Abstract
Filamentous fungi are a large group of diverse and economically important microorganisms. Large-scale gene disruption strategies developed in budding yeast are not applicable to these organisms because of their larger genomes and lower rate of targeted integration (TI) during transformation. We developed transposon-arrayed gene knockouts (TAGKO) to discover genes and simultaneously create gene disruption cassettes for subsequent transformation and mutant analysis. Transposons carrying a bacterial and fungal drug resistance marker are used to mutagenize individual cosmids or entire libraries in vitro. Cosmids are annotated by DNA sequence analysis at the transposon insertion sites, and cosmid inserts are liberated to direct insertional mutagenesis events in the genome. Based on saturation analysis of a cosmid insert and insertions in a fungal cosmid library, we show that TAGKO can be used to rapidly identify and mutate genes. We further show that insertions can create alterations in gene expression, and we have used this approach to investigate an amino acid oxidation pathway in two important fungal phytopathogens.
A powerful asset for functional genomic analysis is the ability to create large annotated single gene mutant collections. For model research organisms such as baker's yeast, Drosophila, Caenorhabditis, Arabidopsis, and mice, whole genome knockout collections (1), transposon lines (2), or insertional mutant collections (3, 4) are well developed. However, in addition to these model organisms there is a vast array of economically important organisms where genome sequences and functional genomic technologies are lacking. For example, filamentous fungi are causal agents of severe human (5, 6) and crop (7, 8) diseases, and many others are being exploited in the fermentation and food industries (9). Few of these fungal genomes have been analyzed, and large-scale approaches to functional analysis are needed.
The model fungus, Saccharomyces cerevisiae, has ≈6000 genes in 12 Mb of DNA sequence (10). Targeted integration (TI) for creating gene-specific mutations is very efficient and requires only 50-bp fragments of target gene homology on either side of a selectable marker (11, 12). In contrast, many filamentous fungi have genome sizes in the range of 30–40 Mb and are estimated to contain at least 10,000 genes (13). Genome studies using expressed sequence tag analysis suggest that more than half of these genes lack homologues in S. cerevisiae (14). TI occurs at very low frequencies (1–20%) for many filamentous fungi, and larger fragments of target gene homology must be used to obtain targeted insertion events (15).
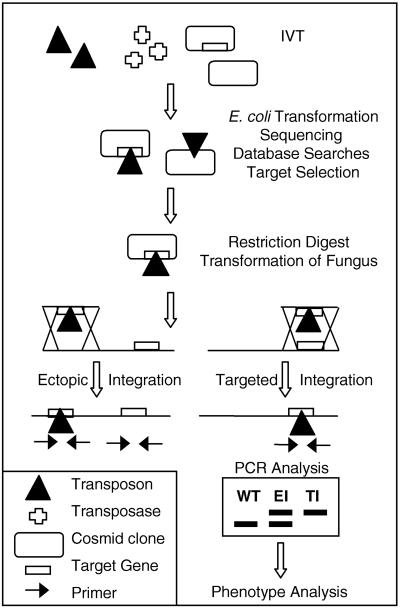
To initiate genome-wide mutagenesis studies in filamentous fungi we developed an approach we call transposon-arrayed gene knockouts (TAGKO) (Fig. 1). In vitro transposition (IVT) (16–19) into cosmid libraries is used to create gene sequencing templates. Subsequent sequencing and analysis from these templates creates an annotated collection of insertional gene disruption vectors. We demonstrate that IVT can be used to create an unbiased set of insertions into an individual cosmid clone or a cosmid library. TI occurs at much higher frequencies with the use of TAGKO generated cosmid vectors than with conventional gene disruption vectors. In addition we demonstrate that insertions into different regions of a gene can cause modulations in gene expression. Finally, we show that TAGKO can be used for comparative analysis of a biochemical pathway in two important crop pathogens, Magnaporthe grisea and Mycosphaerella graminicola.
Figure 1.
