions were performed: one on nuclear pore and DNA repair proteins (A) and the second on a larger 982-protein data set (B). The predictor variables (rows) were broken into six principal components (columns). Table values are the percentage of each predictor variable composing a principal component. For visual clarity, values <10% are not displayed. Each component was regressed against ERC to determine its individual contribution, and the bar above a component shows the percent of ERC variance explained. Components significantly associated with ERC have black bars (P < 0.01).
We then examined a larger, more diverse data set consisting of all pairwise comparisons between 982 proteins, chosen for their completeness of annotation and presence in all 18 species. The predictor variables explained a much smaller amount of ERC variance in this data set (0.69%) because the vast majority of comparisons are between unrelated genes. However, the analysis indicated similar global forces associated with ERC (Fig. 5B). The greatest amount of variance was explained by coevolution of expression and genetic interaction (third principal component: 0.33% of variance; P < 2.2 × 10−16). It is notable that the contribution of coevolution of expression is much greater in this genome-wide data set, suggesting that it could be a major factor in many pathways. Physical interaction and shared function were again associated with ERC (first principal component: 0.28% of variance; P < 2.2 × 10−16), and we speculate that they could explain much more because the annotation of physical interaction and function is largely incomplete, even in the yeast genome.
Discussion
We performed a proteome-wide study in 18 budding yeast species to define and discriminate the forces behind evolutionary rate covariation. A popular conception has been that ERC arises due to coevolution between interacting protein sites (Goh et al. 2000; Pazos and Valencia 2001). However, the totality of evidence presented here indicates that ERC in the yeast proteome is more complex and most reliably reflects shared function between proteins. We demonstrated that direct physical interaction is not required to produce ERC between proteins, since noninteracting but functionally related metabolic enzymes are just as correlated as physically interacting proteins. It has been argued that coevolution and compensatory changes are not major contributors to the signature of ERC between physically interacting proteins (Hakes et al. 2007; Lovell et al. 2010), but evidence to the contrary has also been presented (Kann et al. 2009). Our major assertion here is that the previous preoccupation with physical interaction may have obscured the true potential of ERC to predict functional classes more broadly. One must not assume that a signal of ERC requires that two proteins are physically interacting, because they are just as likely to be only functionally related. This greatly expands the potential impact that ERC can have on genome annotation.
A multivariate analysis allowed us to estimate the relative contributions of biological variables to ERC on a proteome-wide scale. In two different data sets, the two major components associated with ERC were (1) cofunctionality, seen as a combination of shared functional annotation and physical interaction, and (2) coevolution of expression level. While we found the physical interaction variable to be intertwined in a principal component with functional annotation, this does not necessarily implicate direct physical interaction as a driving force; it just could not be separated from functional annotation in this analysis. The novel finding from the multivariate analysis was that coevolution of expression level was a major variable associated with ERC. Whether this association results from a causative relationship remains to be determined.
We propose that ERC results mainly from fluctuation in the evolutionary pressures shared by functionally linked proteins. Such fluctuation could result from changes in either constraint (negative selection) or in adaptive evolution (positive selection). We would argue that fluctuation in constraint is the greater contributor to ERC because it acts on all proteins. Indeed, we observe ERC proteome-wide, including between many highly conserved proteins that would only on rare occasion experience positively selected substitutions. In addition, Elyashiv et al. (2010) have used polymorphism data to argue that most amino acid fixations between S. cerevisiae and Saccharomyces paradoxus were driven by relaxed constraint, rather than positive selection. Episodes of positive selection could contribute to ERC within adaptively evolving pathways, such as immunity or reproduction; however, only limited functional classes undergo frequent positive selection (Kosiol et al. 2008). In summary, we propose that the greater contributor to proteome-wide ERC is pathway-specific fluctuation in selective constraint, which thereby produces correlated rates, primarily through nearly neutral substitutions.
We still do not understand why certain groups of functionally related proteins correlate, while others do not. For example, we observed significantly elevated ERC in 62% of protein complexes, while many other well-annotated complexes showed no elevated ERC at all. What determines whether a group of functionally related proteins will demonstrate this signature? Some possibilities to consider will be network properties, biological function, dispensability, and expression pattern. Despite theses limitations, the study of ERC does provide insight into biological function and should serve as an additional tool for the functional analysis of genomes.
