Abstract
Phylogenomics offers the potential to fully resolve the Tree of Life, but increasing genomic coverage also reveals conflicting evolutionary histories among genes, demanding new analytical strategies for elucidating a single history of life. Here, we outline a phylogenomic approach using a novel class of phylogenetic markers derived from ultraconserved elements and flanking DNA. Using species-tree analysis that accounts for discord among hundreds of independent loci, we show that this class of marker is useful for recovering deep-level phylogeny in placental mammals. In broad outline, our phylogeny agrees with recent phylogenomic studies of mammals, including several formerly controversial relationships. Our results also inform two outstanding questions in placental mammal phylogeny involving rapid speciation, where species-tree methods are particularly needed. Contrary to most phylogenomic studies, our study supports a first-diverging placental mammal lineage that includes elephants and tenrecs (Afrotheria). The level of conflict among gene histories is consistent with this basal divergence occurring in or near a phylogenetic “anomaly zone” where a failure to account for coalescent stochasticity will mislead phylogenetic inference. Addressing a long-standing phylogenetic mystery, we find some support from a high genomic coverage data set for a traditional placement of bats (Chiroptera) sister to a clade containing Perissodactyla, Cetartiodactyla, and Carnivora, and not nested within the latter clade, as has been suggested recently, although other results were conflicting. One of the most remarkable findings of our study is that ultraconserved elements and their flanking DNA are a rich source of phylogenetic information with strong potential for application across Amniotes.
Phylogenomics offers the possibility of a fully resolved Tree of Life (Delsuc et al. 2005; Dunn et al. 2008). Yet the intuitively appealing prospect that the signal of the true species history will overwhelm the random noise inherent to phylogenetic data is tempered by studies showing that treating all genes as if they share a single history can lead to highly supported, but incorrect phylogenies (Mossel and Vigoda 2005). In fact, as more and more DNA sequences are collected, researchers have found that genes often show conflicting histories (Pollard et al. 2006) that must be resolved into a single species history through emerging analytical methods that accommodate this source of phylogenetic uncertainty (Edwards et al. 2007; Knowles 2009). A major contributor to conflicting gene histories is coalescent stochasticity, which describes the random, independent sorting of genes within the boundaries of species histories (Kingman 1982). Coalescent stochasticity leads to discordant phylogenetic patterns among gene trees, especially when speciation events have occurred in quick succession (Maddison 1997). Because rapid radiations are common in nature (Schluter 2000), establishing the Tree of Life depends on a framework that expands genomic coverage while resolving conflict among gene histories. Although it is increasingly feasible to sequence entire genomes, identifying portions of the genome that are orthologous and independently sorting is highly desirable from the perspective of analyses that take coalescent stochasticity into account.
Given their long history of study (Murphy et al. 2001a,b; Bininda-Emonds et al. 2007), extensive genomic resources (Nikolaev et al. 2007), and rapid radiation (Bininda-Emonds et al. 2007; Stadler 2011), placental mammals provide an ideal group for testing approaches that resolve discordant gene histories using phylogenomic data. In the last decade, gene-by-gene sequencing and conventional phylogenetic methods have resolved many mammalian relationships (Murphy et al. 2004). Surprisingly, phylogenomic studies have not demonstrably improved resolution of the mammal Tree of Life, and in some cases, the results of phylogenomic studies have been contradictory (Cannarozzi et al. 2007). The lack of increased phylogenomic resolution can be attributed in part to rapid, ancient diversification (Murphy et al. 2004), conditions in which integrating models of discordant gene histories into the estimation of a single species history becomes critically important (Degnan and Rosenberg 2006), but where such models are rarely applied, often due to computational constraints.
Available evidence indicates that several mammalian divergences occurred extremely rapidly. The basal divergence among the three placental mammal superorders—Afrotheria (e.g., elephants, tenrecs), Xenarthra (e.g., sloths, armadillos), and Boreotheria (e.g., carnivores, bats, rodents, and primates)—is thought, based on fossil-calibrated divergence times, to have been completed within 2 to 5 million years (Springer et al. 2003; Murphy et al. 2007). Even more spectacular is the explosive radiation of orders within the Laurasiatheria (e.g., dogs, bats, horses, cows, dolphins), with several basal splits occurring within 1 to 3 million years of one another (Springer et al. 2003). As predicted by coalescent theory (Degnan and Salter 2005), these are the same divergences where individual gene trees give conflicting answers about evolutionary relationships (Nishihara et al. 2006, 2009; Murphy et al. 2007; Churakov et al. 2009), underscoring the need for an approach that incorporates a model of the process at the root of this discord—coalescent stochasticity. This is especially relevant given the recent discovery of a phylogenetic anomaly zone where rapid speciation results in the most common gene tree being in conflict with the true species history (Degnan and Rosenberg 2006), a situation that will mislead phylogenetic inference unless coalescent stochasticity is taken into account (Kubatko and Degnan 2007).
Here, we outline a generalizable framework for resolving phylogenomic discord using a novel class of markers: ultraconserved elements (UCEs) and flanking DNA (Supplemental Fig. S1). UCEs were first described in humans (Bejerano et al. 2004) and have since been found in other tetrapods (Stephen et al. 2008) and more distantly related species such as worms and yeast (Siepel et al. 2005). Many UCEs are thought to play an important role in regulation and development (Sandelin et al. 2004; Woolfe et al. 2004), and their function is a topic of intense study. There are several reasons why UCEs are expected to be good phylogenomic markers. A recent review identified key problems for phylogenomic analysis, among them incorrect identification of orthologs and saturation of nucleotide substitutions such that multiple substitutions at a given base position obscure true phylogenetic signal (Philippe et al. 2011). Addressing orthology, vertebrate UCEs have little overlap with most types of paralogous genes (Derti et al. 2006) and are found in largely transposon-free parts of the genome (Simons et al. 2006). With regard to saturation, UCEs and flanking sequence are expected to evolve slowly compared with other types of DNA, potentially making saturation less prevalent than with other marker types, a hypothesis we assess below.
