Abstract
Many of the most virulent bacterial pathogens show low genetic diversity and sexual isolation. Accordingly, Mycobacterium tuberculosis, the deadliest human pathogen, is thought to be clonal and evolve by genetic drift. Yet, its genome shows few of the concomitant signs of genome degradation. We analyzed 24 genomes and found an excess of genetic diversity in regions encoding key adaptive functions including the type VII secretion system and the ancient horizontally transferred virulence-related regions. Four different approaches showed evident signs of recombination in M. tuberculosis. Recombination tracts add a high density of polymorphisms, and many are thus predicted to arise from outside the clade. Some of these tracts match Mycobacterium canettii sequences. Recombination introduced an excess of non-synonymous diversity in general and even more in genes expected to be under positive or diversifying selection, e.g., cell wall component genes. Mutations leading to non-synonymous SNPs are effectively purged in MTBC, which shows dominance of purifying selection. MTBC mutation bias toward AT nucleotides is not compensated by biased gene conversion, suggesting the action of natural selection also on synonymous changes. Together, all of these observations point to a strong imprint of recombination and selection in the genome affecting both non-synonymous and synonymous positions. Hence, contrary to some other pathogens and previous proposals concerning M. tuberculosis, this lineage may have come out of its ancestral bottleneck as a very successful pathogen that is rapidly diversifying by the action of mutation, recombination, and natural selection.
Some of the most deadly bacterial pathogens are recently emerged clones showing little genetic diversity (Achtman 2008). These so-called monomorphic lineages are thought to have endured population bottlenecks associated with the acquisition of new functions, e.g., the capacity to multiply within different host cells. While new functionalities allow a lineage to explore a new niche, the associated population bottleneck poses problems from an evolutionary point of view (Ohta 1992; Edmonds et al. 2004). First, standing deleterious polymorphisms in the clone hitchhike with the adaptive allele and become fixed (Smith and Haigh 1974). Second, the population contraction associated with the bottleneck results in less efficient selection leading to the accumulation of further slightly deleterious changes (Nei et al. 1975). Finally, niche change results in increased isolation from closely related strains lowering the rates of recombination, which further enhances the accumulation of deleterious changes (Felsenstein 1974). As a result, lineages such as Shigella flexneri, Yersinia pestis, or Bordetella pertussis show extensive evidence of genome degradation, such as accumulation of pseudogenes in housekeeping functions, hundreds of Insertion Sequences, and high rates of evolution (Jin et al. 2002; Parkhill et al. 2003; Chain et al. 2004; Warnecke and Rocha 2011). Mycobacterium tuberculosis is the causative agent of tuberculosis, the most deadly of human bacterial pathogens, and has arisen recently from a bottleneck. Accordingly, non-SNP-based typing methods have repeatedly suggested purely clonal evolution in the Mycobacterium tuberculosis complex (MTBC) (Smith et al. 2006). Recent works even suggested that MTBC evolves essentially under genetic drift (Hershberg et al. 2008).
Yet, the genomes of M. tuberculosis have moderate numbers of Insertion Sequences, few pseudogenes, and no obvious other signals of extensive genome degradation (Cole 1998; Gordon et al. 1999; Tsolaki et al. 2004). MTBC strains have endured several recent serial losses of genetic material that led to genomes of ∼4.4 Mb in length, which is slightly smaller than the closely related Mycobacterium canettii (4.5 Mb). Many of these deletions are regarded as pathoadaptive, e.g., they allowed the bacterium to invade new hosts (Brodin et al. 2002; Aranaz et al. 2003; Cousins et al. 2003; Mostowy et al. 2005), rather than being the result of an increased genetic drift. Also, the 90%–96% coding density and the 65% G+C content of M. tuberculosis strains are similar or even higher that those of the closely related species M. canettii, Mycobacterium ulcerans, and Mycobacterium marinum (respectively, 88%, 72%, and 82% coding density and 65%, 65%, 66% G+C). Decrease in G+C content and density of coding sequences are expected in lineages with recent abrupt changes in the efficiency of natural selection (Mira et al. 2001). They are not found in M. tuberculosis, which might then represent an example of a lineage that emerged from a bottleneck as a highly successful pathogen without a concomitant heavy genetic load.
M. tuberculosis evolved into six major lineages strongly associated with particular geographic regions (Filliol et al. 2006; Gagneux et al. 2006; Gutacker et al. 2006). While this is consistent with clonal descent, it is not a demonstration of it. For example, the genome of Helicobacter pylori shows evidence of very high rates of recombination but still shows a strong biogeographic signal that allows tracing of human migrations (Falush et al. 2003). The analyses of housekeeping genes from Mycobacterium prototuberculosis, closely related to MTBC, suggested that the extant tubercle bacilli genome is a composite assembly of mosaic blocks by horizontal genetic transfer (HGT) prior to the bottleneck that formed MTBC (Gutierrez et al. 2005). Whole-genome comparisons across mycobacterial species led to the identification of 235 and 137 such horizontally transferred genes based, respectively, on sequence composition (Becq et al. 2007) and phylogenetic methods (Veyrier et al. 2009). These events predate the diversification of MTBC and have thus evolved with the rest of the genome since MTBC's last common ancestor. There is published evidence of interchromosomal recombination ahead of a few loci of the M. tuberculosis genome (Hughes et al. 2002; Liu et al. 2006). Nevertheless, this evidence has been regarded as largely anecdotal, and MTBC is systematically considered as strictly clonal (Smith et al. 2006; Comas and Gagneux 2011).
While MTBC lineages are often considered to be monomorphic, molecular typing techniques such as IS6110-RFLP, spoligotyping, and Variable Number of Tandem Repeat-Mycobacterial Interspersed Repetitive Unit (VNTR-MIRU) revealed a certain level of genetic diversity among strains (Otal et al. 1991; Kamerbeek et al. 1997; Supply et al. 2006). The public health need to classify strains into closely related groups and the need to estimate accurate genetic relationships among isolates have led to the analysis of several variations at the genome scale level. This allowed the selection of different characteristic single nucleotide polymorphisms (SNPs) (Filliol et al. 2006; Gutacker et al. 2006; Dos Vultos et al. 2008; Comas et al. 2010) and large sequence polymorphisms (LSPs) (Brosch et al. 2002; Alland et al. 2007) in the last decade. These studies confirmed the extensive genetic diversity and genome plasticity of the Mycobacterial genome. With the advent of high throughput Next Generation Sequencing technologies (NGS), genome sequencing has moved into a new era, opening the possibility of understanding the genetic basis for strain-specific differences in pathogenicity. This is particularly important for lineages that have emerged recently after a change in niche, where a large number of genomes is necessary for detailed evolutionary analyses.
To perform a genome-scale analysis of how genetic diversity builds up in the lineages, we analyzed the SNPs of the genomes of 24 MTBC strains from all major lineages. The advantage of the use of SNPs data when compared with other approaches such as spoligotyping and VNTR-MIRU is that SNPs are distributed throughout the genome, in intragenic and intergenic regions; they are reliable; they can be precisely assessed; and they have a low reverse mutation rate and homoplasy index (Comas et al. 2009). They are thus ideal to investigate the genomic imprint of natural selection and genetic recombination.
