Abstract
In the process of clone-based genome sequencing, initial assemblies frequently contain cloning gaps that can be resolved using cloning-independent methods, but the reason for their occurrence is largely unknown. By analyzing 9,328,693 sequencing clones from 393 microbial genomes, we systematically mapped more than 15,000 genes residing in cloning gaps and experimentally showed that their expression products are toxic to the Escherichia coli host. A subset of these toxic sequences was further evaluated through a series of functional assays exploring the mechanisms of their toxicity. Among these genes, our assays revealed novel toxins and restriction enzymes, and new classes of small, non-coding toxic RNAs that reproducibly inhibit E. coli growth. Further analyses also revealed abundant, short, toxic DNA fragments that were predicted to suppress E. coli growth by interacting with the replication initiator DnaA. Our results show that cloning gaps, once considered the result of technical problems, actually serve as a rich source for the discovery of biotechnologically valuable functions, and suggest new modes of antimicrobial interventions.
Since 1995, whole genomes of microbial species are being sequenced at an ever-increasing pace (Kyrpides 2009). Thousands of bacterial and archaeal genomes have already been sequenced to completion, with thousands of additional genomes being sequenced every year (Liolios et al. 2010). The availability of sequenced microbial genomes provides access to the huge wealth of biotechnologically beneficial functions encoded in microbial genes, and, indeed, numerous examples for the discovery and utilization of such functions have been described. Genome analyses have led to the discovery of new antimicrobial drug targets (Payne et al. 2007), as well as to the finding of new, clinically successful vaccines (Rappuoli 2007). The study of microbial genes of unknown functions was the basis for the discovery of the CRISPR system (Jansen et al. 2002; Makarova et al. 2002), which was subsequently shown to provide adaptive immunity against phage in bacteria (Barrangou et al. 2007), and is now being used to protect beneficial lactic acid bacteria in the dairy industry against common phages (Sorek et al. 2008; Horvath and Barrangou 2010). Cellulose-degrading enzymes with biofuel implications (Warnecke et al. 2007) and new restriction enzymes (Zheng et al. 2009) are additional examples for industrially valuable functions encoded in microbial genomes. Of obvious specific interest are new genes and genomic elements that have potential implications for discovery of antimicrobials.
Despite the substantial progress in understanding the arsenal of useful functions encoded within microbial genomes, many additional functions are probably still obscure. Indeed, some 30%–50% of all genes in every sequenced microbial genome are of unknown function (Markowitz et al. 2010). Summarized across all currently sequenced genomes, the number of such enigmatic genes exceeds 1.2 million, highlighting the great potential for biotechnologically and clinically useful functions yet to be discovered.
Here we identified thousands of microbial genes that are toxic to E. coli. Our approach is based on a by-product of the Sanger whole-genome shotgun (WGS) sequencing method, in which multiple copies of the organism's genome are randomly sheared into overlapping fragments of DNA, and plasmids containing the cloned fragments are transformed into an E. coli cell. The ends of the cloned fragments are then sequenced, and overlapping sequences are used for genome assembly (Supplemental Fig. S1). However, in the course of many genome sequencing projects, a small fraction of the genome fails to be cloned into E. coli, resulting in sequence gaps. We have previously shown that genes toxic to the E. coli host can cause these gaps, and that many of these toxic genes are protein constituents of the bacterial ribosome (Sorek et al. 2007). The toxicity of such genes was hypothesized to stem from their incompatibility with the E. coli molecular machinery, or from intolerance of the host to increased dosage of the ribosomal protein gene in addition to the endogenous homolog (Sorek et al. 2007).
In contrast to this defined inhibitory action of common, ribosomal protein genes, numerous cloning gaps harbor “hypothetical” genes that have no functional annotation, or do not span protein-coding genes at all, making the reason for their occurrence obscure. In this study, we experimentally demonstrate that such gaps encode a vast array of toxic products, including novel toxins, restriction enzymes, toxic small RNAs, and toxic DNA motifs. We compiled PanDaTox (the Pan Genomic Database for Genomic Elements Toxic to Bacteria), which contains detailed information on thousands of genes and non-coding elements predicted to be toxic to bacteria based on our gap analysis (http://www.weizmann.ac.il/pandatox).
Results
Pan-genomic discovery of toxic genes of unknown functions
To explore the functional toxicity repertoire of cloning-resistant genes, we analyzed 393 microbial genomes (360 bacteria, 28 archaea, and five eukaryotes) for which the raw sequencing data were accessible and that had sufficient clone coverage (Methods). We used the original sequencing data to map the clone positions on these genomes and detected genes that were not fully covered by any single clone (Supplemental Fig. S2). Since the probability of a given gene to be fully covered by a single clone depends on its size and on the sequencing library (e.g., a 4-kb gene will never be fully covered in a library of 3-kb clones), we developed a statistical framework to assign a P-value for unclonability, based on multiple random coverage simulations for each gene (Supplemental Methods). Genes with p < 0.01 (corrected for multiple testing) were further analyzed as cloning-resistant.