After locating UCEs in amniotes and anchoring them in mammal genomes, the second part of our approach tackles the problem of coalescent stochasticity by integrating data from individual loci into recently developed phylogenetic algorithms for estimating species histories from collections of discordant gene histories (Liu et al. 2009). One of the major hurdles to estimating species trees from phylogenomic data has been the reliance of analytical methods on computationally intensive explorations of parameter space (Edwards et al. 2007), which become impractical with large phylogenomic data sets. We use a recent coalescent-based species-tree method based on ranks of coalescent events (STAR) (Liu et al. 2009) to capture relevant information about the species history from gene trees. STAR produces an analytical solution to the species tree but shows similar performance to more computationally intensive Bayesian species-tree methods (Liu et al. 2009). STAR is also robust to violations of a molecular clock, so it is particularly well suited to UCEs, which are likely to show deviations from molecular rate uniformity among taxa (for more detail on available species-tree methods, see Methods).
Results
Identification of UCEs in placental mammals
We used the 100% conserved regions of reptilian (including avian) UCEs to design 2560 in silico probes, which we aligned to existing mammal genomes (Supplemental Table S1). We excised UCE regions and 1000 bp of variable flanking sequence, which we hereafter refer to as “orthologous loci” (we expected few paralogs, but nonetheless screened for paralogs by removing probes with matches to multiple genomic locations). We minimized missing data by requiring all taxa in a data set to have all loci (although within a locus we permitted some missing data; average 6.5% of base pairs per alignment, mostly sequence at the margins of loci in outgroup taxa). Consequently, data sets with more taxa had fewer loci.
Construction of phylogenomic data sets
To explore the extremes of genomic and taxonomic coverage, we assembled two general-purpose data sets with different levels of genomic and taxonomic inclusion (Supplemental Table S1). One data set focused on high taxonomic coverage with 183 loci (total 94,607 bp) and 29 taxa. The other data set had high genomic coverage with 917 loci (total 908,702 bp) found in 19 taxa. To address two outstanding questions in placental mammal phylogeny that involve particularly rapid speciation, we also constructed two specialty data sets that aimed for high genomic coverage. We used the first specialty data set, 591 loci comprising six taxa and one outgroup, to assess the basal relationships of placental mammal superorders. We used the second specialty data set, 683 loci comprising six taxa and one outgroup, to investigate relationships within the rapidly radiating Laurasiatheria, focusing on the phylogenetic position of bats.
Poor taxon sampling can play an important role in phylogenetic inference (Philippe et al. 2011), although recent research by Wiens and Morrill (2011) has called into question some of the more dire predictions of poor taxonomic sampling that were found in a previous simulation study (Lemmon et al. 2009). As with all studies focusing on genome-scale data, our data sets contained many more loci than species. However, our most taxonomically inclusive data set, comprising 29 species, is the largest to date using information from vertebrates with readily available whole-genome information. In cases in which we used fewer species, we selected taxa to break up particularly long branches and mitigate the effects of long-branch attraction (i.e., we selected the two most divergent species within a group, e.g., two deeply divergent bat species).
Variability and saturation of core UCE and flanking DNA compared with exons
UCE loci had an average length of 400–750 bp depending on the data set (Supplemental Fig. S2) and were generally separated by wide physical distances (>2 Mb) (Supplemental Fig. S3), indicating that UCE loci were unlikely to be physically linked and were therefore likely to be segregating independently at the timescales considered. One basic feature of UCE loci is that they show increasing variability moving away from the core UCE toward the flanking regions (Stephen et al. 2008). Even though we defined UCE probe regions as 100% conserved between lizard and bird, there was usually some variability in the core UCE section in the other taxa included in the data sets. Analysis of a subset of loci (n = 20) from the 183-locus data set indicates that both core and flanking UCE regions contain variability and that variability is higher in flanking regions (Fig. 1A). Additionally, flanking regions had a higher ratio of informative sites to variable sites than the core UCE. Both core and flanking UCE regions had lower variability and a lower ratio of informative sites than the 20 loci analyzed for deep-level mammal phylogeny by Springer et al. (2007). However, this difference was largely driven by high variability at the third position of codons, whereas first and second positions had only marginally elevated variability compared with the UCE flanking regions (Fig. 1A). Additionally, core UCEs and flanking regions had significantly lower saturation indexes compared with those of exons, especially at third positions of codons (Fig. 1B), although no marker or codon position showed severe effects of saturation according to the significance tests used by Xia et al. (2003) (Supplemental Fig. S4).
Figure 1.
Variability and saturation of UCE core and flanking regions compared to exons. (A) High variability in exons from Springer et al. (2007) is driven largely by third positions of codons, whereas variability in first and second position codons is more similar to UCE flanking regions. (B) UCEs have low saturation indexes, whereas saturation is highest among third positions of codons of exons. Box plots show the mean (line within box), 25th to 75th percentiles (box), fifth to 95th percentiles (whiskers), and outliers (dots).
Phylogeny of placental mammals from UCE-fueled species trees
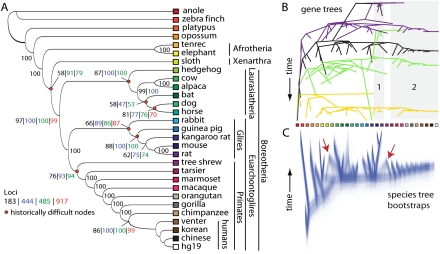
Using a method of species-tree analysis (STAR) (Liu et al. 2009) in which a species history is estimated from independent, often discordant gene histories, we recovered two species trees from our general-purpose data sets that were wholly concordant with one another (Fig. 2A; Supplemental Figs. S5, S6) despite the fact that individual gene histories showed considerable discordance (Fig. 2B; Supplemental Fig. S7). Notwithstanding topological conflict at the gene level, species-tree bootstrap replicates largely agreed (Fig. 2C). Bootstrap support values for the high genomic coverage data set tended to be higher (Fig. 2A). Topologies from the STAR trees largely agreed with topologies produced from Bayesian analysis of the same data sets where genes were concatenated and assumed to share the same evolutionary history (Supplemental Figs. S5, S6). One major difference was that, in the Bayesian concatenation analysis, the tree shrew grouped with Glires instead of with primates. The STAR tree, on the other hand, placed tree shrew in its accepted position as an outgroup to primates (Janecka et al. 2007).
Figure 2.