Methods
Proteome-wide orthologous groups
We analyzed the predicted amino acid sequences for 18 fungal species from both the Fungal Genome Research database (http://fungalgenomes.org/) (Fitzpatrick et al. 2006) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). These species were: S. cerevisiae, S. paradoxus, Saccharomyces mikatae, Saccharomyces bayanus, Saccharomyces kluyveri, Candida glabrata, Kluyveromyces thermotolerans, Kluyveromyces waltii, Kluyveromyces lactis, Kluyveromyces polysporus, Ashbya gossypii, Candida albicans, Candida dubliniensis, Candida tropicalis, Candida guilliermondii, Candida lusitaniae, Lodderomyces elongisporus, Debaryomyces hansenii, and Scheffersomyces stipitis. Starting with the proteins from S. cerevisiae, we determined the best single ortholog, if present, from all other 17 species using the program InParanoid (Remm et al. 2001). Inparanoid was configured to find orthologous sequences using the reciprocal best BLAST hit criterion with a similarity score cut-off of 50 bits. In cases with multiple orthologs due to duplications since speciation (in-paralogs), we conservatively designated only the best two-species pair as orthologs (i.e., 100% confidence orthologs as assigned by InParanoid). The resulting 4459 orthologous groups of proteins were aligned using MUSCLE (Edgar 2004).
Calculating evolutionary rate covariation
For each amino acid alignment we estimated branch lengths using the “aaml” program from the phylogenetic analysis using the maximum likelihood (PAML) package (Yang 2007). Branch lengths were estimated under an empirical model of amino acid substitution rates (Whelan and Goldman 2001) with rate variability between sites modeled as a gamma distribution approximated with four discrete classes (for computational efficiency) plus a class for invariable sites (aaml model “Empirical+F”) (Yang 1996). These lengths were all estimated on the same species tree topology as reported by Fitzpatrick et al. (2006). The resulting branch lengths can be directly used to calculate a correlation coefficient (r) between any two proteins; however, this direct approach provided limited power to discern physically interacting from noninteracting protein pairs. The alternative is to analyze the relative rates of evolution along each branch compared to the expected length in a hypothetical proteome-wide tree. We transformed the raw branch rates into relative rates using the projection operator method of Sato et al. (2005). This greatly improved the power to distinguish physically interacting pairs from controls; power improved from 0.66 to 0.74. To improve gene coverage, pairwise comparisons were made in such a way that allowed for missing sequences by considering only those species shared between each protein pair. Pairs were required to have a minimum of 12 shared species. Any missing species were pruned from the tree topology using the BioPerl::Trees package (Stajich 2007). We were not interested in correlations driven by a single outlier data point, so we set a protein-specific limit of evolutionary rate at two standard deviations from the mean. This limit biased high correlations to those consistently involving multiple branches instead of one outlier branch.
Gene annotation and power analysis
All annotated physical and genetic interactions were downloaded from the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org). Because high-throughput methods to detect physical interactions have high false positive rates, we created a more confident reference set by including only those inferred by at least two independent experimental methods. We estimated the statistical power of ERC to distinguish interacting from noninteracting protein pairs using the area under the receiver operator characteristic (ROC) curve using the R-project module “ROCR” (Sing et al. 2005). In this measure, a perfect predictor will have an area of 1, and a method that has no predictive value will have an area of 0.5. In the subdivided analysis, we selected the highest ERC value for each protein pair out of all four combinations of subdivisions. The proteins involved in specific metabolic pathways were taken from the Saccharomyces Biochemical Pathway Overview available from SGD (September 2010). There were a few annotated physical interactions in the ergosterol, glycolysis, and tricarboxylic acid cycle pathways, so those specific protein pairs were excluded from analysis.
Coevolution of expression level
Coevolution of expression level was calculated using codon bias as a proxy for expression level in each of the 18 species. Codon bias was measured using the codon adaptation index (CAI) estimated by correspondence analysis of codon usage using the program “codonw,” written by John Peden (http://codonw.sourceforge.net/) (Sharp and Li 1987). High-frequency and presumably preferred codons were determined for each species using a set of highly expressed genes from Saccharomyces cerevisiae and their corresponding orthologs in the other species. This highly expressed set encoded mostly ribosomal proteins and metabolic enzymes. Expression level correlations were calculated after transforming the values with the projection operator used above for ERC (Sato et al. 2005).
Multivariate analysis
Predictor variables were compiled from annotation in SGD. Because physical interaction data contain many false positives and because binary variables can produce misleading results in a principal components analysis, we configured the physical interaction variable to reflect the relative confidence in that interaction using the formula: (1 – 0.5t × 0.3(m-1) × 0.1c), where t is the number of times this interaction is reported in SGD, m is the number of independent experimental methods that found this interaction, and c is an indicator variable reporting whether the protein pair is in the same protein complex or not. This formula transforms the separate binary variables onto a more continuous range. The resulting variable is zero for no annotated interaction and approaches one as more independent evidence for an interaction is present. Although there are many possible coefficients for this formula, we found the conclusions of the principal components regression to be robust to alternate encodings. For example, when we ran the analysis with each factor (complex, physical interaction, etc.) as separate binary variables, the main contributing factors to ERC were more complex but unchanged (data not shown). The genetic interaction variable was either scored “one” for an annotated interaction or “zero” for none. The similarity of functional classifications for a protein pair was quantified by the semantic similarity of their Gene Ontology annotations (Ashburner et al. 2000). Semantic similarity gives a numerical score to the relatedness of the annotation between a pair of genes and, hence, represents how similar they are in function or localization. Semantic similarity, as we calculated with the program GOSemSim, also takes into account the hierarchical structure of the Gene Ontology (Yu et al. 2010). Principal components regression was performed using singular value decomposition in the R Module “pls,” written by Ron Wehrens and Bjørn-Helge Mevik. All predictor variables were scaled by their sample standard deviation before principal components analysis.