Transposon-arrayed gene knockouts. The IVT reaction includes a transposon, the corresponding transposase, and recipient DNA. The transposon carries the hygromycin phosphotransferase gene that confers resistance to hygromycin in Escherichia coli, M. grisea, M. graminicola, and other filamentous fungi (35). The recipient DNA consists of cosmid clones. The cosmid vector contains homing endonuclease sites flanking the cloning site and the β-lactamase gene. The IVT products are transformed into E. coli, and individual transposon insertion sites are determined by sequencing. Cosmids containing an appropriate transposon insertion are digested with the homing endonuclease to release full-length inserts for transformation. Ectopic and TI events are distinguished by PCR analysis, and mutants are subjected to phenotype analysis. EI, ectopic integration.
Materials and Methods
Fungal Strains and Assays.
M. grisea strain Guy11 (20) and M. graminicola strain PG1 (isolated from an infected wheat leaf and verified by internal transcribed spacer sequencing) were used as wild-type (WT) fungi in this study. M. grisea and M. graminicola were grown in complete medium and minimal medium (MM) as described (21). Oatmeal agar (22) was used for M. grisea morphology growth assays. M. grisea and M. graminicola mutant and WT strains were grown in the presence of 95 different nitrogen sources in a Phenotype Microarray plate (PM3; Biolog, Haywood, CA) according to the manufacturer's recommendations. Growth experiments were also performed in microtiter plates containing MM or MM with l-phenylalanine (MM-N + phe) or l-tyrosine (MM-N + tyr) as the sole nitrogen source followed by transfer to complete medium to assess viability. All growth experiments were repeated with at least two independent transformants per construct. Appressorium formation and rice infection assays for M. grisea were performed as previously described (23). Images of leaf samples were collected with a Spot RT digital camera (Diagnostics Instruments, Sterling Heights, MI).
Nucleic Acids.
Cosmid vector pcosKA5 was constructed as follows. Homing endonuclease I-CeuI and PI-PspI sites were introduced into pBluescript KS+ (Stratagene) SalI and ClaI sites with the use of the linkers 5′-AGCGGCCCGTAACTATAACGGTCCTAAGGTAGCGAAGGCCGCT-3′ and 5′-AGCGGCCTTCGCTACCTTAGGACCGTTATAGTTACGGGCCGCT-3′ for I-CeuI, and 5′-AGCGGCCAAAATCCTGGCAAACAGCTATTATGGGTATTATGGGTGGCCGCT-3′ and 5′-AGCGGCCACCCATAATACCCATAATAGCTGTTTGCCAGGATTTTGGCCGCT-3′ for PI-PspI to create pKAhome. The XhoI-EcoRV fragment from pKAhome was introduced into the ApaI site of the pKAhome to create a multiple cloning site containing an I-CeuI, PI-PspI, XhoI, I-CeuI, and PI-PspI cassette. This cassette was inserted into the BamHI site of pWE15 (Stratagene), yielding pWEhome. The construct pcosKA5 was created by removing the neomycin resistance gene from pWEhome by SalI/SmaI restriction digestion. Cosmid libraries of M. grisea and M. graminicola were constructed in pcosKA5 as described (24). Cosmid libraries were quality checked by pulsed-field gel electrophoresis, restriction digestion analysis, and PCR identification of single genes (data not shown). DNA from 10 cosmid pools (≈20,000 colony-forming units) was prepared by a modified alkaline lysis method (25). The Frigg transposon was constructed by cloning the hph gene controlled by the TrpC promoter (26) into the HpaI site of pMM2611 (27). Thyra was constructed by insertion of the TrpC/hph gene into the SalI site of EZ∷tnpMOD (Epicentre Technologies, Madison, WI). The TrpC/hph gene was cloned into the EcoRV sites of GPS3 (New England Biolabs) to create Sif. The β-lactamase gene was inactivated by truncation in all transposons. Transposon DNA for IVT was prepared by standard methods. A linear Mu transposon (Finnzymes, Helsinki) was amplified and cloned into pGEM-T (Promega). The kanamycin resistance gene was deleted by NruI/pflMI double digestion, and the TrpC/hph cassette was inserted (blunt) to yield Vor. IVT was performed according to manufacturer's