Evolutionary history of placental mammals resolved from conflicting gene histories. (A) Summary of STAR species trees generated from 183-locus and 917-locus data sets (Supplemental Table S1), in addition to the 444-locus data set that included UCEs from Stephen et al. (2008) and a 485-locus data set that included 41 exons (see Discussion). Note that STAR trees contain no branch length information. (B) Discord among four representative gene trees from the 183-locus data set. In general, gene trees were highly discordant, although some similarities emerged, such as the sister relationship between rat and mouse (shaded box 1) and monophyly of primates (shaded box 2). Discord among all gene trees is depicted in Supplemental Figure S7. (C) Widespread consensus among 1000 species-tree bootstrap replicates of the same 183-locus data set. STEAC trees (see Methods) are depicted because the branch lengths allow for better visualization of branching patterns, but STAR results supported the same topology. Cones emanating from terminal tips of species trees (red arrows) indicate disagreement among bootstrap replicates, for example, in the placement of the sloth and tree shrew. Colored squares indicate terminal taxa from A.
Assessing the need for species trees with coalescent simulation
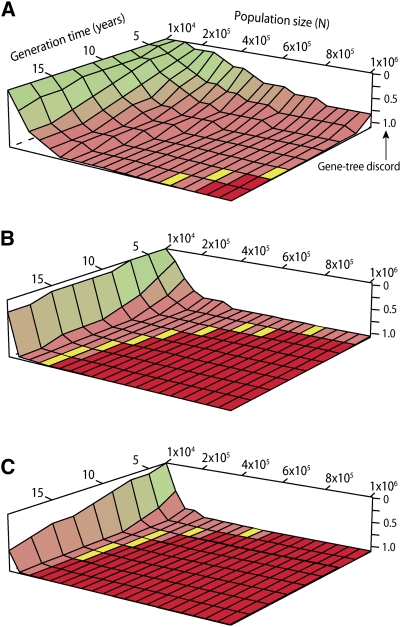
Next, using a coalescent simulation, we assessed the specific need for a species-tree approach to resolve the basal divergence of placental mammal superorders. Simulating gene histories on several possible species histories reflecting current knowledge about early placental mammal divergence (Bininda-Emonds et al. 2007) revealed that discord among gene histories is expected to be high over a large span of realistic demographic values for generation time, population size, and divergence time (Fig. 3). Plotting observed values of gene-tree discord from a recent study (Table 1; Nishihara et al. 2009) on this theoretical state-space suggests that the basal divergence event exists in a region of especially high gene-tree discord and may even lie in a phylogenetic anomaly zone where concatenation will mislead phylogenetic inference (Fig. 3). Values of gene-tree discord estimated from our own data (Table 1) are even higher and thus place the divergence event even closer to the anomaly zone.
Figure 3.
Basal divergence of placental mammals near the phylogenetic “anomaly zone.” Expected regions of gene-tree agreement (green) and discordance (pink) under a range of possible demographic parameters at the time of the divergence of the three placental mammal superorders. The phylogenetic “anomaly zone” where concatenation will fail (red) expands as speciation intervals shorten from 5 Mya (A), to 1 Mya (B), to 0.5 Mya (C). Empirical estimates of gene-tree discord (Table 1) from retroposons (Nishihara et al. 2009) are shown with yellow tiles, whereas estimates observed in our study would occur well within the anomaly zone. Speciation intervals for this divergence are thought to be closer to 2 Mya (Murphy et al. 2004).
Table 1.
Concordance among species trees and gene trees for basal divergence of placental mammals
UCE species tree supports Afrotheria as the first-diverging placental mammal lineage
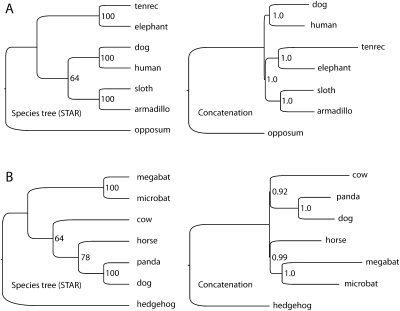
The STAR tree from the 683-locus data set created expressly to explore the basal relationships of placental mammal superorders indicated that Afrotheria was the first superorder to diverge, with 64% bootstrap support (Fig. 4A). Alternate topologies had less than half the bootstrap support: 31% of bootstrap replicates supported a sister relationship between Afrotheria and Xenarthra, and 5% supported a scenario with Xenarthra diverging first (Table 1). As expected based on the coalescent simulations, gene trees were highly discordant for the basal relationships. Of 591 gene trees, 433 (73%) were resolved for the relationships between Afrotheria, Xenarthra, and Boreotheria. Of the resolved gene trees, a plurality supported Afrotheria as diverging first (26% compared with 25% for Afrotheria and Xenarthra as sister taxa and 23% for Xenarthra diverging first) (Table 1). In contrast to the STAR tree, the Bayesian tree built from concatenated data supported Afrotheria and Xenarthra as sister taxa with high PP.
Figure 4.
Species trees and concatenated trees from high genomic coverage data sets for two rapid radiations in placental mammals. (A) Species tree from a 591-locus analysis identifies Afrotheria as the first-diverging lineage of placental mammals, whereas alternate topologies had less than half the bootstrap support (Table 1). A Bayesian analysis based on concatenated data places Afrotheria and Xenarthra together with high PP. Data on gene-tree discordance from Figure 2 suggest that this may be because the basal divergence of placental mammals lies close to the phylogenetic anomaly zone. (B) Species tree of taxa in the Laurasiatheria based on 683 loci places bats (Chiroptera) in a traditional location sister to Perissodactyla, Cetartiodactyla, and Carnivora. Bayesian analysis based on concatenated data produced an unusual tree with bats grouping sister to Perissodactyla, but with Carnivora grouping with Cetartiodactyla. The species tree and concatenated analysis of the 183 locus data set produced a different topology, more supportive of the hypothesized clade Pegasoferae (see text), suggesting that a robust understanding of this divergence event will require further investigation incorporating additional taxa and loci. Note that STAR trees do not contain branch length information.
UCE species tree and the phylogenetic placement of bats
With respect to the phylogenetic position of bats within the Laurasiatheria, our highest genomic coverage data set produced a STAR tree that supports bats as sister to a group containing Perissodactyla, Cetartiodactyla, and Carnivora, with 64% bootstrap support (Fig. 4B). The Bayesian tree with concatenated data produced an unusual topology with high PP for all nodes showing bats grouping sister to Perissodactyla, with this group sister to a monophyletic group containing Cetartiodactyla and Carnivora.