Results
Whole-genome SNP discovery and phylogeny of MTBC strains
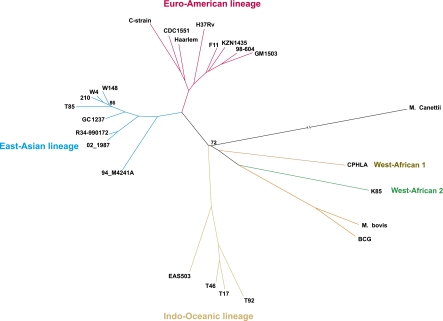
The genomes of three M. tuberculosis strains—GC1237, R34-990172, and W4—were sequenced at high coverage rates (67×, 411×, and 386×, respectively) to obtain a better sampling of the East Asian clade, which includes the fast disseminating Beijing strains (Bifani et al. 2002; von Groll et al. 2010). The GC1237 strain is pan-susceptible to all first-line anti-tuberculosis drugs (rifampin, isoniazid, streptomycin, ethombutol, and pyrazinamide) and is responsible for an ongoing epidemic in the Gran Canaries Island (Caminero et al. 2001; Alonso et al. 2011). We compared these three strains with 21 others publicly available (Supplemental Table S1). We excluded SNPs identified in the ∼10% of the genome composed of repetitive regions because mapping in these regions is error-prone. This includes the highly GC-rich and polymorphic PE/PPE gene family, esx genes, and Insertion Sequences. Gene conversion resulting from frequent intrachromosomal homologous recombination has been reported between members of each of these families (Ho et al. 2000; McEvoy et al. 2007; Karboul et al. 2008; Uplekar et al. 2011). Hence, exclusion of these regions prevents spurious inclusion of intrachromosomal recombination events in the analyses of recombination presented below. A total number of 12,475 polymorphic sites were thus identified and used to generate phylogenetic trees by maximum likelihood (ML) using RaxML (Fig. 1; Stamatakis 2006), Tree-Puzzle (Supplemental Fig. S2A; Schmidt et al. 2002), and by Bayesian methods using BEAST (Supplemental Fig. S2B; Drummond and Rambaut 2007). All nodes of the ML tree generated by RaxML have very good support values. The tree was rooted using the complete genome sequence of M. canettii (NC_015848, CIPT140010059) after removing the recombination tracts found in the ClonalFrame analysis below. All methods positioned the root between the West African clade/BCG/M. bovis and the remaining MTBC, in agreement with previous works (Wirth et al. 2008; Comas et al. 2010). We also did all analyses below with the root placed either between the Indian Ocean lineage and the rest of the MTBC or between the so-called ancient and modern lineages. All results remained qualitatively unchanged upon displacement of the root. The tree is consistent with previous analyses showing the geographical expansion of the MTBC lineages using partial or complete genome data (Gagneux and Small 2007; Comas et al. 2010). Thus, it is a suitable basis to map the origin of SNPs in the lineage.
Figure 1.
Phylogenetic tree constructed using SNPs purged of recombination tracts. The maximum likelihood tree was constructed using RaxML v7.2.8. Only nodes with bootstrap support values below 100% are indicated.
Local diversification of MTBC genomes
To study the patterns of SNP distribution, we first analyzed the 55 HGT regions, including 256 genes, identified by sequence composition methods (Becq et al. 2007). These regions were acquired before MTBC diversification and were thus expected to accumulate SNPs like the rest of the genome. Instead, these regions contain 5.3% of the SNPs, which is significantly more than expected by chance given their size (4.6%, P-value < 0.001, binomial test). Analysis of the 137 genes reported to have been horizontally transferred using phylogenetic methods (Veyrier et al. 2009) also showed an excess of SNPs (P-value < 0.001, same test). Faster evolution in these regions could be caused by their atypical composition but also by the higher probability of generating adaptive changes in recently acquired genes. We explore these hypotheses in a subsequent section.
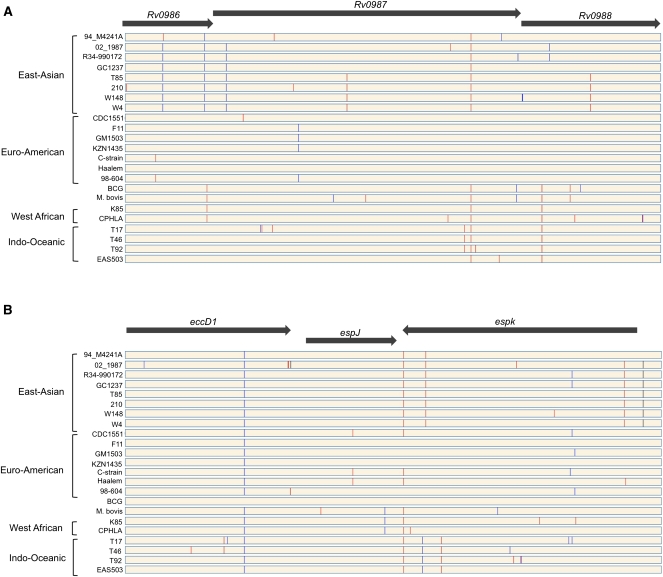
To identify other regions of SNP clustering, we estimated SNP density throughout the genomes using a sliding window of 5 kb. The resulting SNP density map shows a non-random distribution of SNPs, with 13 regions having statistically significant clusters (red bars in Fig. 2). We could not find any common feature between all of these regions (Supplemental Fig. S3). Nevertheless, two of them are of obvious biological relevance. One region (Fig. 3A) corresponds to a previously reported virulence operon including the genes Rv0986–Rv0988 that are included in one of the abovementioned HGT regions (Rosas-Magallanes et al. 2006; Veyrier et al. 2009). This region is part of the in vivo–expressed genomic island (iVEGI) that includes a set of genes highly expressed only in vivo, among which are the two genes Rv0986 and Rv0988 (Talaat et al. 2004). HGT regions have a significant excess of SNPs even after exclusion of this operon (P-value < 0.0001, same test). Thus, this region evolves particularly fast within the fast-evolving HGT regions.
Figure 2.
SNP density map constructed using Circos (Krzywinski et al. 2009). (Yellow bars) The density of SNPs in non-overlapping 5-kb regions. (Red bars) Regions with significantly high SNP density (binomial test, p < 0.05 after sequential Bonferroni correction).
Figure 3.
Graphical representation of the distribution of SNPs in two regions of SNPs clustering. (A) Virulence operon horizontally transferred to the ancestor of M. tuberculosis. (B) Genomic region part of the ESX-1 locus. Synonymous (blue lines) and non-synonymous (red lines) SNPs were identified according to the genome of the reference strain H37Rv. (Black lines) SNPs identified in intergenic regions.
Another region with high density of SNPs (Fig. 3B, eccD1-espK) was found in the ESX-1 locus (RD1 region), which includes a type VII secretion system (Bitter et al. 2009). This secretion system is needed for the secretion of the EsxA (also known as ESAT6)/EsxB (also known as CFP10) proteins involved in the pathogenicity of M. tuberculosis (Guinn et al. 2004). Its deletion is one of the major causes of attenuated virulence of the BCG vaccine (Majlessi et al. 2005). In addition, the deletion of the RD1 region in the M. tuberculosis H37Rv strain results in an attenuated phenotype (Lewis et al. 2003). ESX-1 is also absent from the genome of the weakly virulent vole bacillus Mycobacterium microti (Pym et al. 2002). The protein EccD1 is part of the cell membrane (Bitter et al. 2009), and the protein EspK is secreted by the ESX-1 system and is part of the capsule (Sani et al. 2010), but its precise function, as that of EspJ, is unknown. Given the small number of polymorphisms in the data set, gene-per-gene analysis of positive selection lacks statistical power. Hence, we cannot at this stage assess if the excess of SNPs in these regions is caused by less purifying selection and/or more positive selection. Yet, the observation that regions of higher SNPs density in the genome are intimately linked to the virulence of M. tuberculosis shows the relevance of this trait in the evolution and diversification of the genome.
Mycobacterium tuberculosis complex is not strictly clonal
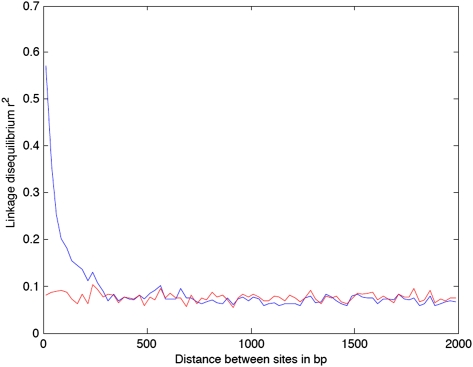
We constructed a Split decomposition network to check for the absence of recombination events between the genomes. This method allows the visualization of the ancestral relationships between elements and does show some conflicting phylogenetic signals as indicated by the presence of cycles in the network (Fig. 4). These cycles are highly supported statistically (Phi = 0) and thus are suggestive of the presence of recombination. To test this hypothesis further, we analyzed the patterns of linkage disequilibrium (LD) between all pairs of SNPs. LD represents the tendency for different alleles to co-occur non-randomly and is higher in the absence of recombination. We found high values of LD for sites that are very close to each other. LD decays quickly as sites further away from each other are considered and stabilizes for distances higher than ∼300 bp (Fig. 5). This pattern is compatible with recombination happening frequently but always involving short fragments of DNA.
Figure 4.
Visualization of conflicting phylogenetic signals by the split decomposition method implemented in SplitsTree4.
Figure 5.
Relationship between linkage disequilibrium (y-axis) and genomic distance between pairs of SNPs (x-axis). (Blue line) LD computed using all SNPs; (red line) LD after excluding SNPs identified in recombination tracts.