recommendations and as described (17). Transformants were grown on Luria Broth agar containing 25 μg/ml hygromycin B and 50 μg/ml ampicillin (Frigg) or 50 μg/ml hygromycin and 50 μg/ml ampicillin (Sif, Thyra, Vor). DNA was prepared as described (21) and cut with HpaI, and Southern analysis was performed as described (24) with the use of M. grisea WT and integration mutant (IM) strains IM-135, IM+3660, IM+6930, and IM+7582. A M. grisea adenylate cyclase (MAC1) probe used for Southern analysis was amplified with the use of the primers CCAGAAGCAAGAGCCCTACA and CATGTTCTTTGCGGCTTTCT. M. grisea total RNA was extracted with an RNAqueous-Midi kit (Ambion, Austin, TX) according to the manufacturer's recommendations. Northern analysis was performed as described (24). A fragment of the actin gene of M. grisea was amplified with the primers CCGTGACTTGACCGACTACC and GACATGGATACCACCGCTCT. Transcript ends were analyzed by 3′ rapid amplification of cDNA ends (28) followed by sequencing.
Sequencing and Sequence Analysis.
Colonies were picked into 384-well trays containing Super Broth (K. D. Medical, Columbia, MD), K. D. 50 μg/ml ampicillin, and 8% glycerol and incubated overnight at 37°C. Ninety-six-well blocks were inoculated with the overnight cultures and incubated overnight with agitation at 37°C, and the 384-well plate was stored at −80°C. Cosmid DNA was prepared as above. Cosmids were sequenced with the use of Big Dye Terminators (Perkin–Elmer) and primers at the ends of each transposon. The average read length from each transposon end was 500 Phred 20 bases (29, 30). Reactions were purified with the use of Sephadex columns and analyzed on an ABI377 or 3700 (Perkin–Elmer) under standard conditions.
DNA sequences containing at least 100 nucleotides with a Phred value of 20 or above (29–30) were selected for analysis. The total number of transposon insertions was calculated by combining the two sequences obtained from the same transposon insertion event when applicable. Crossmatch (31) was used to identify insertions into the cosmid vector and M. grisea repeated sequences. M. grisea genomic DNA sequences were analyzed with the blastn algorithm for an M. grisea expressed sequence tag data set (Paradigm Genetics) and with the ungapped blastx algorithm for protein databases (32). The protein databases were derived from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov, nr, December 6, 2000), the S. cerevisiae genome (http://genome-www.stanford.edu/Saccharomyces, orf trans.fasta), the Candida albicans genome (http://sequence-www.stanford.edu/group/candida, Ca-Assembly6.orf trans.gz), and a filamentous fungal protein database. The filamentous fungus protein database was constructed from information from the European Bioinformatics Institute (http://www.ebi.ac.uk, SWALL, 21-Dec-2000:fungi, not Saccharomycetes, not Schizosaccharomycetales).
Fungal Transformation.
Transformation of M. grisea and M. graminicola was performed essentially as described (33, 34), but with β-glucanase (InterSpex Products, San Mateo, CA). Transformants were purified and DNA was extracted as described (33). PCR was used to identify homologous recombination events. IMs are named according to the distance between the start codon and the insertion; i.e., IM-135 has an integration event 135 bp upstream of the start codon. Primers were as follows: CTTCAGCCGTCTCCAAAGTC and CTTGTTTGCCCACCAAATCT for IM+3660; GCCCAATCGCTAACAACATC and AGTATTGGCAGCGACTCACC for IM-135; CATATGGGGTCTGTGGAAGC and CCCTTGAAGGCAAGAGTTGT for IM+6930; GGCCACCTTGTTGTTTTGTT and CAAAAGGGCCAGCTGTATGT for IM+7582; ATAACTTTCGCGGCACCTT and TCCTCTTGCAACTCCCAGTC for M. grisea IM+840; CCCGGTATGCGATAATCTTC and CGGAATCAGATGACTTGCTG for M. grisea IM-190; GACCGAACAGGTGATGTGAA and AGCAAAATCGGCACACTCTT for M. graminicola IM-20; and TCGCTTCGTCTTCACATCAC and TCCTTGTCATCGACACTCCA for M. graminicola IM+411. Primer pairs were designed to flank the transposon insertion site. For each disruption experiment two independent TI events and one ectopic integration event were characterized.