Discussion
Accurate phylogenomic inference has two important facets: estimating gene histories and inferring species histories from gene histories. The phylogenomic framework we use here addresses both of these challenges to infer the evolutionary history of placental mammals. With regard to gene trees, we introduce a novel class of genetic marker anchored by ultraconserved elements (UCEs) and show that UCE flanking regions have similar variability to first and second base positions of codons in exons (if slightly less information per locus) and significantly less saturation (Fig. 1B). Most of the information content of exons lies in the third positions of codons (Fig. 1A). UCEs have less informative variability (the same is true of first/second codon positions), but core UCEs and flanking regions also have much lower saturation scores. So although the lower variability of UCEs does not offer a strong advantage over exons, low saturation does make UCEs appealing markers for inferring gene trees for ancient evolutionary divergences, where multiple hits can create homoplasy, which obscures phylogenetic signal (Whitfield and Lockhart 2007).
With regard to species trees, we note that species-tree analysis, like the STAR method we use, can be conducted using most types of genetic markers. Retroposons are an exception, although there is no reason why the theoretical framework could not be extended to accommodate them. However, the principal benefit of using UCEs in species-tree analysis, compared with other types of markers like exons, is that the core region is highly conserved. This conservation allows UCEs to be rapidly characterized in high numbers in a broad array of species across the Tree of Life and with few of the problems of paralogy that plague other types of phylogenomic markers (Philippe et al. 2011). The sheer quantity of discrete UCEs shared among evolutionarily distant species thus addresses the issue of coalescent stochasticity, which requires many independent loci, in a way that would be more difficult to do with other marker types like exons. Additionally, purifying selection on UCEs (Katzman et al. 2007) could reduce the incidence of incomplete lineage sorting during short speciation intervals by reducing the effective population size (e.g., Hobolth et al. 2007; McVicker et al. 2009), which would also be an advantage over other marker types, but this remains to be tested.
When analyzed with a species-tree method, UCEs and flanking DNA sequence data successfully recovered points of broad agreement in placental mammal phylogeny, including many relationships that have been considered contentious during the last 20 years (Novacek 1992). These include tree shrews (Order Scandentia) as a close outgroup to primates (Janecka et al. 2007), a Glires clade composed of rabbits (Lagomorpha) and rodents, and the hedgehog (Order Eulipotyphyla) as the first-diverging member of Laurasiatheria (Murphy et al. 2001a). For several nodes, bootstrap values improved when we increased genomic coverage from 183 to 917 loci (e.g., monophyly of Glires and the sister relationship between chimpanzee and humans; but see the relationship between dog and horse for a case where bootstrap support decreased). Also, bootstrap support in our study was much higher for many nodes than a previous species-tree analysis of placental mammals that also used the STAR method (Liu et al. 2009) in conjunction with 20 loci (largely exons) from previous mammal phylogenetic studies (Springer et al. 2007). Although the topologies of the two trees were similar, the tree of Liu et al. (2009) had low bootstrap support for most controversial relationships, including 46% support for a first-diverging Afrotheria lineage (91% in our study), 27% support for tree shrews grouping with primates (94% in our study), and 45% for a monophyletic Glires clade (87% in our study). This suggests that even at phylogenomic scales, more loci can be beneficial for resolving difficult evolutionary histories and for improving the confidence of phylogenetic inference.
Despite broad concordance among species-tree bootstrap replicates (Fig. 2C), individual gene trees showed pervasive discord among loci, although they frequently recovered some close relationships, such as that between rat and mouse as well as the relationships among primates (Fig. 2B). Trees generated from concatenated data sets, which treat conflicting genes as though they have the same evolutionary history, were generally similar to the species trees (Supplemental Figs. S5, S6) but with two important differences. First, PPs were extremely high in the concatenated trees, especially the 917-locus data set where PPs were 1.0 for all nodes. Second, the concatenated tree for the 183-locus data set placed treeshrew sister to Glires with a PP of 1.0, instead of placing it as the outgroup to primates (Janecka et al. 2007). Overcredibility of PPs on phylogenies using concatenated data has been described previously (Suzuki et al. 2002), and inflated PP is especially problematic in data sets featuring mixed phylogenetic signals (Mossel and Vigoda 2005; Kubatko and Degnan 2007). Conflicting signal is likely the case with a rapidly radiating group like placental mammals. In contrast, species trees avoid this pitfall by taking the discordant phylogenetic signals of independently sorting loci into account for both topology and support values.
Other sources of UCE loci corroborate and augment the bootstrap support for the phylogeny we report. We identified UCEs through alignments of bird and lizard in an effort to isolate loci that were conserved across most reptilians. Other ways of detecting UCEs may provide more loci for specific research questions. For example, Stephen et al. (2008) found a large pool of UCEs from mammal alignments. This set of UCEs was largely nonoverlapping with UCEs from this study: We found 897 UCEs shared between data sets, which corresponds to 7% of the Stephen et al. (2008) UCE loci or 38% of the UCE loci identified here. Our study thus represents a major, novel source of UCEs in amniotes (in addition to those of Janes et al. 2011). When we processed the UCEs from Stephen et al. (2008) using our pipeline for probe design and alignment (including the requirement that all loci are present in all 29 species), we identified 261 additional loci, which we analyzed separately and in combination with our 183 locus data set (Supplemental Fig. S8). The combined data set of 444 (183 + 261) loci resulted in topologies nearly identical to our earlier analyses, but with higher bootstrap support at most nodes (Fig. 2A; Supplemental Fig. S8).
When we adapted our probe design and alignment pipeline for exons (mining either the 16-Mb alignment of vertebrate exons from Stephen et al. 2008 or the probe set designed to target the 50-Mb human exome from Coffey et al. 2011 for conserved, nonduplicated sequences), we identified far fewer loci conserved across all species relative to the number of UCEs we identified (41 exons compared with 444 UCEs). When we analyzed the 41 exons located in all 29 species in conjunction with the 444 UCEs, we recovered a topology identical to the topology recovered from UCEs alone (Fig. 2A; Supplemental Fig. S9). We also used the 41 exon loci alone, to recover a tree that was generally similar to the UCE and UCE+exon tree, although this exon-only tree showed several inconsistencies. For example, the outgroup taxa opossum and platypus were incorrectly joined as sister taxa (Supplemental Fig. S9). Whether these inconsistencies result from the smaller number of exon loci conserved across species or properties of locus type itself (exon vs. UCE) requires further investigation. Initial explorations show that the number of loci alignable across many divergent species decays more rapidly in exons than UCEs (Supplemental Fig. S10). We are currently conducting a separate, detailed study comparing the informativeness of UCEs and exons at deep phylogenetic scales. Our analyses here suggest that UCEs may be easier to collect and align in high numbers across phylogenetically divergent taxa. However, the high information content of exons (particularly at third codon positions) (Fig. 1) suggests that studies would do well to combine both sources of data, if possible.