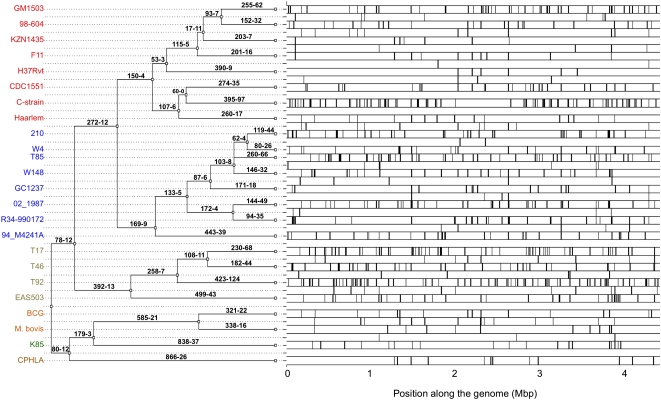
Given the evidence for recombination, we made a scan for conversion tracts in the MTBC genomes using geneconv (Sawyer 1989), which is fast, has low false-positive rates (Posada and Crandall 2001), and provides statistical assessment of recombination tracts. Many significant (p < 0.05) recombination tracts were identified covering over 150 kb of the genomes. Intriguingly, geneconv indicated that around half (47%) of the recombination tracts arise by recombination with DNA from clades outside MTBC. Such tracts have 2.4 times higher SNP density than tracts from recombination within MTBC, suggesting that they are important drivers of MTBC diversification. We then used the more recent and powerful ClonalFrame software (Didelot and Falush 2007) to reconstruct the clonal genealogy and the recombination events that affected the MTBC genomes (Fig. 6). The reconstructed genealogy was perfectly compatible with the phylogenies obtained using maximum likelihood and Bayesian methods. Recombination was found to have occurred on all branches of this genealogy (Fig. 6; see Supplemental Fig. S4 for some examples). Terminal branches in the tree account for 74% of the tree. Yet, recombination events in terminal branches of the reconstructed genealogy account for 85.8% of the total. It is possible that the difference between these two frequencies is caused by less efficient identification of ancient recombination tracts. In any case, this shows that recombination is still ongoing in contemporary MTBC strains. While Figure 1 does not show abnormally long terminal branches, some draft genome strains, such as C-strain and T92, have longer terminal branches than their sister taxa. Longer branches are expected when recombination is intense in one branch (Hudson 1983), as seems to be the case for these two strains (Fig. 6). We analyzed the repair genes in these strains and found them to have SNPs in key homologous recombination and 3R genes (repair, replication, recombination) (C-strain: uvrB, recA, dinF, and dnaQ; T92: recD, recB, recC, mfd, uvrC, ruvA, and ung). Further sequencing and experimental data are necessary to show that these strains do recombine more and that this is caused by changes in these genes.
Figure 6.
Clonal genealogy reconstruction from the 24 genomes (left) and distribution of the recombination events identified by ClonalFrame (right). Each branch of the genealogy is labeled with the number of mutations (left) and recombination events (right) inferred to have occurred on that branch.
Sequencing errors introduce singletons that might inflate the number of recombination events in the terminal branches. Such errors are expected to be more frequent in the draft genomes. Therefore we reanalyzed the available Illumina reads of five genomes (T92, T85, T17, GM1503, and W148) in order to compare the identified SNPs on the bases of their quality control and coverage rates to those included in this study, which were taken from the published contigs (see Methods). This allowed estimation of the rate of discordant SNPs and the frequency with which these are included in recombination tracts. If we consider that all discordant SNPs are due to sequencing errors or misassembling and that every single error in a recombination tract leads to the identification of a spurious recombination event, we obtain an upper value of 6.8% for the rate of false-positive recombination tracts in the terminal branches. These tests indicate that sequence or assembling errors should be of very small effect in the inferred recombination rate. Finally, we recomputed LD after removing the recombination tracts predicted by ClonalFrame and found that essentially no LD was left (Fig. 5). Hence, all methods give consistent information about the presence of recombination in MTBC.
Approximately a third of the recombined segments are found in intergenic regions (Supplemental Table S2), which is more than would be expected by chance alone since ∼90% of the positions of the M. tuberculosis genome are coding (p < 0.001, χ2 test). We estimated that recombination events occurred five times less frequently than mutations (ρ/θ = 0.22 with 95% credibility interval CI = [0.18;0.27]) and involved small recombination tracts (average δ = 54 bp, CI = [50;58]). This low number is consistent with the results obtained in the LD analysis above. The 95% credibility interval of r/m, which represents the relative impact of recombination on sequence diversification, was 0.426–0.565 (mean = 0.486), indicating that, unlike mutation, recombination affects several nucleotides at each occurrence. Using the same method, the r/m value was estimated as 0.63–1.13 and 0.56–1.01 in Clostridium difficile and Chlamydia trachomatis, respectively (He et al. 2010; Joseph et al. 2011). These genomes have thus slightly higher relative input of observable diversity caused by recombination, because they have much larger tract lengths, averaging several hundred base pairs (Didelot et al. 2010; Joseph et al. 2011). As expected, the free-living genomes of Bacillus cereus are more deeply affected by recombination (r/m = 2.37–2.45) (Didelot et al. 2010).
The average amount of polymorphism introduced by recombination events was very high (4.3%, CI = [3.9%;4.8%]), i.e., approximately two orders of magnitude above the average pairwise distance between the 24 genomes (π = 0.075%). This is a strong indication that the identified recombination events involved donors outside of the M. tuberculosis complex. We therefore compared the sequences of the imported fragments with the genomes of other distant Mycobacteria (Mycobacterium Kansassii, Mycobacterium avium, and M. marinum). We did not find sequences identical to recombination tracts in any of these distant genomes, as expected given the requirement of high sequence similarity in homologous recombination (Vulic et al. 1997; Majewski and Cohan 1998). On the other hand, for 40 out of 964 imports, we found an exact match of the sequence in the genome of M. canettii. For an additional 229, we found matches in M. canettii with >98% sequence identity. Note that the M. canettii sequence is not used to detect recombination tracts in the ClonalFrame analysis. Hence, there is no intrinsic bias toward M. canettii in our analysis. The high diversity of recombination tracts, the inference of their origin outside MTBC, and the match with sequences from a closely related genome strongly suggest that M. tuberculosis lineages recombine with closely related strains of other species.
To quantify the likelihood of missing recombination events within MTBC, we expunged the multiple alignment from the conversion tracts found by ClonalFrame. We then selected random subsequences of 100 nt, twice the average size of the observed conversion tracts, in the multiple alignment. Finally, we randomly picked two taxa and quantified the number of differences between them in each subsequence (Supplemental Fig. S5). This simulation was repeated 100,000 times. We found that 96.75% of the pairs of 100-nt subsequences had no differences and 3.18% had only one difference. Hence, only 0.07% of the pairwise comparisons have the minimal number of SNPs of the conversion tracts we observe (two SNPs). If recombination between MTBC were random, this suggests that we miss ∼99.93% of all events. Since the most divergent sequences in our analysis correspond to MTBC strains of different geographical areas, which are less likely to meet, even this high number is an under-estimate. However, because recombination between identical sequences does not introduce SNPs, the r/m ratio given above is expected to be much less affected by recombination within MTBC. In short, both the SplitsTree analysis, which is insensitive to recombination between MTBC and more distant lineages, geneconv, and the ClonalFrame analyses show many recent recombination events. We expect that the vast majority of recombination events take place between closely related strains and that these events will pass unnoticed by our analyses. Together, these results strongly argue against the paradigm that MTBC strains are strictly clonal.
The evolution of the composition of MTBC genomes
HGT regions have atypical sequence composition reflecting recent acquisition by the genome (Rosas-Magallanes et al. 2006; Becq et al. 2007; Veyrier et al. 2009), even if this acquisition predates MTBC diversification. Hence, high SNP density in HGT regions could result from mutational pressure, i.e., amelioration toward the composition of the rest of the genome (Lawrence and Ochman 1997). To test this hypothesis, we analyzed the composition of these regions and their nucleotide substitution patterns. Substitution matrices were computed from synonymous SNPs at fourfold degenerate sites and accounting for nucleotide composition at these sites, as in Rocha et al. (2006b). The G+C composition at these positions is lower at HGT regions (70%) than in the rest of the genome (82%). To test if rapid evolution of HGT regions was caused by mutation pressure toward the native genome G+C content, we computed the nucleotide composition at equilibrium given the substitution matrices inferred from these regions (Supplemental Table S3). The results show only very slightly lower equilibrium values for the HGT regions (55% G+C) than for the rest of the genome (56% G+C). Moreover, both values are much lower than the extant G+C content (82%). Thus, HGT regions are not “ameliorating” toward the genome composition. Instead, unchecked long-term application of these mutational patterns would lead to G+C-poorer regions in both HGT and non-HGT regions. This clearly shows that mutation pressure toward the native genome G+C composition is not causing the observed excess of SNPs in HGT regions.