Data access
The full genome-wide ERC matrix is available for download at http://www.csb.pitt.edu/faculty/clark/data.html.
Acknowledgments
We thank Huifeng Jiang and Zhenglong Gu for providing data sets and Brian Lazzaro for discussion. This work was supported by a National Institutes of Health (NIH) postdoctoral fellowship (GM084592) to N.L.C., a NIH grant (GM53085) to E.A., and a NIH grant (GM36431) to C.F.A.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.132647.111.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD 1982. Codon selection in yeast. J Biol Chem 257: 3026–3031 [PubMed] [Google Scholar]
- Clark NL, Aquadro CF 2010. A novel method to detect proteins evolving at correlated rates: identifying new functional relationships between coevolving proteins. Mol Biol Evol 27: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ 2009. Coevolution of interacting fertilization proteins. PLoS Genet 5: e1000570 doi: 10.1371/journal.pgen.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robichon-Szulmajster H 1958. Induction of enzymes of the galactose pathway in mutants of Saccharomyces cerevisiae. Science 127: 28–29 [DOI] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO 2006. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol 23: 327–337 [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol 17: 68–74 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyashiv E, Bullaughey K, Sattath S, Rinott Y, Przeworski M, Sella G 2010. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res 20: 1558–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6: 99 doi: 10.1186/1471-2148-6-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Wall DP, Eisen MB 2004. Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci 101: 9033–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE 2000. Co-evolution of proteins with their interaction partners. J Mol Biol 299: 283–293 [DOI] [PubMed] [Google Scholar]
- Hakes L, Lovell SC, Oliver SG, Robertson DL 2007. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc Natl Acad Sci 104: 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan D, Pazos F, Valencia A 2008. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc Natl Acad Sci 105: 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann MG, Shoemaker BA, Panchenko AR, Przytycka TM 2009. Correlated evolution of interacting proteins: Looking behind the mirrortree. J Mol Biol 385: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A 2008. Patterns of positive selection in six mammalian genomes. PLoS Genet 4: e1000144 doi: 10.1371/journal.pgen.1000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Hartl DL 2004. Regulatory evolution across the protein interaction network. Nat Genet 36: 1059–1060 [DOI] [PubMed] [Google Scholar]
- Li WH, Tanimura M, Sharp PM 1987. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol 25: 330–342 [DOI] [PubMed] [Google Scholar]
- Lovell SC, Robertson DL 2010. An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol 27: 2567–2575 [DOI] [PubMed] [Google Scholar]
- Madaoui H, Guerois R 2008. Coevolution at protein complex interfaces can be detected by the complementarity trace with important impact for predictive docking. Proc Natl Acad Sci 105: 7708–7713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J 1982. Use of the singular value decomposition in regression analysis. Am Stat 36: 15–24 [Google Scholar]
- Moyle WR, Campbell RK, Myers RV, Bernard MP, Han Y, Wang X 1994. Co-evolution of ligand-receptor pairs. Nature 368: 251–255 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A 2001. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng 14: 609–614 [DOI] [PubMed] [Google Scholar]
- Pazos F, Ranea JA, Juan D, Sternberg MJ 2005. Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J Mol Biol 352: 1002–1015 [DOI] [PubMed] [Google Scholar]
- Platt A, Reece RJ 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J 17: 4086–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Sato T, Yamanishi Y, Kanehisa M, Toh H 2005. The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21: 3482–3489 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH 1987. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941 [DOI] [PubMed] [Google Scholar]
- Stajich JE 2007. An introduction to BioPerl. Methods Mol Biol 406: 535–548 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Travers SA, Fares MA 2007. Functional coevolutionary networks of the Hsp70-Hop-Hsp90 system revealed through computational analyses. Mol Biol Evol 24: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic, and proteomic effects. Trends Genet 24: 390–397 [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Yang Z 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol 11: 367–372 [DOI] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S 2010. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26: 976–978 [DOI] [PubMed] [Google Scholar]