Results and Discussion
Targeted Integration in Filamentous Fungi.
When filamentous fungi are transformed with DNA carrying a selectable marker, integration occurs at homologous or ectopic sites. The frequency of integration at a homologous site is variable and may depend on the size of the homologous DNA region of the transforming DNA (15). Transformation of M. grisea was performed with different sized DNA fragments containing the MAC1 gene (36) and 10 other DNA fragments generated with the use of TAGKO (Fig. 1). We measured the frequency of gene targeting, analyzing between 24 and 96 transformants per construct. TI frequencies were generally low when the homologous region contained less than 5 kb of homologous DNA (Fig. 2), was introduced as a closed circle, or lacked homologous sequences at the ends (unpublished results). However, TI frequencies increased with increasing length of homologous DNA (Fig. 2). Although TI frequencies were somewhat locus dependent, TAGKO-generated DNA fragments generally yielded higher TI frequencies than previously published vectors for M. grisea (Fig. 2). The results strongly suggest that increased DNA fragment size increases gene targeting frequencies.
Figure 2.
TI is a function of homologous DNA length. TI frequency vs. homologous DNA length is plotted for three different data sets. ▴, Five published gene knockouts in M. grisea strain Guy 11. MA, MAC1 (36); P, PMK1 (23); A, ABC1 (37); M, MPG1 (21); N = NUT1 (38). Published TI frequencies are 16%, 11%, 21%, 3%, and 22%, respectively. □, MAC1 TAGKO cosmid subjected to KpnI, EcoRI and SpeI, restriction digestion yielding, three MAC1 constructs with the same insertion but different homologous DNA lengths of 5.5 kb, 13.5 kb, and 20.2 kb, respectively. ○, Ten different TAGKO constructs created by IVT into a cosmid library pool.
Transposon Mutagenesis.
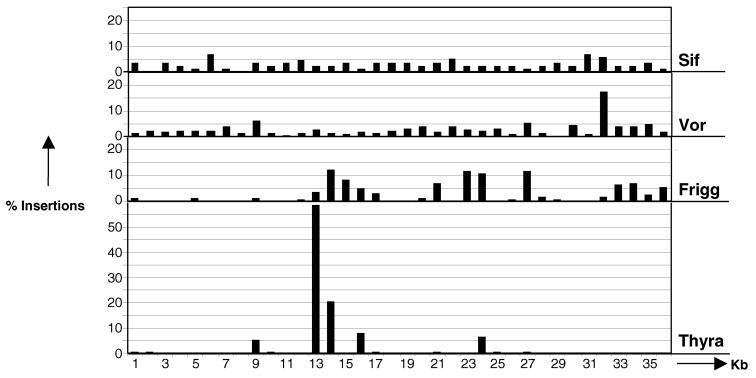
A potential limitation for transposon mutagenesis is the preferential or biased insertion of transposons into certain sequences in a targeted DNA region. We surveyed the insertion site biases for four different transposons engineered to carry a hygromycin phosphotransferase gene (Thyra, Frigg, Sif, and Vor) in a 36-kb recipient DNA fragment (Fig. 3). Insertion sites were determined by sequencing from the transposon ends. Monte Carlo simulations (39) showed that 89 random insertions must be recovered to mutagenize each kilobase of a 36-kb cosmid sequence (P = 0.95). The target cosmid insert is 52% AT (typical for many filamentous fungi) and does not contain long homopolymeric tracts or simple sequence repeats (data not shown). Frigg and Thyra have strongly biased in vitro insertion patterns (Fig. 3). The bias does not correspond to AT-rich or GC-rich DNA, coding or noncoding DNA, or sites of DNA bending as determined by computer analysis (data not shown). Sif and Vor displayed much less bias, the former being superior. Based on this analysis we selected Sif for whole genome analysis.