The sequence of divergence among the placental mammal superorders Afrotheria, Xenarthra, and Boreotheria (= Laurasiatheria + Euarchontoglires) (see Fig. 2A) has remained controversial despite extensive study (Hallström et al. 2007; Murphy et al. 2007; Nikolaev et al. 2007; Wildman et al. 2007; Churakov et al. 2009; Nishihara et al. 2009). Phylogenomic studies using concatenated data sets have, by and large, found strong support for a sister relationship between Afrotheria and Xenarthra (Hallström et al. 2007; Murphy et al. 2007; Wildman et al. 2007), which has been used to validate the importance to placental mammal evolution of a major North–South split caused by the break-up of Pangaea into Gondwana and Laurasia during the Cretaceous (Wildman et al. 2007). Retroposon studies, on the other hand, have found roughly equal support for all three possible topologies (Churakov et al. 2009; Nishihara et al. 2009), except for one study that found homogeneous support for Xenarthra diverging first, although only two retroposons were reported (Kriegs et al. 2006). Meanwhile, morphologists have long considered Xenarthra to be the first-diverging lineage of placental mammals (McKenna and Bell 1997).
In contrast to these previous studies, we found that species trees from three data sets support Afrotheria as the first-diverging superorder of placental mammals (Figs. 1A, 3A). In the most taxonomically inclusive data set, bootstrap support for Afrotheria diverging first jumped from 58% to 91% when we augmented the number of loci from 183 to 444 with loci from Stephen et al. (2008) (Fig. 2A). We also found the same topology when we combined 444 UCEs with 41 exons, although bootstrap support was somewhat lower (79%). Analysis of 41 exons alone joined Afrotheria and Xenarthra as sister taxa (Supplemental Fig. S9). Long-branch attraction is unlikely to play a role in the result showing that Afrotheria diverged first because the two longest branches are those leading to Xenarthra and Afrotheria, which did not group together in the species trees. Interestingly, although analyses of the concatenated 183-locus and 444-locus data sets also supported Afrotheria as diverging first, the concatenated 591-locus (high genomic coverage) data set produced a strongly supported sister relationship between Afrotheria and Xenarthra (Fig. 4A). The differing results for the species tree versus concatenated tree suggests that the sister relationship between Afrotheria and Xenarthra, reported by many phylogenomic studies (Hallström et al. 2007; Murphy et al. 2007; Wildman et al. 2007), might be generally attributable to a failure to account for conflicting gene histories, in addition to other possible sources of error, including long-branch attraction (for further discussion, see Nishihara et al. 2007). A scenario in which Afrotheria was the first placental mammal lineage to diverge would cast doubt on the biogeographic hypothesis of a North–South split in early placental mammal evolution caused by the break-up of Pangaea in the Cretaceous (Wildman et al. 2007).
Results from our coalescent simulation provide an explanation for why retroposon studies have found highly heterogeneous signal among gene trees bearing on the divergence of placental mammal superorders (Murphy et al. 2007; Churakov et al. 2009; Nishihara et al. 2009). Our finding that the divergence events lie close to or within a phylogenetic anomaly zone (Degnan and Rosenberg 2006) also cautions that concatenated data sets are probably not appropriate for answering this particular question, and concatenated data sets may even produce misleading results. We note that although retroposons are less likely to show homoplasy than DNA sequence data (although they may not always be free of homoplasy [Han et al. 2011]) and therefore are excellent markers for accurately recording gene trees (Shedlock et al. 2004), retroposons are not immune to the effects of coalescent stochasticity. Unfortunately, retroposons are also rarely found in high enough numbers to inform very rapid speciation events if, as coalescent simulation studies suggest, greater than 500 loci may be necessary to accurately estimate divergences in the anomaly zone (Liu et al. 2009). For comparison, the number of loci used in studies investigating placental mammal divergence with retroposons has ranged from the single digits (Kriegs et al. 2006; Nishihara et al. 2006) to as many as 68 (Nishihara et al. 2009).
Our results are also relevant to one of the most enduring mysteries of mammal phylogenetics: the evolutionary affinities of bats (Chiroptera) in the superorder Laurasiatheria. Twenty years ago, even the monophyly of Chiroptera was debated, and most researchers thought bats' evolutionary affinities were more with primates (Novacek 1992). Later molecular phylogenies rejected this hypothesis and defined four major groups of mammals, with bats placed within the Laurasiatheria (Madsen et al. 2001) among carnivores, some ungulates, and some insectivores, although the rapid radiation of this group made further phylogenetic resolution difficult. Recently, a surprising clade (Pegasoferae) uniting Chiroptera with Carnivora (e.g., dog) and Perissodactyla (e.g., horse) to the exclusion of Cetartiodactyla (e.g., cow, alpaca, dolphin) emerged from a study of retroposons (Nishihara et al. 2006). However, this result was based on only three retroposon gene trees, with one retroposon supporting a conflicting topology. Our species tree based on 693 loci lends some support to a more traditional placement of Chiroptera as sister to a clade containing Carnivora, Perissodactyla, and Cetartiodactyla, not nested within it (Fig. 4B). At 64%, bootstrap support for this topology was not high, and the 183-locus and 444-locus STAR trees supported an arrangement favoring Pegasoferae, albeit with even lower bootstrap support (Fig. 2A). The mixed support from various data sets and analytical techniques suggests that basal relationships in the Laurasiatheria are a particularly difficult phylogenetic problem that will likely require even more loci and taxa to address. Our results do not find strong support for Pegasoferae, but neither do they find strong support for any particular placement of bats with the Laurasiatheria.