The previous results show that A+T-enriching SNPs are more abundant than expected in both HGT regions and in the rest of the genome. This follows a pattern previously found in a large number of genomes (Balbi et al. 2009; Hildebrand et al. 2010). Since it is unlikely that all bacterial genomes are becoming AT-rich, it is commonly thought that this mutational bias is moderated by the action of natural selection or biased gene conversion (Rocha and Feil 2010). Accordingly, GC-rich monomorphic lineages with very low effective population size, e.g., Yersinia pestis or Burkholderia mallei, exhibit extreme AT-enriching substitution patterns close to the ones of Buchnera (Hershberg and Petrov 2010). Our data show 56% of equilibrium G+C content at non-HGT-derived fourfold degenerate sites. Hence, the mutational patterns of M. tuberculosis are clearly not as extreme as those of the abovementioned lineages with presumably very low effective population sizes. The availability of the conversion tracts found by ClonalFrame allows testing the hypothesis that biased gene conversion purges AT SNPs. The SNPs carried by recombination tracts lead to an inferred fourfold degenerate G+C composition at equilibrium of 62% G+C. This is also well below the extant G+C composition of the genome at these positions. Thus, biased gene conversion does not enrich the G+C content of the genome. Instead, it introduces SNPs that have been moderated by natural selection for a larger period of time and have thus fourfold degenerate sSNP composition intermediate between that of extant genomes (82%) and those given by the mutation matrix outside HGT regions (56%) (Castillo-Ramírez et al. 2011). This is in agreement with the action of natural selection in moderating G+C content by purging selectively AT-enriching SNPs, in which case, it suggests that synonymous polymorphisms are not neutral in MTBC.
Natural selection moderates nsSNP and sSNP in MTBC
Overall, two-thirds of the SNPs in our data set are non-synonymous (nsSNP), and one-third are synonymous (sSNP). Yet, the ratio of nsSNP/sSNP varies along the tree, such that terminal branches have a ratio ∼25% higher than the inner branches close to the root (1.9 and 1.5, respectively, p < 0.001, χ2 test). This trend remains significant after removal of the recombination tracts of ClonalFrame (p < 0.005, same test). Previous analyses of smaller MTBC data sets failed to find this pattern (Hershberg et al. 2008), but similar results were found in several other species, e.g., Bacillus cereus, Staphylococcus aureus, Streptococcus, Rickettsia, Shigella, Salmonella, and Clostridium difficile (Rocha et al. 2006a; Blanc et al. 2007; Holt et al. 2008). The simplest explanation for this pattern is that many of the standing non-synonymous changes are slightly deleterious and are purged by natural selection. Because this takes time, internal branches, which represent older polymorphisms, show a smaller ratio of nsSNP/sSNP. These results show that nsSNPs are more intensely purged by natural selection than sSNPs in MTBC.
We then analyzed the nsSNP/sSNP ratio in relation to HGT regions and conversion tracts. After removing recombination tracts, HGT regions have an excess of nsSNPs relative to the rest of the genome (nsSNP/sSNP ratios of 2.5 and 1.8, respectively, P-value < 0.0001, χ2 test). Using the same method, we obtained the same result when we analyzed the HGT regions previously identified by phylogenetic methods (Veyrier et al. 2009) (nsSNP/sSNP ratios of 2.3 and 1.8, respectively, P-value < 0.0001, χ2 test). This is consistent with increased genetic diversity in these regions caused by positive selection and/or relaxed purifying selection. We also found an over-representation of nsSNP in recombination tracts relative to the rest of the genome (2.4 and 1.8, respectively, p < 0.0001, same test). This is surprising since recombination tracts over-represent old polymorphisms and are thus expected to contain a higher fraction of sSNP (Castillo-Ramírez et al. 2011). This further suggests an important role for recombination in providing adaptive genetic diversity in MTBC.
Recent experimental work has reached the surprising conclusion that the mutation rate of M. tuberculosis per unit of time is nearly constant (2.4 × 10−10, 3 × 10−10, and 3.5 × 10−10 per day, for the states of active disease, latency, and reactivated infection in the macaque model) (Ford et al. 2011). As a result, the rate of accumulation of SNPs in neutral regions of the genome should allow dating of the last common ancestor of the M. tuberculosis lineages. We used the ClonalFrame results to exclude recombination tracts and computed the number of tip-to-root synonymous SNPs. We found an average of 419 tip-to-root synonymous SNPs (sSNPs) for a total of ∼900,000 synonymous positions (computed as one-fourth of all gene positions) (Nei and Gojobori 1986). Using these values and assuming sSNPs to evolve neutrally, we computed an expected date for the last common ancestor of M. tuberculosis. The values are 5200 yr/4200 yr/3600 yr before the present using the mutation rate for the three different physiological states. This value is too low since data from both paleogenomics and population genomics approaches show that MTBC arose 10,000–40,000 yr ago (Gutierrez et al. 2005; Wirth et al. 2008; Donoghue 2009; Djelouadji et al. 2011). These results are consistent with previous observations in other bacteria showing that the short-term and long-term evolutionary rates can differ by orders of magnitude (Morelli et al. 2010). The slower-than-expected accumulation of sSNPs in MTBC is in agreement with our analyses of the mutation matrices: They both suggest significant moderation of synonymous substitutions by natural selection.
Cell envelope–related genes diversify at high rates by recombination
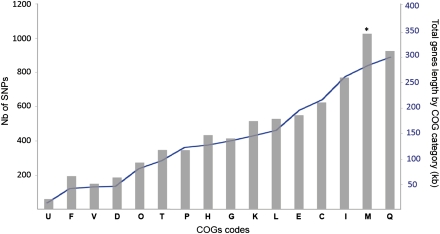
If diversification of the genomes were significantly moderated by natural selection, we would expect genes under diversifying selection (e.g., genes with virulence-associated functions) to have an excess of SNPs relative to other genes. Given the low level of divergence between the strains, genewise tests of positive selection lack power and were not useful to test this hypothesis (data not shown). Instead, we identified the functional classes that have diversified to a larger extent in the MTBC. We analyzed the distribution of SNPs according to the different classes of the Clusters of Orthologous Groups (COG) (Tatusov et al. 1997, 2003). After accounting for the relative size of COGs, we found that genes playing a role in the class cell wall/membrane/envelope biogenesis (class M) are significantly enriched in SNPs (p = 0.0015) (Fig. 7; Supplemental Table 4). No other COG class significantly over-represents or under-represents SNPs. The class Q (secondary metabolites biosynthesis, transport, and catabolism) includes more SNPs than class M, but it is also larger, and its relative over-representation of SNPs is not statistically significant (p > 0.1 after sequential Bonferroni correction).
Figure 7.
Distribution of SNPs according to the Clusters of Orthologous Groups (COG) classification. (U) Intracellular trafficking and secretion; (F) nucleotide transport and metabolism; (V) defense mechanisms; (D) cell cycle control, mitosis, and meiosis; (O) post-translational modification, protein turnover, chaperones; (T) signal transduction mechanisms; (P) inorganic ion transport and metabolism; (H) coenzyme transport and metabolism; (G) carbohydrate transport and metabolism; (K) transcription; (L) replication, recombination, and repair; (E) amino acid transport and metabolism; (C) energy production and conversion; (I) lipid transport and metabolism; (M) cell wall/membrane/envelope biogenesis; (Q) secondary metabolites biosynthesis, transport, and catabolism. (*) Class with significant over-representation of SNPs (p = 0.0015, Binomial test and sequential Bonferroni correction).