Our analysis retrieved 15,927 unclonable genes, as well as an additional 25,894 genes with coverage significantly reduced compared with the coverage expected by chance (Supplemental Table S1). Unclonable genes with COG functional annotations largely followed the pattern we previously observed, belonging to a very narrow set of gene families, i.e., the same orthologous gene could not be cloned into E. coli from many different genomes (Supplemental Fig. S3; Sorek et al. 2007). However, 10,509 uncharacterized genes, many of them species-specific, were also found to be unclonable or with significantly reduced coverage (p < 0.01) (Supplemental Fig. S4). These genes are of potential biotechnological interest, because they might represent novel functional modules inhibiting the growth of bacteria.
To examine whether gap-residing genes of unknown function are toxic to E. coli, we attempted to clone 56 such protein-coding genes from 25 different genomes (chosen based on availability of genomic DNA material to amplify from) into an inducible expression system that strongly suppresses the expression of the cloned gene in the absence of the expression inducer (IPTG) (Supplemental Table S2). We successfully cloned 55 of the 56 selected genes. Our inability to clone the remaining gene implies that it is highly toxic, so that even a minute leakage of its expression was lethal to the host cells.
We next tested whether the protein products of the 55 genes we cloned in the inducible expression system are toxic to E. coli. For this, we activated their expression within E. coli using increasing amounts of IPTG as described in Sorek et al. (2007). Forty-four of the genes (80%) inhibited the growth of E. coli following induction of their expression, indicating that the products of these genes are toxic to the host (Supplemental Table S2). These results verify our cloning-analysis method in detecting thousands of microbial genes that are potentially toxic to E. coli.
New restriction enzymes
We set out to study the repertoire of toxicity mechanisms in the toxic genes set. It was previously shown that restriction endonucleases, when cloned into an E. coli sequencing host without the presence of their companion DNA methyltransferase, can kill the host due to cleavage of its DNA (Zheng et al. 2009). This concept was used to identify restriction enzymes in five bacterial genomes based on shotgun sequencing data (Zheng et al. 2009). Indeed, 64 genes annotated as belonging to restriction/modification systems were found in our data to be unclonable or had statistically significant decrease in their clone coverage (Supplemental Table S3).
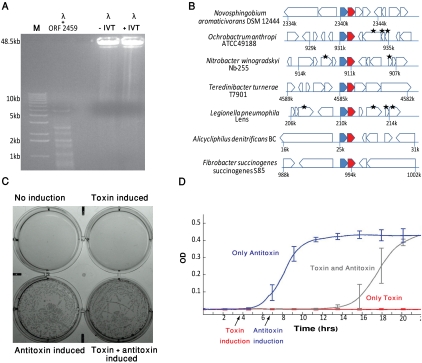
Because restriction enzymes evolve rapidly (Stern and Sorek 2011), they frequently lack homology with known sequences and are thus annotated as uncharacterized genes. We therefore hypothesized that some of the uncharacterized unclonable genes in our set are, in fact, new restriction endonucleases. To test this hypothesis, we used in vitro transcription/translation to synthesize protein products for the 44 genes whose toxicity to cells was verified (above), and tested their ability to cleave DNA by incubating them with lambda-phage genomic DNA. One of these proteins was able to cleave DNA into a fragment pattern typical for restriction enzymes (Fig. 1A). This gene is annotated as a “hypothetical protein” in the genome of Synechococcus elongatus PCC 7942 (locus tag Synpcc7942_2459) and has very few homologs in other microbial genomes, none of which is functionally characterized. It is adjacent to a DNA-cytosine methyltransferase in that genome, characteristic of bona fide restriction enzymes. The methyltransferase is predicted to modify the recognition sequence CCWGG (Roberts et al. 2010), further suggesting that the new restriction enzyme we found cleaves at this sequence motif. These results demonstrate that our set of toxic genes is a potent source for discovery of previously uncharacterized restriction enzymes.
Figure 1.
Functional interrogation of toxic genes. (A) Endonuclease activity of a gap-residing gene. ORF #2459 in Synechococcus elongatus PCC 7942 was translated in vitro, and the translated product was incubated for 2 h with 1 μg of phage lambda genomic DNA (48.5 kb long). The band pattern on the agarose gel indicates digestion of the lambda DNA. Negative control lanes: lambda DNA incubated with in vitro translation reaction only (+IVT) or not incubated (−IVT). (B) shosA/shosT is a bi-gene operon that undergoes extensive horizontal transfer. The genomic context surrounding the gene pair in seven genomes is shown to be different for every genome, indicative of horizontal gene transfer. Genes are depicted as block arrows. (Blue and red filled arrows) The predicted antitoxin (shosA) and toxin (shosT), respectively. Genes marked by an asterisk are transposase and integrase genes, which are hallmarks of horizontal gene transfer. (C) ShosA/ShosT is a toxin–antitoxin system. The predicted toxin and antitoxin were cotransformed into Escherichia coli BL21(DE3)pLysS on compatible plasmids: the toxin cloned under the control of an IPTG-responsive promoter, and the antitoxin under the control of an arabinose-responsive promoter. Induction of toxin expression (100 μM IPTG) and induction of antitoxin (0.3% arabinose) were performed. Cells do not grow when toxin is induced unless antitoxin is coinduced. Cells without any inducer also do not grow, most probably due to toxin leakage from the IPTG-inducible promoter. (D) ShosT has a bacteriostatic effect. The bacterial OD600 was measured over time when only antitoxin was induced (dark blue); only toxin was induced (red); and when antitoxin expression was induced 2.5 h after toxin induction (gray). Each experiment was performed in six repetitions (biological triplicate and technical duplicate).