The overall approach to inferring phylogenies using UCEs is especially promising because the in silico probes used in our study can be adapted easily to an in vitro design to capture DNA (Gnirke et al. 2009) from virtually any species across the amniote Tree of Life. For example, probes synthesized from the in silico set we designed successfully captured >1000 loci in various reptile species, including sufficient flanking sequence to resolve deep-level phylogenetic relationships (Faircloth et al. 2012). Similar probe sets could be designed for other phylogenetic groups (Siepel et al. 2005). Increasingly sophisticated hierarchical methods for bar-coding individuals (Kenny et al. 2011) will enable targeted capture and sequencing of orthologous DNA from many individuals at hundreds or thousands of loci in a partial, massively parallel sequencing run without the laborious intermediate steps of marker discovery, variability screening, individual PCRs, and haplotype phasing. We envision that this approach, when applied to nonmodel organisms, could stimulate a shift in the way researchers collect and analyze broad-scale phylogenetic data, which will build from and complement whole-genome data produced by the Genome 10K Project (http://genome10k.soe.ucsc.edu/).
Methods
Identification of UCEs
We identified ultraconserved elements (UCEs) by screening whole-genome alignments of the chicken (Gallus gallus) and Carolina anole (Anolis carolinensis) prepared by the UCSC genome bioinformatics group using a custom Python script to identify runs of at least 60 bases having 100% sequence identity. We stored metadata for these regions in a relational database (RDB). Because the zebra finch to chicken genome–genome alignment was not yet available from UCSC, we aligned each 100% conserved region from the chicken–lizard alignments to the zebra finch (UCSC taeGut1) genome using a custom Python program and BLAST (Altschul et al. 1997), and we stored metadata for each match having an e-value ≤ 1×10−15 in the RDB. We removed duplicates from the group of matches containing data from chicken, lizard, and zebra finch, and we defined the remaining set of 3154 sequences as UCEs.
Design of in silico probes from UCEs
Our approach to designing molecular probes from UCEs was that our in silico workflow should be rapidly adaptable to in vitro designs for maximal applicability to organisms without existing genomes. Therefore, instead of simply aligning UCEs to genome-enabled organisms and extracting the whole UCE and flanking sequence, our in silico approach mimicked a commercially available sequence capture workflow (Gnirke et al. 2009) by tiling probes across UCEs and then reassembling UCEs and flanking sequence on a per-species basis (described below).
We designed in silico probes by selecting UCEs from the RDB, adding sequence to those shorter than 120 bp in length to make them 120 bp by selecting equal amounts of 5′ and 3′ flanking sequence from a repeat-masked chicken genome assembly, and recording the length of flanking sequence, if any, added to each. When UCEs were >180 bp, we tiled 120 bp in silico probes across UCEs at 2× density (i.e., probes overlapped by 60 bp). If UCEs were <180 bp in total length, we selected a single probe from the center of the UCE. We conducted a BLAST search of in silico probes against themselves to identify and remove duplicates arising as a result of probe design, and we selected a reduced set of 2560 in silico molecular probes from the RDB having zero duplicate matches, <10 masked bases, and <50 added bases (25 to each side) for downstream use. These probes represented 2386 UCEs in chicken, lizard, and zebra finch.
Alignment of in silico probes to amniote genomes
We aligned in silico probes to available genomic sequence from placental mammals and several outgroups (Supplemental Table S1) using LASTZ (available at http://www.bx.psu.edu/miller_lab). We retained only those probes aligning to genomes having ≥92.5% identity across ≥100 bp of the 120-bp probe sequence, and we ignored probes that matched in multiple locations within any genomic sequence, to filter out potential paralogs. We created a table of unique in silico probes located in each species and stored these data in a separate relational database (RDB2). From RDB2, we selected the sets of probes present in all members of our data sets described below.
Generating the data sets
Because data sets that were taxonomically broader resulted in fewer aligning loci, we constructed two data sets for general phylogenetic reconstruction of placental mammals, each having differing levels of taxonomic and genomic coverage (Supplemental Table S1). To obtain high genomic coverage data sets for two particularly difficult phylogenetic hypotheses, we also created a seven-taxon data set to elucidate the phylogenetic position of bats within Laurasiatheria and a seven-taxon data set to address the early radiation of placental mammals into superorders Afrotheria, Xenarthra, and Boreotheria. We favored species for inclusion in a data set if they allowed for more loci and if they were as divergent as possible from other species in the same group, to minimize long branches (e.g., we chose dog and human for the seven-taxon data set exploring basal placental mammal relationships because they are representatives of the two divergent groups within Boreotheria).
Assembling orthologous loci from UCEs, variability, and saturation
For each species within a data set, we excised the alignment of each in silico probe, plus 500 bp of flanking sequence upstream (5′) and downstream (3′) for a total of 1000 bp of flanking sequence because preliminary investigation revealed that this distance would likely contain alignable regions with variation. Using these sequences, we assembled in silico probes back into their respective UCEs using a custom Python program that integrated LASTZ—to match probes to their UCE—and MUSCLE (Edgar 2004) to assemble multiple probes designed for the same UCE. After assembly, we referred to each UCE as a locus. For each locus, we aligned the data across species within a data set using a custom Python program and MUSCLE. We used a moving average across a 20-bp window to trim the ends of all alignments ensuring ends contained at least 50% sequence identity and that nonaligning sequence was removed. We culled loci with missing species within each data set.
To illustrate that loci are likely to be independently segregating, we computed the physical distances between loci in chicken, mouse, and human, because these genome builds (UCSC galGal3, mm9, hg19) are likely the most accurate that also span the taxonomic range we investigated. We calculated physical distance between loci (x-bar ;± ;95% CI) as the difference in start and stop positions between adjacent loci on each chromosome using a custom Python program.
We compared saturation in 20 randomly chosen UCE loci drawn from the 183-locus data set and the 20 nuclear loci used in Springer et al. (2007) using the saturation index (ISS) of Xia et al. (2003). We analyzed the core UCE region and UCE flanking regions separately. For the 16 coding exons from the Springer et al. (2007) data set, we also calculated saturation of third positions of codons separately. We determined the proportion of variable sites and the ratio of informative to variable sites for UCEs and the Springer et al. (2007) loci using MEGA5 (Tamura et al. 2011). Here, we used the 13 Springer et al. (2007) loci that had complete data for a subset of lineages shared with our 29-taxon data set (opossum, sloth, elephant, hedgehog, cow, alpaca, bat, dog, horse, rabbit, mouse, marmoset, and human). For the exons that were unambiguously in-frame for their entire length (n = 6), we also calculated variability statistics separately for third versus first/second codon positions.