Figure 3.
In vitro transposition site distribution. Comparison of transposition insertion site distribution in the MAC1 cosmid insert (≈36 kb) for transposons Thyra, Frigg, Sif, and Vor. The percentage of transposition events (y axis) within 1-kb segments (x axis) of the cosmid is shown. The number of insertion events (n) analyzed for each transposon is as follows: SIF, n = 89; Vor, n = 237; Frigg, n = 209; Thyra, n = 336.
Cosmid DNA pools prepared from a 10× genome-equivalent M. grisea cosmid library were mutagenized in vitro with the Sif transposon, and the DNA sequences adjacent to the transposon insertions were analyzed. A total of 38,882 sequences representing 25,179 insertions were analyzed. The vector (6 kb) constitutes ≈15% of the cosmid, and the actual number of insertions into the vector (4,702/25,179 = 18.7%) supports the random transposon insertion model. Of the 20,477 insertions characterized as M. grisea DNA insertions, 0.1% showed matches to ribosomal DNA and 6.8% to M. grisea transposon repeat sequences. Comparison to an expressed sequence tag set (8,044 unique M. grisea expressed sequence tags) showed homology to 2,680 different genes. A total of 31.8% of the insertions showed strong (e < 10−20) or moderate (10−20 ≤ e ≤ 10−05) matches to the National Center for Biotechnology Information nonredundant protein database. We found 16.4% and 18.6% matches to proteins from the genome databases of Saccharomyces cerevisiae and Candida albicans, respectively, and ≈20.9% of the insertions revealed homologies to filamentous fungal proteins. Interestingly, 49.1% (2,102/4,285) of the genes showing homology to filamentous fungal proteins were exclusive to this data set, showing no homology (e > 10−05) to either S. cerevisiae or C. albicans protein data sets. The p-hydroxyphenylpyruvate dioxygenase (HPD4) protein (see below) was a member of this exclusive set. We conclude that many of the insertions produced by TAGKO identify uncharacterized genes.
Generation of Allele Diversity.
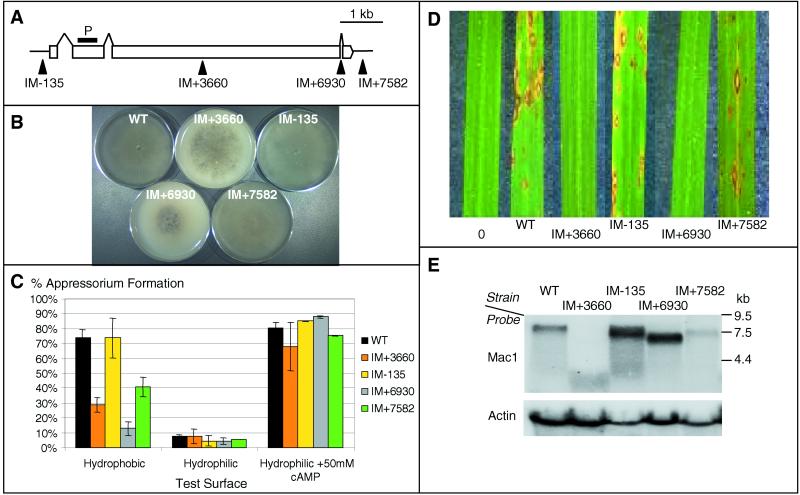
Insertional mutations should create different allele phenotypes. We investigated a collection of insertions in the MAC1 gene (Fig. 4A). cAMP signaling is required for the formation of infection structures (appressoria) in M. grisea (36). Appressoria differentiate when the tip of a spore germ tube contacts a hydrophobic surface. Appressoria generate turgor pressure needed to penetrate the rice leaf cuticle (21). IM+3660 (exon insertion) and IM+6930 (intron insertion) show reduced growth, sporulation, appressorium formation, and pathogenicity consistent with a MAC1 null phenotype (Fig. 4 A–D and ref. 36). However, IM+3660 and IM+6930 show dramatically different MAC1 mRNA levels (Fig. 4E). As anticipated, IM+3660 produces no full-length MAC1 mRNA, as detected by RNA blotting. IM+6930 produces abundant mRNA that does not contain the 3′ exon (data not shown). The resulting null phenotype of IM+6930 suggests that this region is required for MAC1 function.