New toxin–antitoxin systems
Toxin–antitoxin (TA) systems are comprised of a stable toxic protein and an unstable antitoxin that neutralizes it. Although originally hypothesized to be responsible mainly for plasmid stabilization (Jensen and Gerdes 1995; Van Melderen and Saavedra De Bast 2009), these systems are now known to be involved also in anti-phage defense (Pecota and Wood 1996; Hazan and Engelberg-Kulka 2004; Fineran et al. 2009; Koga et al. 2011), antibiotics resistance via a dormant “persistence” behavior (Lewis 2010), and altruistic cell suicide in bacterial communities (Hazan et al. 2004; Amitai et al. 2009), and are therefore attracting growing biotechnological attention (Gerdes et al. 2005). Type II toxin–antitoxin systems, in which both toxin and antitoxin are proteins, are the most widespread across bacteria, and are classified into 11 families based on protein sequence similarity (Shao et al. 2011).
We hypothesized that our set of toxic genes might contain novel families of toxin–antitoxin systems. To search for novel TA systems, we searched, among the 44 genes whose toxicity to cells was verified, for those genes that consistently appeared in a bi-gene operon and showed a high rate of horizontal gene transfer, which is typical of known TA systems (Pandey and Gerdes 2005). One such gene was identified and selected for further functional assays (Fig. 1B). This gene, found in the genome of Nitrobacter winogradskyi Nb-255 (locus tag Nwi_0828), is preceded by a small protein (the putative antitoxin) that has a predicted DNA binding domain, which is a common characteristic of antitoxins. We found nine homologs of this bi-gene operon in various microbial genomes, and in three of the four cases for which we had clone coverage data, the putative toxin showed unclonability or statistically significant reduction in clone coverage. We denote these putative toxin/antitoxin pairs shosT and shosA, respectively (identified based on shotgun sequencing).
To test whether shosT/shosA indeed form a new TA system, we further cloned shosT from Novosphingobium aromaticivorans DSM 12444 in an expression vector under the control of an IPTG-responsive promoter, and the cognate shosA in a second expression vector under the control of an arabinose-responsive promoter. E. coli cells holding both these vectors were unable to grow when the expression of shosT alone was induced, but thrived when both shosT and shosA were expressed simultaneously (Fig. 1C). Furthermore, induction of shosA expression 2.5 h after shosT induction resulted in cell growth recovery, indicating that the toxin has a bacteriostatic effect (Fig. 1D). Since neither shosT nor shosA is homologous to any component of previously identified TA systems, the system we discovered forms a completely new family of type II TA systems. The mechanism by which shosT inhibits growth is yet to be determined.
Toxic small RNAs
Whereas the vast majority of cloning gaps in the 393 genomes analyzed could be explained by the presence of toxic protein-coding genes, some gaps did not span any gene at all. We recorded 873 instances of uncloned intergenic regions, where the two genes flanking the region were covered by at least one clone each (Supplemental Table S4; Methods). Of these, 46 intergenic gaps probably harbor previously unannotated short toxic protein-coding genes based on the presence of a conserved ORF (Supplemental Table S4; Supplemental File S1). Gene annotation software frequently fails to identify short protein-coding genes in microbial genomes (Overbeek et al. 2007).
However, the lack of obvious conserved ORFs in the vast majority of intergenic gaps prompted us to hypothesize that some of these gaps might contain non-coding RNA (ncRNA) species that inhibit bacterial growth. To test this hypothesis, we first searched these uncloned intergenic regions for sequence signatures of a sigma 70/38 promoter and a downstream rho-independent transcriptional terminator as evidence for the presence of a transcribed ncRNA (Methods). We then clustered these putative ncRNAs into groups based on sequence similarity and looked for orthology groups in which the same intergenic region was reproducibly unclonable in two or more genomes (Fig. 2A; Supplemental Table S5). As additional evidence for ncRNAs, we searched for conservation patterns indicative of RNA secondary structures (Fig. 2B). Altogether, we were able to cluster 69 of the intergenic toxic regions into 17 orthology groups, each of these putatively representing a toxic small RNA (tsRNA) species (Supplemental Tables S4, S5).
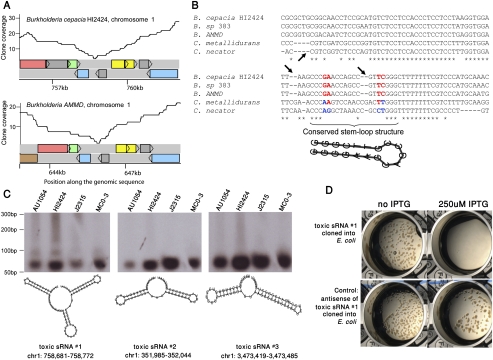
Figure 2.