Analysis of gene trees and species trees
We estimated gene trees under maximum likelihood in PhyML 3.0 (Guindon et al. 2010) using their most likely substitution model as estimated with MrAIC 1.4.4 (Nylander 2004). We estimated species trees from these gene trees using the STAR (species trees based on average ranks of coalescences) method implemented with the R package Phybase (Liu and Yu 2010). STAR calculates a species tree topology analytically based on average ranks of coalescent events in a collection of gene trees (Liu et al. 2009). STAR performs similarly to probabilistic coalescent-based species-tree methods (e.g., BEST), which are unsuited from a practical perspective for the large data sets used here. STAR also performs well when gene trees deviate from equal evolutionary rates, likely the case in the deep and taxonomically diverse phylogeny we investigated (Liu et al. 2009). In initial explorations, STAR and another analytical species-tree estimation method that uses average coalescence times—STEAC—produced identical topologies with similar bootstrap support. After generating a single STAR tree, we performed 1000 nonparametric bootstrap replicates by resampling nucleotides within loci as well as resampling the loci within the data set (Seo 2008) using a custom Python program, and we generated a cloudogram of gene trees and species-tree bootstraps for the 183 locus data set with DensiTree (Bouckaert 2010). For the visualization of species-tree bootstrap replicates (Fig. 3C), we used STEAC trees because, unlike STAR trees, they contain branch length information, which aided visualization. We analyzed concatenated alignments using MrBayes 3.1 (Huelsenbeck and Ronquist 2001), grouping genes with the same substitution model as estimated with MrAIC 1.4.4 into different partitions. We tried several partitioning schemes: (1) no partitioning with one GTR+I+Γ substitution model; (2) partitioning according to the MrAIC substitution model; and (3) partitioning according to the MrAIC substitution model with unlinked molecular rates. We observed no topological differences among results from these partitioning schemes. However, scheme 3 had trouble reaching convergence for all data sets except the 183-locus data set. We thus present results from partitioning scheme 2. All MrBayes analyses consisted of two independent runs (four chains each) of 10,000,000 iterations each, with trees sampled every 100 iterations, for a total of 100,000 trees, from which we sampled the last 50,000 after checking for convergence with the log of posterior probability within and between the independent runs for each analysis.
Calculation of gene-tree probabilities for basal divergence of superorders
We calculated gene-tree probabilities for a five-taxon species tree representing the divergence of the placental mammal lineages Afrotheria, Xenarthra, Euarchontoglires, and Laurasiatheria, and a marsupial outgroup (opossum) with COAL (Degnan and Salter 2005). We created a species tree in coalescent units (t/2N, where t is the divergence time in generations and N is the effective population size) under a range of possible demographic parameters (generation time = 3–20 yr, population size = 10,000–1,000,000 individuals) and divergence times (0.5, 1, and 5 Mya) (Bininda-Emonds et al. 2007). We summarized gene-tree discordance with the mathematical variance in the frequencies of different gene-tree topologies, which we then rescaled to range between 0 and 1 (low to high discord, respectively). We compared these theoretical values with empirical estimates of gene-tree discord from our 591-locus, seven-taxon data set and retroposon results from the literature (Table 1; Nishihara et al. 2009).
Data access
We provide all Python code, RDB and RDB2, BED files representing both the UCEs and tiled probes, LASTZ alignments, instructions, etc., under open-source (BSD and Creative Commons) licenses at http://dx.doi.org/10.5060/D21N7Z2Z. Users should be aware that we maintain updated code/workflows for many of the steps outlined in the Methods.
Acknowledgments
We thank M. Springer for sharing the 20-locus mammal data set and J. Mattick for sharing UCE and exon data. S.P. Hubbell, S. Edwards, J. Degnan, M. Sheehan, M. Alfaro, B. Carstens, and three anonymous reviewers provided helpful comments. One reviewer suggested the point about selection potentially improving the phylogenetic utility of UCEs. H. Hoekstra provided access to the Odyssey cluster supported by the Harvard FAS Sciences Division Research Computing Group to conduct phylogenetic analysis. A research grant from Amazon Web Services (Amazon.com) also supported phylogenetic computation. We thank the many scientists, institutions, and funding agencies that have contributed genomic data available via the UCSC Genome Browser (see complete list at http://genome.ucsc.edu/goldenPath/credits.html and Supplemental Material).
Authors' contributions: J.E.M., B.C.F., N.G.C., R.T.B., and T.C.G. designed the study; B.C.F. designed ultraconserved probes and created data sets and performed phylogenetic analysis; N.G.C. performed phylogenetic analysis; J.E.M. performed gene-tree frequency analysis; P.A.G. provided analytical resources; J.E.M., B.C.F., N.G.C., R.T.B., and T.C.G. wrote the manuscript. J.E.M., B.C.F., N.G.C., and T.C.G. contributed equally to the study. All authors discussed the results and commented on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.125864.111.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent W, Mattick J, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A 2007. The delayed rise of present-day mammals. Nature 446: 507–512 [DOI] [PubMed] [Google Scholar]
- Bouckaert RR 2010. DensiTree: Making sense of sets of phylogenetic trees. Bioinformatics 26: 1372–1373 [DOI] [PubMed] [Google Scholar]
- Cannarozzi G, Schneider A, Gonnet G 2007. A phylogenomic study of human, dog, and mouse. PLoS Comput Biol 3: e2 doi: 10.1371/journal.pcbi.0030002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Kriegs JO, Baertsch R, Zemann A, Brosius J, Schmitz J 2009. Mosaic retroposon insertion patterns in placental mammals. Genome Res 19: 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey AJ, Kokocinski F, Calafato MS, Scott CE, Palta P, Drury E, Joyce CJ, LeProust EM, Harrow J, Hunt S 2011. The GENCODE exome: Sequencing the complete human exome. Eur J Hum Genet 19: 827–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA 2006. Discordance of species trees with their most likely gene trees. PLoS Genet 2: e68 doi: 10.1371/journal.pgen.0020068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Salter LA 2005. Gene tree distributions under the coalescent process. Evolution 59: 24–37 [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6: 361–375 [DOI] [PubMed] [Google Scholar]
- Derti A, Roth FP, Church GM, Wu C-t 2006. Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat Genet 38: 1216–1220 [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK 2007. High-resolution species trees without concatenation. Proc Natl Acad Sci 104: 5936–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, McCormack JE, Crawford NG, Brumfield RT, Glenn TC 2012. Ultraconserved elements anchor thousands of genetic markers for target enrichment spanning multiple evolutionary timescales. Syst Biol. doi: 10.1093/sysbio/SYS004 [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hallström BM, Kullberg M, Nilsson MA, Janke A 2007. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol 24: 2059–2068 [DOI] [PubMed] [Google Scholar]