Figure 4.
Analysis of M. grisea TAGKO mutants in MAC1. (A) Transposon insertion sites in MAC1. P designates the probe used for RNA analysis. (B) Fungal growth assays. Mutant and WT strains were grown on oatmeal agar and photographed after 7 days. (C) Appressorium formation on various surfaces or in the presence of cAMP. The experiment was repeated three times with 250 germinated spores counted for each strain. (D) Rice pathogenicity assays. Sample leaves were photographed 7 days after inoculation. (E) RNA blot analysis of MAC1 mRNA levels. Thirty micrograms of total RNA was used for each strain.
Upstream (IM-135) or downstream (IM+7582) insertions in MAC1 displayed wild-type growth and colony morphology (Fig. 4B). The insertion in IM-135 is 135 bp upstream of the MAC1 start codon and has WT levels of appressorium formation and pathogenicity but elevated mRNA levels (Fig. 4 C and E). Southern analysis confirmed a single integration event in IM-135. The increased mRNA levels show that this insertion up-regulates the MAC1 mRNA. In contrast, IM+7582 shows reduced appressorium formation, slightly reduced pathogenicity (Fig. 4 C and D), and dramatically reduced MAC1 mRNA levels (Fig. 4E). cDNA sequencing shows that the WT MAC1 mRNA terminates 901 bp downstream from the stop codon, whereas the IM+7582 message terminates 439 bp downstream from the stop codon (data not shown). We conclude that TAGKO has the potential to generate allelic variants with different levels of gene expression and function.
Cross-Validation of Gene Function with TAGKO.
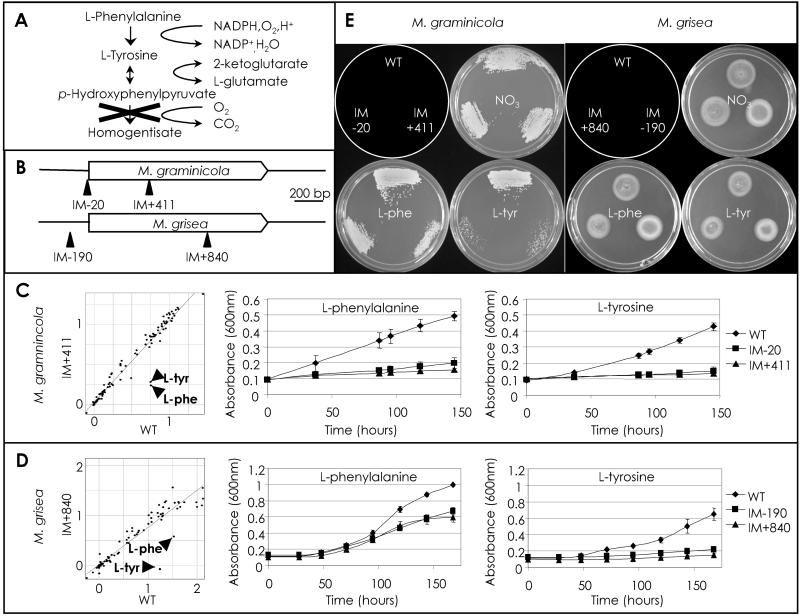
The cereal fungicide market is primarily focused on rice and wheat to control diseases such as rice blast (caused by M. grisea) and wheat blotch (caused by M. graminicola) (40). A useful approach to discovering fungicide target candidates is to identify pathways that, when blocked, lead to the rapid arrest of growth and/or cell death. Tyrosine and phenylalanine are metabolized through a pathway involving p-hydroxyphenylpyruvate dioxygenase (HPD4 gene) (Fig. 5A) (41–43). Inhibitors of this pathway are known, and a block in this pathway leads to the generation of toxic intermediates (44). Multiple TAGKO insertions in both the M. graminicola and M. grisea HPD4 genes (Fig. 5B) were identified and used to generate insertional mutant strains. Mutant strains of both fungi (M. graminicola: IM-20, IM+411; M. grisea: IM-190, IM+840) were tested for their ability to use a variety of nitrogen sources in a 96-well-based assay, and all mutant strains showed impaired growth in the presence of tyrosine and phenylalanine (Fig. 5 C and D). Growth of both fungi was severely limited when tyrosine was used as the sole nitrogen source (MM-N + tyr). To determine whether blockage of HPD4 function could lead to cytotoxicity, cells were removed from the presence of tyrosine or phenylalanine and tested for recovery on complete medium (Fig. 5E). When compared with recovery from nitrate or phenylalanine-containing media, M. graminicola