Toxic small RNAs reside in intergenic gaps. (A) Reproducibility of cloning deficiency of an intergenic region in Burkholderia. Coverage plots of a syntenic 6-kb region in chromosome 1 of two Burkholderia species. Colored rectangles represent genes. This region harbors toxic small RNA #1 (tsRNA #1). (B) Multiple alignment of the cloning-deficient intergenic region. The orthologous region to those shown in panel A was aligned between five closely related organisms. Arrows depict deletions that are not divisible by 3, indicating that this region does not code for protein. Compensatory, stem-preserving mutations are colored red/blue, indicative of conserved RNA structure. (C) Northern blot analysis of three tsRNAs found in intergenic gaps in Burkholderia. Radio-labeled probes for each of the tsRNAs were hybridized to the total RNA of four Burkholderia species: Burkholderia cenocepacia AU1054, B. cenocepacia HI2424, B. sp. J2315, and B. sp. MC0-3. Predicted secondary structures of small RNAs were calculated using RNAfold (Hofacker et al. 1994). Coordinates are given relative to the B. cenocepacia HI2424 genome. Two bands of tsRNA #1 probably indicate post-transcriptional sRNA processing of the long form (94 bases) into a shorter form (∼60 bases). (D) Toxicity of tsRNA #1 in E. coli. tsRNA #1 from B. cenocepacia HI2424 was cloned into E. coli under the control of IPTG-inducible promoter. (Top left) Colonies of E. coli grow when the expression of the tsRNA is suppressed. (Top right) Growth inhibition is observed following activation of tsRNA expression. (Bottom) Growth inhibition is not observed when the antisense of the small RNA is expressed in the same system.
To verify that unclonable intergenic gaps are, indeed, transcribed into short RNAs in their genome of origin, we selected three such putative tsRNAs for further validation. These putative tsRNAs were conserved in the lineage of beta-proteobacteria and specifically occurred in intergenic gaps in the bacterium Burkholderia cenocepacia, an opportunistic pathogen that can lead to necrotizing pneumonia or death in cystic fibrosis and immunocompromised patients (LiPuma 2003). Using Northern blot analysis with total RNA derived from four Burkholderia species, we showed that these tsRNAs are indeed expressed in all four species tested (Fig. 2C). To further examine whether the expression of these tsRNAs is toxic to E. coli, we cloned them under the control of an IPTG-inducible promoter and activated their expression within the E. coli cell. Indeed, bacterial growth was completely abolished following induction of tsRNA expression, confirming that these non-coding RNA species are capable of inhibiting E. coli growth (Fig. 2D).
Growth of E. coli was not affected when in vitro expressed tsRNAs were added to its growth medium, indicating that these tsRNAs cannot penetrate the bacterial cell from the outside (Methods). We were also unable to isolate tsRNAs from the growth media when B. cenocepacia was grown in the presence of Pseudomonas aeruginosa or E. coli, indicating that tsRNAs are not secreted by B. cenocepacia in the experimental conditions tested (Methods).
To further explore the biological roles of these tsRNAs in beta-proteobacteria, we searched for conserved sequence signatures within each of the three tsRNAs under study. In all three cases, a highly conserved sequence motif was found in a region predicted to be structurally open (Supplemental Fig. S5). These conserved motifs, sized 11 bp, harbored a sequence that was complementary to the consensus ribosomal binding site (RBS) in Burkholderia (Supplemental Fig. S5). The RBS targeting motif in a structurally open domain of a short RNA is a strong characteristic of regulatory small RNAs (sRNAs) in bacteria, which are known to regulate gene expression by short base-pairings with the 5′ untranslated region (5′ UTR) in the mRNA of the regulated genes (Waters and Storz 2009). Such sRNAs, in complex with the bacterial protein Hfq, usually inhibit the translation of the targeted mRNA and/or lead it to degradation (Waters and Storz 2009). Therefore, the tsRNAs we detected presumably hold a conserved function in gene regulation in Burkholderia. The reason for their toxicity in E. coli is yet to be determined, although it could be hypothesized that they might target and misregulate essential genes within the E. coli cell. Together, these results show that investigation of cloning gaps in intergenic regions can lead to discovery of novel non-coding RNA genes.
Toxic DNA binding motifs
To investigate whether some features of the cloned DNA itself, regardless of expression, can be toxic to the E. coli sequencing host, we performed a motif enrichment search in those intergenic gaps in which no sRNAs or short ORFs were predicted. The most prominent motif found was the 9-mer TTATCCACA, which was enriched by 64-fold over what was expected by chance (p < 4×10−35, Fisher's exact test). This motif is identical to the E. coli DnaA box, a short DNA element crucial for replication initiation (Crooke et al. 1993). Multiple DnaA box sequences are found at the origin of replication (oriC) and serve as the binding sites for the DNA replication initiator protein DnaA (Fig. 3; Fuller et al. 1984; Samitt et al. 1989). This binding initiates a chain of structural changes at the oriC, and this, along with recruitment of additional proteins, leads to an eventual formation of the replication fork (for review, see Katayama et al. 2010).