- Han K-L, Braun EL, Kimball RT, Reddy S, Bowie RCK, Braun MJ, Chojnowski JL, Hackett SJ, Harshman J, Huddleston CJ, et al. 2011. Are transposable element insertions homoplasy free? An examination using the avian tree of life. Syst Biol 60: 375–386 [DOI] [PubMed] [Google Scholar]
- Hobolth A, Christensen OF, Mailund T, Schierup MH 2007. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet 3: e7 doi: 10.1371/journal.pgen.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ 2007. Molecular and genomic data identify the closest living relative of Primates. Science 318: 792–794 [DOI] [PubMed] [Google Scholar]
- Janes DE, Chapus C, Gondo Y, Clayton DF, Sinha S, Blatti CA, Organ CL, Fujita MK, Balakrishnan CN, Edwards SV 2011. Reptiles and mammals have differentially retained long conserved noncoding sequences from the Amniote ancestor. Genome Biol Evol 3: 102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D 2007. Human genome ultraconserved elements are ultraselected. Science 317: 915. [DOI] [PubMed] [Google Scholar]
- Kenny EM, Cormican P, Gilks WP, Gates AS, O'Dushlaine CT, Pinto C, Corvin AP, Gill M, Morris DW 2011. Multiplex target enrichment using DNA indexing for ultra-high throughput SNP detection. DNA Res 18: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J 1982. The coalescent. Stochastic Process Appl 13: 235–248 [Google Scholar]
- Knowles LL 2009. Estimating species trees: Methods of phylogenetic analysis when there is incongruence across genes. Syst Biol 58: 463–467 [DOI] [PubMed] [Google Scholar]
- Kriegs J, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J 2006. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol 4: 537 doi: 10.1371/journal.pbio.0040091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko L, Degnan J 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol 56: 17–24 [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Brown JM, Stanger-Hall K, Lemmon EM 2009. The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Syst Biol 58: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yu L 2010. PHYBASE: An R package for phylogenetic analysis. Bioinformatics 26: 962–963 [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Pearl DK, Edwards SV 2009. Estimating species phylogenies using coalescence times among sequences. Syst Biol 58: 468–477 [DOI] [PubMed] [Google Scholar]
- Maddison WP 1997. Gene trees in species trees. Syst Biol 46: 523–536 [Google Scholar]
- Madsen O, Scally M, Douady CJ, Kao DJ, DeBry RW, Adkins R, Amrine HM, Stanhope MJ, de Jong WW, Springer MS 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409: 610–614 [DOI] [PubMed] [Google Scholar]
- McKenna MC, Bell SK 1997. Classification of mammals above the species level. Columbia University Press, New York [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5: e1000471 doi: 10.1371/journal.pgen.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel E, Vigoda E 2005. Phylogenetic MCMC algorithms are misleading on mixtures of trees. Science 309: 2207–2209 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ 2001a. Molecular phylogenetics and the origins of placental mammals. Nature 409: 614–618 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW 2001b. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348–2351 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pevzner PA, O'Brien SJ 2004. Mammalian phylogenomics comes of age. Trends Genet 20: 631–639 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res 17: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S, Montoya-Burgos JI, Margulies EH 2007. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet 3: e2 doi: 10.1371/journal.pgen.0030002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Hasegawa M, Okada N 2006. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci 103: 9929–9934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Okada N, Hasegawa M 2007. Rooting the eutherian tree: The power and pitfalls of phylogenomics. Genome Biol 8: R199 doi: 10.1186/gb-2007-8-9-r199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Maruyama S, Okada N 2009. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc Natl Acad Sci 106: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novacek M 1992. Mammalian phylogeny: Shaking the tree. Nature 356: 121–125 [DOI] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrAIC.pl. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. http://www.abc.se/∼nylander. [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol 9: e1000602 doi: 10.1371/journal.pbio.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard DA, Iyer VN, Moses AM, Eisen MB 2006. Widespread discordance of gene trees with species tree in Drosophila: Evidence for incomplete lineage sorting. PLoS Genet 2: e173 doi: 10.1371/journal.pgen.0020173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Bailey P, Bruce S, Engström PG, Klos JM, Wasserman WW, Ericson J, Lenhard B 2004. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5: 99 doi: 10.1186/1471-2164-5-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. 2000. The ecology of adaptive radiation. Oxford University Press, New York. [Google Scholar]
- Seo TK 2008. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol Biol Evol 25: 960–971 [DOI] [PubMed] [Google Scholar]
- Shedlock A, Takahashi K, Okada N 2004. SINEs of speciation: Tracking lineages with retroposons. Trends Ecol Evol 19: 545–553 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LDW, Richards S 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Pheasant M, Makunin IV, Mattick JS 2006. Transposon-free regions in mammalian genomes. Genome Res 16: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Murphy W, Eizirik E, O'Brien S 2003. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc Natl Acad Sci 100: 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Burk-Herrick A, Meredith R, Eizirik E, Teeling E, O'Brien SJ, Murphy WJ 2007. The adequacy of morphology for reconstructing the early history of placental mammals. Syst Biol 56: 673–684 [DOI] [PubMed] [Google Scholar]
- Stadler T 2011. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci 108: 6187–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, Mattick JS 2008. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Glazko GV, Nei M 2002. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc Natl Acad Sci 99: 16138–16143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Lockhart PJ 2007. Deciphering ancient rapid radiations. Trends Ecol Evol 22: 258–265 [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Morrill MC 2011. Missing data in phylogenetic analysis: Reconciling results from simulations and empirical data. Syst Biol 60: 719–731 [DOI] [PubMed] [Google Scholar]
- Wildman DE, Uddin M, Opazo JC, Liu G, Lefort V, Guindon S, Gascuel O, Grossman LI, Romero R, Goodman M 2007. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci 104: 14395–14400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K 2004. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi: 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y 2003. An index of substitution saturation and its application. Mol Phylogenet Evol 26: 1–7 [DOI] [PubMed] [Google Scholar]