mutants recovered poorly from tyrosine-inhibited growth. M. grisea mutants showed similar levels of recovery in the two cases. We conclude that the M. grisea and M. graminicola HPD4 genes function in an orthologous pathway involved in the metabolism of tyrosine and phenylalanine, but that phenotypic differences may indicate differential sensitivities to pathway intermediates. Fungal-specific components of these pathways may yet be useful as fungicide targets. TAGKO can thus be used to cross-validate pathways for use in fungicide discovery.
Figure 5.
Analysis of M. graminicola and M. grisea HPD4 TAGKO mutants. (A) The l-phenylalanine (l-phe) and l-tyrosine (l-tyr) degradation pathway. X indicates the pathway block by inactivation of HPD4. (B) M. graminicola and M. grisea HPD4 alleles showing the location of the TAGKO insertions. (C) Graphs showing growth of M. graminicola WT vs. IM+411 at 7 days in the presence of 95 different nitrogen sources (l-Phe and l-Tyr are indicated) and time courses of M. graminicola WT, IM-20, and IM+411 in MM-N + Phe and MM-N + Tyr, respectively. (D) Graphs showing growth of M. grisea WT vs. IM+840 at 7 days in the presence of 95 different nitrogen sources (l-Phe and l-Tyr are indicated) and time courses of M. grisea WT, IM-190, and IM+840 in MM-N + Phe and MM-N + Tyr, respectively. (E) Plate images showing growth recovery on complete medium of M. graminicola WT, IM-20, and IM+411 (after 8 days) and M. grisea WT, IM-190, and IM+840 (after 5 days) after incubation in MM, MM-N + Phe or MM-N + Tyr, as indicated. The schematic plate diagram depicts the location of each strain.
Summary
In vitro transposition into large insert libraries, followed by DNA sequencing, can be used to rapidly generate a library of insertional mutagenesis vectors for functional genomic approaches. Although the genome sequences for M. grisea and M. graminicola are unknown, sufficient DNA sequence information is obtained from each insertion to allow annotation. The approach described here for analyzing fungal genomes may be a useful addition or alternative to traditional skim sequencing strategies. An advantage of TAGKO is the generation of clones useful for insertional mutagenesis.
We chose cosmid libraries to generate large inserts for improved gene targeting. Although cosmid libraries have been used routinely for analyzing fungal genomes (45, 46), we envision that additional types of libraries may be equally suited to this protocol. For example, for filamentous fungi with higher frequencies of gene targeting, plasmid libraries may be better suited. In other fungi, including M. grisea, bacterial artificial chromosomes libraries have been constructed to facilitate mapping (47). Bacterial artificial chromosomes can be used for fungal transformation (47), and thus in vitro transposon mutagenesis of a minimal tile of clones may also be useful. It is conceivable that the strategy described here could be used in other organisms amenable to DNA-based transformation and gene targeting.