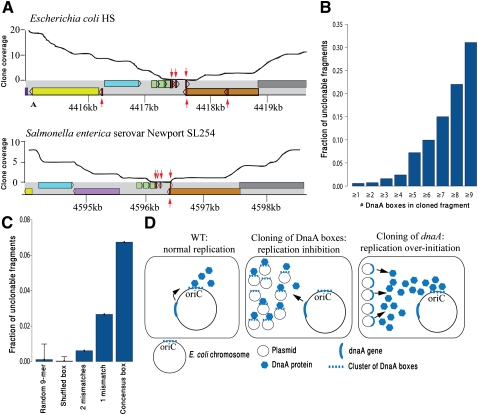
Figure 3.
Toxic DNA binding sites. (A) Reproducibility of an intergenic gap in Enterobacteria. Coverage plots of a syntenic 5 kb in E. coli (refseq: NC_009800) and Salmonella enterica (refseq: NC_011080) are presented. Colored rectangles represent genes, with homologous genes sharing the same color. (Red arrows) DnaA boxes with up to one mismatch relative to the consensus sequence; (upper arrows) DnaA boxes on the forward strand; (lower arrows) DnaA boxes on the reverse strand. This region harbors the DnaA-titrating datA locus (Kitagawa et al. 1998). (B) Unclonability increases when multiple clustered DnaA boxes exist in a cloned DNA fragment. DnaA boxes distant up to 20 bp apart were clustered together. The horizontal axis denotes the minimal cluster size. (C) Unclonability increased with predicted affinity of the DnaA box to DnaA. Consensus DnaA boxes (TTATCCACA) are more unclonable than boxes with one or two mismatches. Data are shown for clusters containing two or more DnaA boxes. (D) A model for dnaA/DnaA-box toxicity. In normal conditions (left), DnaA is produced from a single locus and binds to the oriC to initiate replication. When DnaA box clusters are cloned on a high-copy plasmid (middle), they titrate cellular DnaA protein and inhibit replication. When additional copies of the dnaA gene are cloned (right), replication over-initiation occurs.
Since the level of DnaA in the cell is tightly controlled to prevent premature replication initiation, addition of high-affinity DnaA boxes on a multi-copy plasmid is expected to titrate available DnaA proteins from the chromosomal oriC, leading to replication inhibition (Fig. 3; Christensen et al. 1999). We hypothesize this to be the mechanism of toxicity leading to the observed unclonability of the DnaA box sequences in our set. Indeed, unclonability was positively correlated with the number of cloned DnaA boxes (Fig. 3B) and was more pronounced when high-affinity DnaA boxes were present (Fig. 3C). Previous attempts to directly clone DNA sequences that contain multiple DnaA boxes were successful only when the DnaA box was mutated (van den Berg et al. 1985).
One of the recurrently unclonable DnaA-box-containing regions was an intergenic region occurring next to the orn gene in E. coli (Fig. 3A). This region was conserved in 13 Enterobacteria and unclonable in nine (69%) of them. Intriguingly, this region was previously identified as datA, a regulatory locus for replication, which titrates exceptionally large amounts of the DnaA protein to prevent over-initiation and ensure a single replication per cell cycle (Kitagawa et al. 1998; Ogawa et al. 2002). Indeed, it was previously shown that increasing the copy number of datA in E. coli results in dramatic reduction in oriC-dependent replication (Morigen et al. 2001), supporting our hypothesis that increased amounts of DnaA boxes in the cell have a growth-inhibiting effect due to replication deficiency.
If the interaction between DnaA and the DnaA boxes is so crucial for regulation of DNA replication, then one would expect that increasing the amount of the DnaA protein in the cell will also have toxic effects, because this will cause multiple premature replication initiation events (Fig. 3D, right panel). We indeed found that dnaA is one of the most highly unclonable genes across bacteria (Supplemental Table S1; Sorek et al. 2007). Moreover, we and others have previously shown that overexpression of DnaA in E. coli results in growth inhibition (Grigorian et al. 2003; Sorek et al. 2007). Together, our results imply that deregulation of the interaction between DnaA and the DnaA box results in growth inhibition in E. coli.
The Pan Genomic Database for Genomic Elements Toxic to Bacteria (PanDaTox)
The diversity of functions found in our vast collection of genes toxic to E. coli dictates that the efforts of a single laboratory cannot expose the full functional repertoire encoded within these toxic genes, and calls for a community-based research effort. To provide the scientific community with data-rich access to these genes, we developed the web-based resource PanDaTox (the Pan Genomic Database for Genomic Elements Toxic to Bacteria; http://www.weizmann.ac.il/pandatox). Using PanDaTox, researchers can search for toxic genes and toxic non-coding elements in their genome of interest; find the homologs of these toxic genes and query whether these homologs are toxic as well; use keywords to search for genes answering specific criteria; and perform sequence-based searches (BLAST) for genes in the database that are similar to a query sequence of choice. The toxicity information for each gene is presented in both numerical and graphical manners, and details on each gene, including its DNA sequence, protein sequence, and various links to external web sources, are shown.