The over-representation of SNPs in the class of “cell envelope” is compatible with previous observations that some of the most striking clusters of polymorphism occur in proteins that come into direct contact with the host (Deitsch et al. 1997; Kennemann et al. 2011). Interestingly, it has recently been shown that human T-cell epitopes are highly conserved in MTBC (Comas et al. 2010), in line with the presence of strong purifying selection. Variation in “cell envelope” might thus not be implicated in antigenic variation but in other processes such as adaptation to local genetic background (Caws et al. 2008). The SNPs identified in genes included in this COG category are distributed throughout 246 genes among which 33 genes have SNPs (in different positions) in all M. tuberculosis strains. Each lineage has its own specificities because 47, 41, and 43 genes accumulate SNPs for the East-Asian, Euro-American and Indo-Oceanic lineages, respectively. This COG class significantly over-represents SNPs even when removing from the analysis the 13 regions where the density of SNPs is higher than in the rest of the genome (P-value < 0.01). If we remove from the analysis the SNPs located in recombination tracts, then this class no longer over-represents SNPs (P-value > 0.1). This result indicates that quick diversification of class M proteins is at least in part driven by recombination rather than by positive selection of newly arising mutations. This illustrates the importance of accounting for recombination when analyzing the functional aspects of the diversification of M. tuberculosis.
Discussion
M. tuberculosis is still a very serious public health problem and a challenge for the scientific community. It is often depicted as a strictly human, slow-growing, intracellular pathogen with a geographically structured population resulting from sexual isolation, very low infectious dose, and small effective population sizes (Sreevatsan et al. 1997; Musser et al. 2000; Gagneux and Small 2007). As a result, it has often been assumed that genetic diversity in MTBC accumulates clonally and neutrally. Yet, many of these arguments are disputable:
There is no significant correlation between optimal growth rate and effective population size in prokaryotes (Vieira-Silva et al. 2011). On the contrary, it has been proposed that Mycobacterium granulomas grow larger at lower growth rates (Segovia-Juarez et al. 2004), suggesting that slow growth is adaptive, not the result of genetic drift.
Just 10 MTBC cells are enough to start an infection with 50% probability (ID50) (Balasubramanian et al. 1994; Dean et al. 2005). If only a few cells are transmitted each time, then population bottlenecks could severely decrease the effective population size. Yet, ID50 is a lower-bound value; the exact number of cells transmitted is unknown and might be much larger.
Ours and previous works have shown that there is substantial genetic diversity within MTBC given its recent ancestry (Gutierrez et al. 2005; Gagneux and Small 2007; Hershberg et al. 2008). Importantly, this has been accompanied by rapid population growth (Wirth et al. 2008).
As mentioned in the introduction, the genomes of MTBC lack the obvious traces of massive genome degradation present in other lineages such as expansion of transposable elements, abundant pseudogenes, and extended loss of housekeeping functions. In fact, the size of MTBC genomes is nearly the same size as that of M. canettii. Paleogenomics data also suggest that MTBC strains have stably maintained their virulence traits for a large period of time, suggesting that the disease might be older than the last common ancestor of extant strains (Brosch et al. 2002; Donoghue et al. 2004; Gutierrez et al. 2005).
There is evidence that different MTBC strains are adapted to different human genetic backgrounds (Hirsh et al. 2004), and this could indicate population fragmentation and lower effective population sizes. Yet, the Beijing strains are virulent in many such backgrounds (Glynn et al. 2002), and even M. bovis, which is responsible for <1% of human tuberculosis in industrialized countries (Thoen et al. 2006), might be more prevalent in particular areas and populations. Even in San Diego, 7% of human tuberculosis was caused by M. bovis between 1994 and 2000 (LoBue et al. 2003).
Many animal hosts, including ∼30% of the human population, carry the tubercle bacilli for very large periods of time. Coresidence and coinfection of different MTBC strains, and mycobacteria outside the MTBC clade, suggest that adaptations to hosts do not preclude coinfections or genetic exchange (Braden et al. 2001; Das et al. 2004; Mendez et al. 2009; Mallard et al. 2010).
Therefore, there may be less ground than commonly thought to the idea that the census population size of M. tuberculosis is particularly small for a human pathogen. Finally, we show in this study that previous claims of strict clonality and very weak purifying selection in MTBC also need revising.
The relatively high polymorphism introduced in the inferred recombination tracts is a strong indication that many such recombination events, which occurred on all branches of the tree, were originally acquired from genomes outside the MTBC. This is consistent with the finding of recombination tracts strictly identical to extant M. canettii sequences, the only Mycobacterium prototuberculosis strain available for comparison at the moment. M. prototuberculosis strains are very close to MTBC and have been shown to engage very often in recombination (Gutierrez et al. 2005). Therefore, the observation that some recombination tracts match M. canettii should be interpreted with care. This strain is thought to have a restricted geographical distribution in the Horn of Africa (van Soolingen et al. 1997), limiting its contact with MTBC strains. However, the lack of rapid and efficient diagnostic tests that allow the differentiation between M. canettii and M. tuberculosis may lead to an under-estimation of its range. Indeed, recent reports have suggested that the number of true cases of tuberculosis caused by M. canettii may be under-represented (Somoskovi et al. 2009; Fabre et al. 2010). Further genomic and epidemiological data are necessary to understand this pattern of transfer and which specific strains of M. prototuberculosis are sharing genetic information with MTBC. It will also be of interest to assess if exchange is reciprocal, i.e., if these strains also receive genetic information from MTBC and what are the consequences of this for the evolution of their virulence.
The high diversity of recombination tracts should not be interpreted as lack of recombination within MTBC strains that is nearly undetectable using available methods. Recombination frequency increases exponentially with genetic similarity (Vulic et al. 1997), and the MTBC is strongly structured by geography. Therefore recombination between MTBC should be much more frequent than recombination of MTBC with other species. As a result, we might be vastly under-estimating the rate of recombination within MTBC. Genetic exchanges between closely related strains can have a considerable role in terms of the population genetics of M. tuberculosis, e.g., they might allow breaking clonal interference and more efficient purge of slightly deleterious mutations. Precise assessment of the amount of recombination within MTBC is not possible currently because we do not know how to model the effects of population structure at large, geography, nor at small, coinfection, scales on the frequency of recombination. Experimental work and more intensive whole-genome sampling will be needed to clarify the extent of recombination between closely related strains of M. tuberculosis.
Strict clonality in MTBC has become a paradigm in the community. It is therefore important to understand why our results contradict the perceived clonality of MTBC. Given its recent origin, the MTBC shows low SNP density, and MLST-like analyses have not been as developed as for other pathogens. Instead, most typing and epidemiological studies of MTBC strains use methods based on deletions of genetic material (spoligotyping, regions of difference [RD], and MIRU-VNTR) (Otal et al. 1991; Kamerbeek et al. 1997; Supply et al. 2006) that are prone to homoplasy (Comas et al. 2009). It is not surprising that these methods have failed to identify frequent recombination. Single crossover requires sequence identity for at least ∼25 nt and high similarity throughout the recombining segment (Vulic et al. 1997). Because the frequency of recombination depends exponentially on the size of the fragment, sequences smaller than 300 nt recombine at low frequency (Biswas et al. 1993). A deletion can be reverted by recombination when incoming DNA sequences are large enough to include the deleted section and include bordering sequences recombining at each edge (double crossover). The small size of recombination fragments renders reversion of deletions in MTBC very unlikely. Hence, standard typing methods are not adapted to detect recombination in MTBC. Only SNP analysis can provide this information. Importantly, this also justifies why large (RD) deletions follow patterns typical of strict clonality: Small conversion tracts cannot recombine them.
Several lines of evidence in our study point to a significant role of natural selection in shaping MTBC genomes. First, the SNP distribution in genomes is not random, suggesting that diversifying selection is at work notably in genes playing a role in cell wall/membrane/envelope biogenesis, which tend to accumulate an excess of SNPs. This result is probably related to the lifestyle of the tubercle bacilli, producing long chronic colonization of hosts, which might result in selection pressure for arms races with the host, e.g., to evade host defenses (Dawkins and Krebs 1979) or diversifying selection to adapt to different host genetic backgrounds (Di Pietrantonio et al. 2011). While we still lack examples of horizontal gene transfer after the last common ancestor of MTBC, our approach was not aimed at identifying regions that could not be mapped in extant genomes; rather, we showed that regions transferred to the ancestor of MTBC also accumulate an excess of SNPs. These regions include genes contributing to the virulence of the M. tuberculosis genome whose diversification might be relevant for the differentiation of MTBC from other Mycobacteria and for the specialization of MTBC lineages. Second, we find evidence for predominantly purifying selection at non-synonymous positions. We show that ancient nsSNPs have been more intensely purged from populations, reflecting the efficiency of natural selection in removing deleterious polymorphisms. Third, our results are consistent with genome-wide purifying selection at synonymous sites, which is thought to require large effective population sizes. The substitution matrix of MTBC is far from the extremes that are the hallmark of lineages with very low effective population sizes. Nevertheless, it has in common with practically all other bacteria a gap between A+T content given by the mutation matrix and the extant genome. Since this gap is not compensated by recombination, we suggest that selective processes preferentially purge AT-enriching mutations. This efficiency of selection in MTBC is consistent with the observed codon usage bias in highly expressed M. tuberculosis genes (Andersson and Sharp 1996). The relatively low codon usage bias in MTBC needs not be ascribed to low effective population size because slow growers have low codon usage biases independently of effective population sizes (Vieira-Silva and Rocha 2010).