Discussion
Hundreds of bacterial genomes have been sequenced for >15 yr using the clone-based, Sanger sequencing method. In this study, we show that sequencing gaps, which used to be considered a technical nuisance, are actually a biotechnological goldmine, containing extensive information on thousands of genes toxic to E. coli cells. We further demonstrate that these highly diverse toxic sequences hold various functions relevant for biotechnological applications. Still, our studies merely scratched the surface of the potential, producing functional assignment for only a minority of the toxic genes we detected; further studies are needed in order to reveal other beneficial functions, not tested in this study, using directed assays on these toxic genes. We expect that PanDaTox will be an important enabling tool for such future studies.
Although microbial genomes are no longer routinely sequenced by clone-based methods, the concept of cloning gaps as reporters of gene toxicity can be further expanded to any genome of choice. It is relatively straightforward to prepare a clone library of any sequenced genome within E. coli or any bacterium of choice, and then use next-generation sequencing of the cloned fragments to detect genes absent from the clone library and hence toxic to the receiving host. We envision that such additional clonability analyses would be especially informative when applied on fungal genomes, based on the expectation that such genomes would contain an arsenal of antimicrobial genes.
Our experiments on 55 protein-coding genes suggest a true-positive predictive rate of 80%, i.e., 80% of genes predicted to be toxic based on clonability information were found to inhibit the growth of bacteria experimentally. While the reasons for the 20% false positives observed in our experiments are currently unclear, several explanations are possible. First, the predicted unclonability of these genes might stem from a nearby intergenic element (tsRNAs or toxic binding site) that affects the clonability of the neighboring gene (“hitchhiker” effect) (see the Supplemental Methods). Second, the toxicity of the gene might be caused by a very strong promoter, since such promoters (e.g., the rRNA operon promoter) are known to limit vector propagation when cloned into plasmids (Boros et al. 1983). Because our experiments involve replacing the native promoter by an inducible promoter, the toxic promoter effect in these cases may be abolished.
Since E. coli was used as the host in all sequencing projects analyzed, the genes we identified are primarily those that are toxic to E. coli. Our data set is therefore biased against genes that are not functional in this organism, or those genes whose targets are not present in this organism.
Our results pinpoint the interaction between DnaA and the DnaA box as a favorable axis for antimicrobial intervention. Compounds that inhibit this interaction, either by binding to the protein or by sequestering the DNA sequence motif, are expected to form potent antibiotics. Since both the protein and the sequence motif are highly conserved (Fujita et al. 1992), antibiotics that target the protein–DNA interface might have a broad range of function. Moreover, it will be difficult for bacteria to escape the effect of such a putative compound by mutating the DnaA box and the interfacing binding domain in DnaA, because the DnaA protein must bind multiple DnaA-box sequences to allow replication initiation.
There is an undisputable urgent need for new antimicrobial compounds in the clinic. The study of the vast and diverse set of toxic elements presented in this study might suggest multiple ways by which growth of bacteria can be inhibited, as implied in the case of the DnaA/DnaA-box interactions. Moreover, it is not unlikely that the set of toxic genes we identified also includes antimicrobial peptides and other toxins produced by specific bacteria to gain selective advantage over competing microbes. Our collection of toxic genes can therefore serve as a lead for future discoveries of antimicrobials having clinical implications.
Methods
Mapping of raw sequencing data to finished genomes was performed as previously described (Sorek et al. 2007), and toxic genes were identified using a statistical framework developed to assess gene clonability (Supplemental Methods). Initial experimental evaluation of gene toxicity was also performed as described (Sorek et al. 2007; Supplemental Methods).
Nuclease activity assay
Toxic genes were transcribed and translated in vitro using the RTS 100 E. coli KY kit (Roche cat # 03-186-156-001) in a total reaction volume of 10 μL, for 6 h at 30°C according to manufacturer's instructions. Then, 1 μL of the reaction volume was added to 1 μg of unmethylated lambda-phage DNA (Promega), in the presence of one of the buffers 1, 2, 3, or 4 (New England BioLabs) in a total reaction volume of 20 μL. This 20-μL reaction was incubated for 2 h at 37°C, and was then run on a 0.7% LE agarose gel to test for a band pattern indicative of nuclease activity. As negative control, the same procedure was repeated without addition of DNA template of the toxic gene into the RTS 100 E. coli KY reaction (+IVT in Fig 1A), or without incubating lambda DNA with the RTS 100 E. coli KY reaction (−IVT in Fig 1A).
Toxin–antitoxin experiments
The shosA antitoxin gene (locus tag saro_2199) and the shosT toxin gene (locus tag saro_2200) were amplified from N. aromaticivorans DSM 12444 chromosomal DNA. The toxin was then directionally ligated into the pRSFDuet-1 vector (Novagen), and the antitoxin was ligated into the pBAD/HisA plasmid (Invitrogen). Since transformation of the toxin gene alone resulted in mutations in the toxin due to toxicity, the two plasmids (carrying the toxin and antitoxin) were cotransformed into E. coli BL21(DE3)pLysS (Stratagene) in the presence of 0.3% arabinose to induce the antitoxin. The clones were screened and verified by direct sequencing with primers on the pRSFduet-1 and pBAD/HisA vectors.
For the toxicity assay on plates, clones were cultured in LB medium with 100 μg/mL ampicillin, 50 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 0.3% arabinose overnight. The next day, a portion of each overnight culture was inoculated into fresh medium (10-fold dilution), and 10 μL was spotted on LB plates supplemented with 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 34 μg/mL chloramphenicol. Toxin, antitoxin, or both were induced by 100 μM IPTG and 0.3% arabinose, respectively, as indicated in Figure 1C.
For the kinetics experiment (Fig. 1D), three different colonies of E. coli BL21(DE3)pLysS containing both the toxin and antitoxin were cultured overnight in 5 mL of liquid LB with 100 μg/mL ampicillin, 50 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 0.3% arabinose. In the next day, cells were diluted into 175 μL of LB with ampicillin, kanamycin, and chloramphenicol (same concentrations) in a final OD600 of ∼0.05 and placed in a 96-well plate (six wells for each of the three colonies) that was inserted in a plate reader (Infinite M200). To avoid liquid evaporation during kinetic measurements, 35 μL of mineral oil was supplemented to each well on top of the LB. Cells were grown and shaken at 37°C, and OD600 was measured every 7 min for 22 h. For the “only antitoxin” curve in Figure 1D, 0.3% arabinose was added at T=0 in technical duplicate for each of the three colonies. For the “only toxin” curve in Figure 1D, 100 μM IPTG was added after ∼4 h (between cycles 35 and 36). For the “toxin and antitoxin” curve in Figure 1D, 100 μM IPTG was added after ∼4 h (between cycles 35 and 36), and 0.3% arabinose was then added ∼2.5 h later (between cycles 57 and 58).
Toxic small RNAs
Computational analysis of toxic small RNAs and DnaA boxes within uncloned intergenic regions was performed as described in the Supplemental Methods. For the experiments with Burkholderia tsRNAs, four B. cenocepacia strains (AU1054, HI2424, J2315, MC0-3) were grown to mid-log phase as follows: Cultures were grown in 10 mL of LB overnight at 37°C. One milliliter of overnight culture was used to inoculate 100 mL of LB media (1:100 dilution) and shaken at 200 rpm at 37°C until OD600 neared 1.0. Cultures were spun in 50-mL sterile centrifuge tubes for 15 min at 4°C at 4000 rpm in an Eppendorf 5810R centrifuge. Supernatants were disposed and cell pellets were resuspended in 6 mL of phosphate-buffered saline [PBS (7.95 g of Na2HPO4, 1.44 g of KH2PO4, 90 g of NaCl, per liter] and mixed with 1 mL of RNAlater solution (Ambion, cat # AM7021). Samples were stored at −20°C. RNA was then extracted using the hot phenol method (Masse et al. 2003).
The tsRNAs were amplified from the genome of B. cenocepacia HI2424 and labeled by Ready-To-Go DNA Labeling Beads (Amersham, cat # 27-9240-01) as instructed by the manufacturer. For Northern blot analyses, the NortherMax Kit (Ambion) was then used for all gel running, blotting, and hybridization procedures, using 10 μg of RNA per lane.
To test the toxicity of tsRNAs to E. coli, amplified tsRNAs were cloned into a PCR-SMART vector (Lucigen) and transformed into E. coli BL21- Gold(DE3)pLysS cells (Stratagene). Toxicity was then tested using increasing concentrations of IPTG as indicated above for toxic protein-coding genes. As negative control, each tsRNA was similarly cloned in the reverse orientation, so that it was expressed (upon IPTG-mediated activation of expression) from its antisense strand. To test whether tsRNAs can act when administered in the growth media, clones holding tsRNAs were grown to OD600 in 10 mL of LB medium, and IPTG was then added to a final concentration of 1 mM. After 4 h of incubation with IPTG, cells were spun down at 4000 rpm (15 min at 4°C), and total RNA was extracted using TRIzol reagent. Expression of sRNAs was verified by gel electrophoresis of the extracted RNA. Ten microliters of RNA at a concentration of 1 μg/μL was then added to E. coli BL21-Gold(DE3)pLysS cells growing in 15 mL of LB medium, with a starting OD600 of 0.1. Growth of cells was monitored for 6 h with and without the addition of RNA.
For the coculturing experiments, E. coli TW8211 or P. aeruginosa was grown overnight in LB medium, diluted 1:100 in 100 mL of fresh LB, and grown to OD600∼1.0. Bacterial cells were collected by centrifugation and resuspended in 2 mL of LB. One milliliter of this was mixed with either B. cenocepacia AU1054 or HI2424 cultures grown to the same OD in parallel. The mixtures were then grown for 2 h with shaking at 37°C. In parallel, individual cultures were also grown and similarly treated. Cells were harvested, and RNA was extracted and purified from either pelleted cells or supernatants using the hot phenol method (Masse et al. 2003).