One might argue that convergent evolution creates homoplasies and that we are mistaking this process with recombination. We find this hypothesis very unlikely. Given the low density of SNPs in MTBC, homoplasies will only arise at high rates under strong selection for a given adaptation in the different lineages and when such an adaptation is only possible through a very small number of mutations. Accordingly, convergent evolution caused by precisely identical point mutations is very rarely observed in experimental evolution and works of population genetics (Woods et al. 2006; Christin et al. 2010). Furthermore, the recombination tracts identified by ClonalFrame have a minimum of two incongruent closely spaced SNPs. Hence, for these tracts to be caused by convergent evolution, one needs to invoke two closely spaced very unlikely events. Finally, among the recombination tracts identified in intragenic regions, 52.3% include sSNPs and 11.2% include only sSNPs, which are not expected to be under strong selection and lead to convergence. While we cannot exclude the presence of homoplasies in our data set caused by natural selection, these are very unlikely to affect significantly our estimates of recombination rates.
Selective diversification of MTBC might explain the association between host response and strain genetic background recently reported (Portevin et al. 2011). Recombination, as an important component of MTBC diversification, may have allowed rapid generation of genetic diversity and more efficient purifying selection of deleterious substitutions that became accidentally fixed at the original or subsequent bottlenecks of the MTBC lineage. This is especially relevant for the tracts imported from lineages outside the MTBC clade as these add more SNPs. Interestingly, both in Streptococcus and in Listeria, there is also evidence for higher rates of recombination in regions under diversifying selection (Orsi et al. 2008; Croucher et al. 2011; Marttinen et al. 2011). We found that a large fraction of recombination events take place in intragenic regions. Such microdiversity could affect gene expression and thereby play a role in the evolution of virulence. Accordingly, we found that recombination is an essential contributor to genetic diversification of proteins involved in “cell wall.” MTBC have been perceived as strictly clonal, monomorphic lineages in which genetic drift dominates. Our results suggest that this view should be moderated. (1) MTBC shows a significant imprint of recombination with other clades and might recombine frequently with other MTBC. (2) Natural selection shapes the diversification of the lineages both at non-synonymous and synonymous sites. (3) Natural selection and recombination moderate the effects of mutations such that certain functions get enriched and others impoverished in genetic diversity. Hence, contrary to other well-known recent pathogens, M. tuberculosis may have emerged from the founding bottleneck with little genetic load and capable of stably diversifying into a successful pathogen.
Methods
Bacterial strains
We used three different sources of sequence data: complete genome sequences, unfinished genomes, and paired-end Illumina reads (Supplemental Table S1). We sequenced the genomes of three Beijing strains: R34-990172, GC1237, and W4. All sequencing data were deposited to the European Nucleotide Archive (ENA) database, and the entire data set is available using the study accession number ERP001173. We analyzed five other Beijing strains from the East-Asian lineage (94_M4241A*, 02_1987*, T85*, and W148*; 210), seven strains from the Euro-American lineage (CDC1551, F11, KZN1435, Haarlem, C-strain, GM1503*, 98-604*), four strains from the Indo-Oceanic lineage (T17*, T46*, T92*, EAS054*), two strains from the West-African lineage (K85*, CPHLA*), and finally, the BCG str. Pasteur 1173P2 and the M. bovis AF2122/97 strain. Supplemental Figure S1 summarizes the steps followed to analyze the data. Strains marked above with an asterisk were made available by the Mycobacterium tuberculosis Diversity Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/).
DNA sequencing
The genomic DNA was sequenced using the Illumina Genome Analyzer GAII, according to the manufacturer's specifications with the paired-end module attachment (GATC Biotech AG). Samples were prepared to produce a mean fragment size of 200 bp and 36 bp reads for the GC1237 genome and 77 bp for the R34-990172 and W4 strains. To optimize downstream analyses, quality control of the read-pairs of the Illumina pipeline were performed using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) by analyzing each set of reads.
SNPs mapping and comparative analyses
Paired-end reads were mapped to the genome sequence of M. tuberculosis H37Rv reference strain using two different programs. MAQ (Mapping and Assembly with Qualities) (Li et al. 2008) performs ungapped alignment of the Illumina reads to the H37Rv reference genome. MAQ uses read-pairs to accurately map reads to repetitive sequences if their mate-pairs are confidently aligned. The MAQ output file was visualized using the MapView viewer (Bao et al. 2009). We also made gapped global alignment of paired-end reads with the inGAP pipeline (Qi et al. 2010), which implements the Burrows-Wheeler transform-based aligner (Li and Durbin 2009). We visualized the mapped reads to the whole-genome sequence of the reference strain and its genome annotation using the inGAP pipeline. We developed several Python modules that use the output of the different aligner programs listed above (MAQ, BWA, MUMmer 3.2, and Mauve 2.3.1) to identify specific and shared SNPs between all compared genomes. For the sequenced genomes in this study, SNPs have been checked by taking into account the quality-control score of each nucleotide position (QC > 20). For the other genomes, all specific SNPs for each strain were manually inspected by taking into account if SNPs were detected by the two aligners, MUMmer 3.2 (Kurtz et al. 2004) and MAUVE (Darling et al. 2004), and by checking if these SNPs were located outside repetitive sequences. On the other hand, and in order to get a more precise estimation of the quality of identified SNPs in our study, we were able to retrieve for five strains (T92, T85, T17, GM1503, and W148) the original set of Illumina reads with their quality-control files. For these strains, the comparison of the two sets of SNPs—(1) the SNPs obtained by analyzing the contig data (this study) and (2) the SNPs obtained after analyzing the Illumina reads—gave us an estimate of the number of spurious SNPs arising from sequencing or assembling errors.
SNP clustering
From all SNPs identified in the 23 genome sequences, the density of SNPs was calculated throughout the Mycobacterial genome using a sliding-window size of 5 kb (step of the sliding window = 5 kb). This analysis led to the construction of a SNP clustering map. The distribution of all of these regions was compared with the genomic location of the previously reported horizontal transfer regions (HGT) (Becq et al. 2007; Veyrier et al. 2009).
Evolutionary analyses
Phylogenies were inferred using RaxML (version 7.2.8) (Stamatakis 2006), Tree-Puzzle (version 5.2) (Schmidt et al. 2002), and BEAST (version 1.6.1) (Drummond and Rambaut 2007). RaxML was used for maximum likelihood (ML)–based estimates of the MTBC phylogeny, and 1000 bootstrap replicates were performed to assess statistical support. The trees were visualized using FigTree (version 1.2.1). For Tree-Puzzle and BEAST phylogenies, please refer to the Supplemental Material. The substitutions leading to each SNP were mapped in the phylogenetic tree using pamp from the PAML package (version 4.4e) (Yang 2007).
Detection of recombination
We used the split decomposition method implemented in SplitsTree4 (version 4.11.3) to compute unrooted phylogenetic networks (Huson and Bryant 2006), which were validated statistically using the Phi test. The Phi test calculates the pairwise homoplasy index (PHI) as the mean of the refined incompatibility scores obtained for nearby nucleotide sites along the sequences (Bruen et al. 2006). For all pairs of SNPs, we also computed the correlation coefficient r2, which is a measure of linkage disequilibrium between sites (Hill 1975). We compared the level of LD with the genomic distance between pairs of sites to test for the presence of recombination tracts in the ancestry of the 24 genomes (Shapiro et al. 2009). Recombination was also detected using geneconv (Sawyer 1989) with default parameters.