Data access
Access to PanDaTox is available at http://www.weizmann.ac.il/pandatox. Sequences of tsRNAs and new annotations of protein-coding genes have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession numbers JQ317270–JQ317274 and JQ323150–JQ323152.
Acknowledgments
We thank Uri Gophna, Zohar Biron-Sorek, Shany Doron, Eran Mick, Adi Stern, Sarah Melamed, and Tal Dagan for stimulating discussions; R. Roberts for information on restriction enzymes and predicted sites; Malka Cymbalista and Shlomit Afgin for web interface design and implementation; and J.M. Tiedje for sharing Burkholderia materials. R.S. was supported by the NIH R01AI082376-01, ISF-FIRST program (grant 1615/09), ERC-StG, and the EMBO-YIP program. O.W. and A. Levy are grateful to the Azrieli Foundation for the award of an Azrieli Fellowship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.133850.111.
References
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet 5: e1000390 doi: 10.1371/journal.pgen.1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Boros I, Kiss A, Sain B, Somlyai G, Venetianer P 1983. Cloning of the promoters of an Escherichia coli rRNA gene. New experimental system to study the regulation of rRNA transcription. Gene 22: 191–201 [DOI] [PubMed] [Google Scholar]
- Christensen BB, Atlung T, Hansen FG 1999. DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J Bacteriol 181: 2683–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A 1993. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol 233: 16–24 [DOI] [PubMed] [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP 2009. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc Natl Acad Sci 106: 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita MQ, Yoshikawa H, Ogasawara N 1992. Structure of the dnaA and DnaA-box region in the Mycoplasma capricolum chromosome: conservation and variations in the course of evolution. Gene 110: 17–23 [DOI] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A 1984. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38: 889–900 [DOI] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A 2005. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol 3: 371–382 [DOI] [PubMed] [Google Scholar]
- Grigorian AV, Lustig RB, Guzman EC, Mahaffy JM, Zyskind JW 2003. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J Bacteriol 185: 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R, Engelberg-Kulka H 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics 272: 227–234 [DOI] [PubMed] [Google Scholar]
- Hazan R, Sat B, Engelberg-Kulka H 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol 186: 3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P 1994. Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol 17: 205–210 [DOI] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170 [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Ozaki T, Moriya S, Ogawa T 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev 12: 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Otsuka Y, Lemire S, Yonesaki T 2011. Escherichia coli rnlA and rnlB compose a novel toxin–antitoxin system. Genetics 187: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC 2009. Fifteen years of microbial genomics: meeting the challenges and fulfilling the dream. Nat Biotechnol 27: 627–632 [DOI] [PubMed] [Google Scholar]
- Lewis K 2010. Persister cells. Annu Rev Microbiol 64: 357–372 [DOI] [PubMed] [Google Scholar]
- Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC 2010. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 38: D346–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiPuma JJ 2003. Burkholderia and emerging pathogens in cystic fibrosis. Semin Respir Crit Care Med 24: 681–692 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV 2002. A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res 30: 482–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K, et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res 38: D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morigen, Boye E, Skarstad K, Lobner-Olesen A 2001 Regulation of chromosomal replication by DnaA protein availability in Escherichia coli: effects of the datA region. Biochim Biophys Acta 1521: 73–80 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yamada Y, Kuroda T, Kishi T, Moriya S 2002. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol Microbiol 44: 1367–1375 [DOI] [PubMed] [Google Scholar]
- Overbeek R, Bartels D, Vonstein V, Meyer F 2007. Annotation of bacterial and archaeal genomes: improving accuracy and consistency. Chem Rev 107: 3431–3447 [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K 2005. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL 2007. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6: 29–40 [DOI] [PubMed] [Google Scholar]
- Pecota DC, Wood TK 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol 178: 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R 2007. Bridging the knowledge gaps in vaccine design. Nat Biotechnol 25: 1361–1366 [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38: D234–D236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samitt CE, Hansen FG, Miller JF, Schaechter M 1989. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J 8: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z 2011. TADB: a web-based resource for Type 2 toxin–antitoxin loci in bacteria and archaea. Nucleic Acids Res 39: D606–D611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452 [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6: 181–186 [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R 2011. The phage–host arms race: shaping the evolution of microbes. Bioessays 33: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg EA, Geerse RH, Memelink J, Bovenberg RA, Magnee FA, van de Putte P 1985. Analysis of regulatory sequences upstream of the E. coli uvrB gene; involvement of the DnaA protein. Nucleic Acids Res 13: 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M 2009. Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet 5: e1000437 doi: 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, et al. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–565 [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Posfai J, Morgan RD, Vincze T, Roberts RJ 2009. Using shotgun sequence data to find active restriction enzyme genes. Nucleic Acids Res 37: e1 doi: 10.1093/nar/gkn883 [DOI] [PMC free article] [PubMed] [Google Scholar]