ClonalFrame analysis
We used ClonalFrame version 1.2 (Didelot and Falush 2007) to reconstruct the clonal genealogy relating the 24 genomes to each other, as well as the identification of the genomic position of homologous recombination events where inheritance did not follow this clonal genealogy. The basic model in ClonalFrame is one of extrapopulation recombination. However, as demonstrated before (Didelot and Falush 2007), this can also pick up a fair proportion of intrapopulation recombination. ClonalFrame was run for 100,000 iterations, the first half of which were discarded as burn-in. Convergence and mixing were found to be satisfactory by manual comparison of four independent runs as well as using the method of Gelman and Rubin (1992). We looked for the origin of recombination tracts in the more distantly related genomes Mycobacterium avium (NC_008595), M. marinum (NC_010612), M. kansassii ATCC 12478 (NZ_ACBV00000000), and M. canettii (NC_015848, CIPT140010059). We extracted from the ClonalFrame output the sequences that had been inferred to be recombinant, and searched for similar sequences using BLAST (Altschul et al. 1990, 1997) against each of the genomes above, as done in Didelot et al. (2009).
Data access
All sequencing data have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under study accession number ERP001173.
Acknowledgments
This work was supported by the European Commission Seventh Framework Programme (FP7/2009-2011, TB-VIR grant no. 200973), ANR-09-MIEN-024-02. The EPCR laboratory is funded by the CNRS and the Institut Pasteur. We thank Ed Feil (Department of Biology & Biochemistry, University of Bath) for comments and criticisms on a previous version of this manuscript. We thank the reviewers for their comments, suggestions, and criticisms.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.129544.111.
References
- Achtman M 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62: 53–70 [DOI] [PubMed] [Google Scholar]
- Alland D, Lacher DW, Hazbon MH, Motiwala AS, Qi W, Fleischmann RD, Whittam TS 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J Clin Microbiol 45: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso H, Aguilo JI, Samper S, Caminero JA, Campos-Herrero MI, Gicquel B, Brosch R, Martin C, Otal I 2011. Deciphering the role of IS6110 in a highly transmissible Mycobacterium tuberculosis Beijing strain, GC1237. Tuberculosis (Edinb) 91: 117–126 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson GE, Sharp PM 1996. Codon usage in the Mycobacterium tuberculosis complex. Microbiology 142: 915–925 [DOI] [PubMed] [Google Scholar]
- Aranaz A, Cousins D, Mateos A, Dominguez L 2003. Elevation of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to species rank as Mycobacterium caprae comb. nov., sp. nov. Int J Syst Evol Microbiol 53: 1785–1789 [DOI] [PubMed] [Google Scholar]
- Balasubramanian V, Wiegeshaus EH, Taylor BT, Smith DW 1994. Pathogenesis of tuberculosis: Pathway to apical localization. Tuber Lung Dis 75: 168–178 [DOI] [PubMed] [Google Scholar]
- Balbi KJ, Rocha EP, Feil EJ 2009. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol 26: 345–355 [DOI] [PubMed] [Google Scholar]
- Bao H, Guo H, Wang J, Zhou R, Lu X, Shi S 2009. MapView: Visualization of short reads alignment on a desktop computer. Bioinformatics 25: 1554–1555 [DOI] [PubMed] [Google Scholar]
- Becq J, Gutierrez MC, Rosas-Magallanes V, Rauzier J, Gicquel B, Neyrolles O, Deschavanne P 2007. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol 24: 1861–1871 [DOI] [PubMed] [Google Scholar]
- Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol 10: 45–52 [DOI] [PubMed] [Google Scholar]
- Biswas I, Gruss A, Ehrlich SD, Maguin E 1993. High-efficiency gene inactivation and replacement system for Gram-positive bacteria. J Bacteriol 175: 3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, et al. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5: e1000507 doi: 10.1371/journal.ppat.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Ogata H, Robert C, Audic S, Suhre K, Vestris G, Claverie JM, Raoult D 2007. Reductive genome evolution from the mother of Rickettsia. PLoS Genet 3: e14 doi: 10.1371/journal.pgen.0030014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braden CR, Morlock GP, Woodley CL, Johnson KR, Colombel AC, Cave MD, Yang Z, Valway SE, Onorato IM, Crawford JT 2001. Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin Infect Dis 33: e42–e47 [DOI] [PubMed] [Google Scholar]
- Brodin P, Eiglmeier K, Marmiesse M, Billault A, Garnier T, Niemann S, Cole ST, Brosch R 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect Immun 70: 5568–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci 99: 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia I, Cabrera P, Lafoz C, Samper S, Takiff H, Afonso O, et al. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med 164: 1165–1170 [DOI] [PubMed] [Google Scholar]
- Castillo-Ramírez S, Harris SR, Holden MTG, He M, Parkhill J, Bentley SD, Feil EJ 2011. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog 7: e1002129 doi: 10.1371/journal.ppat.1002129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NT, Thuong NT, Stepniewska K, Huyen MN, Bang ND, Loc TH, et al. 2008. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog 4: e1000034 doi: 10.1371/journal.ppat.1000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci 101: 13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Weinreich DM, Besnard G 2010. Causes and evolutionary significance of genetic convergence. Trends Genet 26: 400–405 [DOI] [PubMed] [Google Scholar]
- Cole ST 1998. Comparative mycobacterial genomics. Curr Opin Microbiol 1: 567–571 [DOI] [PubMed] [Google Scholar]
- Comas I, Gagneux S 2011. A role for systems epidemiology in tuberculosis research. Trends Microbiol 19: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Homolka S, Niemann S, Gagneux S 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE 4: e7815 doi: 10.1371/journal.pone.0007815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins DV, Bastida R, Cataldi A, Quse V, Redrobe S, Dow S, Duignan P, Murray A, Dupont C, Ahmed N, et al. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int J Syst Evol Microbiol 53: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331: 430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT 2004. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Narayanan S, Hari L, Mohan NS, Somasundaram S, Selvakumar N, Narayanan PR 2004. Simultaneous infection with multiple strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis 8: 267–270 [PubMed] [Google Scholar]
- Dawkins R, Krebs JR 1979. Arms races between and within species. Proc R Soc Lond B Biol Sci 205: 489–511 [DOI] [PubMed] [Google Scholar]
- Dean GS, Rhodes SG, Coad M, Whelan AO, Cockle PJ, Clifford DJ, Hewinson RG, Vordermeier HM 2005. Minimum infective dose of Mycobacterium bovis in cattle. Infect Immun 73: 6467–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Moxon ER, Wellems TE 1997. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol Mol Biol Rev 61: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Darling A, Falush D 2009. Inferring genomic flux in bacteria. Genome Res 19: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D 2010. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186: 1435–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietrantonio T, Correa JA, Orlova M, Behr MA, Schurr E 2011. Joint effects of host genetic background and mycobacterial pathogen on susceptibility to infection. Infect Immun 79: 2372–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djelouadji Z, Raoult D, Drancourt M 2011. Palaeogenomics of Mycobacterium tuberculosis: Epidemic bursts with a degrading genome. Lancet Infect Dis 11: 641–650 [DOI] [PubMed] [Google Scholar]
- Donoghue HD 2009. Human tuberculosis—an ancient disease, as elucidated by ancient microbial biomolecules. Microbes Infect 11: 1156–1162 [DOI] [PubMed] [Google Scholar]
- Donoghue HD, Spigelman M, Greenblatt CL, Lev-Maor G, Bar-Gal GK, Matheson C, Vernon K, Nerlich AG, Zink AR 2004. Tuberculosis: From prehistory to Robert Koch, as revealed by ancient DNA. Lancet Infect Dis 4: 584–592 [DOI] [PubMed] [Google Scholar]
- Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, Rasolofo V, Tonjum T, Sola C, Matic I, Gicquel B 2008. Evolution and diversity of clonal bacteria: The paradigm of Mycobacterium tuberculosis. PLoS ONE 3: e1538 doi: 10.1371/journal.pone.0001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 doi: 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL 2004. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci 101: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre M, Hauck Y, Soler C, Koeck JL, van Ingen J, van Soolingen D, Vergnaud G, Pourcel C 2010. Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect Genet Evol 10: 1165–1173 [DOI] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299: 1582–1585 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 1974. The evolutionary advantage of recombination. Genetics 78: 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbon MH, Bobadilla del Valle M, Fyfe J, Garcia-Garcia L, Rastogi N, Sola C, et al. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: Insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol 188: 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, et al. 2011. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 43: 482–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, Small PM 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7: 328–337 [DOI] [PubMed] [Google Scholar]
- Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, et al. 2006. Variable host–pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci 103: 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman AG, Rubin DB 1992. Inference from Iterative simulation using multiple sequences. Stat Sci 7: 457–511 [Google Scholar]
- Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: A systematic review. Emerg Infect Dis 8: 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiol 145: 881–892 [DOI] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, Musser JM 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis 193: 121–128 [DOI] [PubMed] [Google Scholar]
- Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog 1: e5 doi: 10.1371/journal.ppat.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci 107: 7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6: e1001115 doi: 10.1371/journal.pgen.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, et al. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6: e311 doi: 10.1371/journal.pbio.0060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet 6: e1001107 doi: 10.1371/journal.pgen.1001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG 1975. Linkage disequilibrium among multiple neutral alleles produced by mutation in finite population. Theor Popul Biol 8: 117–126 [DOI] [PubMed] [Google Scholar]
- Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci 101: 4871–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TB, Robertson BD, Taylor GM, Shaw RJ, Young DB 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, et al. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat Genet 40: 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR 1983. Properties of a neutral allele model with intragenic recombination. Theor Popul Biol 23: 183–201 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R, Murray M 2002. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg Infect Dis 8: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, et al. 2002. Genome sequence of Shigella flexneri 2a: Insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res 30: 4432–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD 2011. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol Direct 6: 28 doi: 10.1186/1745-6150-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karboul A, Mazza A, Gey van Pittius NC, Ho JL, Brousseau R, Mardassi H 2008. Frequent homologous recombination events in Mycobacterium tuberculosis PE/PPE multigene families: Potential role in antigenic variability. J Bacteriol 190: 7838–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, et al. 2011. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci 108: 5033–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12 doi: 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H 1997. Amelioration of bacterial genomes: Rates of change and exchange. J Mol Evol 44: 383–397 [DOI] [PubMed] [Google Scholar]
- Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 187: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gutacker MM, Musser JM, Fu YX 2006. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol 188: 8169–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoBue PA, Betacourt W, Peter C, Moser KS 2003. Epidemiology of Mycobacterium bovis disease in San Diego County, 1994–2000. Int J Tuberc Lung Dis 7: 180–185 [PubMed] [Google Scholar]
- Majewski J, Cohan FM 1998. The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics 148: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majlessi L, Brodin P, Brosch R, Rojas MJ, Khun H, Huerre M, Cole ST, Leclerc C 2005. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J Immunol 174: 3570–3579 [DOI] [PubMed] [Google Scholar]
- Mallard K, McNerney R, Crampin AC, Houben R, Ndlovu R, Munthali L, Warren RM, French N, Glynn JR 2010. Molecular detection of mixed infections of Mycobacterium tuberculosis strains in sputum samples from patients in Karonga District, Malawi. J Clin Microbiol 48: 4512–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J 2011. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40: e6 doi: 10.1093/nar/gkr928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy CR, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM 2007. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 87: 393–404 [DOI] [PubMed] [Google Scholar]
- Mendez MP, Landon ME, McCloud MK, Davidson P, Christensen PJ 2009. Co-infection with pansensitive and multidrug-resistant strains of Mycobacterium tuberculosis. Emerg Infect Dis 15: 578–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet 17: 589–596 [DOI] [PubMed] [Google Scholar]
- Morelli G, Didelot X, Kusecek B, Schwarz S, Bahlawane C, Falush D, Suerbaum S, Achtman M 2010. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet 6: e1001036 doi: 10.1371/journal.pgen.1001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Inwald J, Gordon S, Martin C, Warren R, Kremer K, Cousins D, Behr MA 2005. Revisiting the evolution of Mycobacterium bovis. J Bacteriol 187: 6386–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Amin A, Ramaswamy S 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: Evidence of limited selective pressure. Genetics 155: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Chakraborty R 1975. The bottleneck effect and genetic variability of populations. Evolution 29: 1–10 [DOI] [PubMed] [Google Scholar]
- Ohta T 1992. Theoretical study of near neutrality. II. Effect of subdivided population structure with local extinction and recolonization. Genetics 130: 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, Sun Q, Wiedmann M 2008. Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol Biol 8: 233 doi: 10.1186/1471-2148-8-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otal I, Martin C, Vincent-Levy-Frebault V, Thierry D, Gicquel B 1991. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol 29: 1252–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35: 32–40 [DOI] [PubMed] [Google Scholar]
- Portevin D, Gagneux S, Comas I, Young D 2011. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog 7: e1001307 doi: 10.1371/journal.ppat.1001307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA 2001. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc Natl Acad Sci 98: 13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Brosch R, Huerre M, Cole ST 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol 46: 709–717 [DOI] [PubMed] [Google Scholar]
- Qi J, Zhao F, Buboltz A, Schuster SC 2010. inGAP: An integrated next-generation genome analysis pipeline. Bioinformatics 26: 127–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Feil EJ 2010. Mutational patterns cannot explain genome composition: Are there any neutral sites in the genomes of bacteria? PLoS Genet 6 e1001104 doi: 10.1371/journal.pgen.1001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ 2006a. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol 239: 226–235 [DOI] [PubMed] [Google Scholar]
- Rocha EP, Touchon M, Feil EJ 2006b. Similar compositional biases are caused by very different mutational effects. Genome Res 16: 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, Neyrolles O 2006. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol 23: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, et al. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-secreted proteins. PLoS Pathog 6: e1000794 doi: 10.1371/journal.ppat.1000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S 1989. Statistical tests for detecting gene conversion. Mol Biol Evol 6: 526–538 [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A 2002. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504 [DOI] [PubMed] [Google Scholar]
- Segovia-Juarez JL, Ganguli S, Kirschner D 2004. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol 231: 357–376 [DOI] [PubMed] [Google Scholar]
- Shapiro BJ, David LA, Friedman J, Alm EJ 2009. Looking for Darwin's footprints in the microbial world. Trends Microbiol 17: 196–204 [DOI] [PubMed] [Google Scholar]
- Smith JM, Haigh J 1974. The hitch-hiking effect of a favourable gene. Genet Res 23: 23–35 [PubMed] [Google Scholar]
- Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat Rev Microbiol 4: 670–681 [DOI] [PubMed] [Google Scholar]
- Somoskovi A, Dormandy J, Mayrer AR, Carter M, Hooper N, Salfinger M 2009. “Mycobacterium canettii” isolated from a human immunodeficiency virus-positive patient: First case recognized in the United States. J Clin Microbiol 47: 255–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci 94: 9869–9874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44: 4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat AM, Lyons R, Howard ST, Johnston SA 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci 101: 4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ 1997. A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: An updated version includes eukaryotes. BMC Bioinformatics 4: 41 doi: 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen C, Lobue P, de Kantor I 2006. The importance of Mycobacterium bovis as a zoonosis. Vet Microbiol 112: 339–345 [DOI] [PubMed] [Google Scholar]
- Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere YO, Aman K, Kato-Maeda M, Small PM 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: Insights from genomic deletions in 100 strains. Proc Natl Acad Sci 101: 4865–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uplekar S, Heym B, Friocourt V, Rougemont J, Cole ST 2011. Comparative genomics of Esx genes from clinical isolates of Mycobacterium tuberculosis provides evidence for gene conversion and epitope variation. Infect Immun 79: 4042–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, Hoogenboezem T, de Haas PE, Hermans PW, Koedam MA, Teppema KS, Brennan PJ, Besra GS, Portaels F, Top J, et al. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: Characterization of an exceptional isolate from Africa. Int J Syst Bacteriol 47: 1236–1245 [DOI] [PubMed] [Google Scholar]
- Veyrier F, Pletzer D, Turenne C, Behr MA 2009. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol Biol 9: 196 doi: 10.1186/1471-2148-9-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S, Rocha EP 2010. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet 6: e1000808 doi: 10.1371/journal.pgen.1000808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Silva S, Touchon M, Abby SS, Rocha EP 2011. Investment in rapid growth shapes the evolutionary rates of essential proteins. Proc Natl Acad Sci 108: 20030–20035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Groll A, Martin A, Stehr M, Singh M, Portaels F, da Silva PE, Palomino JC 2010. Fitness of Mycobacterium tuberculosis strains of the W-Beijing and Non-W-Beijing genotype. PLoS ONE 5: e10191 doi: 10.1371/journal.pone.0010191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulic M, Dionisio F, Taddei F, Radman M 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci 94: 9763–9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Rocha EP 2011. Function-specific accelerations in rates of sequence evolution suggest predictable epistatic responses to reduced effective population size. Mol Biol Evol 28: 2339–2349 [DOI] [PubMed] [Google Scholar]
- Wirth T, Hildebrand F, Allix-Beguec C, Wolbeling F, Kubica T, Kremer K, van Soolingen D, Rusch-Gerdes S, Locht C, Brisse S, et al. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 4: e1000160 doi: 10.1371/journal.ppat.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci 103: 9107–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]