Despite biased insertion patterns for three of the transposons tested, we were able to recover insertions in most parts of the M. grisea MAC1 gene and demonstrate alterations in phenotype and gene expression. The ability to generate different alleles is an important advantage over traditional gene knockout strategies. Transposons active in vitro could be engineered to carry promoters, gene fusions, or additional genetic markers for enhancing functional genomic approaches.
Here we have used TAGKO to mutate and analyze genes in two phytopathogenic fungi. On a larger scale, TAGKO libraries in several fungal pathogens should allow functional comparison of multiple target genes and pathways. In addition to target discovery, functional genomic studies in economically important filamentous fungi can aid in the identification of pathways and enhanced processes for microbial biotechnology.
Acknowledgments
We thank Marie Coffin for assistance with the Monte Carlo simulation experiment.
Abbreviations
- TAGKO
transposon-arrayed gene knockout
- TI
targeted integration
- IVT
in vitro transposition
- IM
integration mutant
- WT
wild type
- MM
minimal medium
- MAC1
Magnaporthe grisea adenylate cyclase gene
- HPD4
p-hydroxyphenylpyruvate dioxygenase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF32553 (M. grisea HPD4)].
References
- 1.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 2.Hamer L, DeZwaan T M, Montenegro-Chamorro M V, Frank S A, Hamer J E. Curr Opin Chem Biol. 2001;5:67–73. doi: 10.1016/s1367-5931(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 3.Maier F J, Schafer W. Biol Chem. 1999;380:855–864. doi: 10.1515/BC.1999.105. [DOI] [PubMed] [Google Scholar]
- 4.Metzger D, Feil R. Curr Opin Biotechnol. 1999;10:470–476. doi: 10.1016/s0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 5.Latge J P. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graybill J R. Braz J Infect Dis. 2000;4:47–54. [PubMed] [Google Scholar]
- 7.Eyal Z. In: Septoria on Cereals: A Study of Pathosystems. Lucas J A, Bowyer P, Anderson H M, editors. Oxfordshire, U.K.: Commonwealth Agricultural Bureaux; 1999. pp. 1–25. [Google Scholar]
- 8.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 9.Timberlake W E, Marshall M A. Science. 1989;244:1313–1317. doi: 10.1126/science.2525275. [DOI] [PubMed] [Google Scholar]
- 10.Mewes H W, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver S G, et al. Nature (London) 1997;387, Suppl. 6632:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 11.Hua S B, Qiu M, Chan E, Zhu L, Luo Y. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- 12.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupfer D M, Reece C A, Clifton S W, Roe B A, Prade R A. Fungal Genet Biol. 1997;21:364–372. doi: 10.1006/fgbi.1997.0982. [DOI] [PubMed] [Google Scholar]
- 14.Nelson M A, Kang S, Bruan E L, Crawford M E, Dolan P L, Leonard P M, Mitchell J, Armijo A M, Bean L, Blueyes E, et al. Fungal Genet Biol. 1997;21:348–363. doi: 10.1006/fgbi.1997.0986. [DOI] [PubMed] [Google Scholar]
- 15.Asch D K, Kinsey J A. Mol Gen Genet. 1990;221:37–43. doi: 10.1007/BF00280365. [DOI] [PubMed] [Google Scholar]
- 16.Devine S E, Boeke J D. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampe D J, Churchill M E A, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 18.Goryshin I Y, Reznikoff W S. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 19.Haapa S, Taira S, Heikkinen E, Savilahti H. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung H, Borromeo E S, Bernardo M A, Notteghem J L. Phytopathology. 1988;78:1227–1233. [Google Scholar]
- 21.Talbot N J, Ebbole D J, Hamer J E. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford M S, Chumley F G, Weaver C G, Valent B. Genetics. 1986;114:1111–1129. doi: 10.1093/genetics/114.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J R, Hamer J E. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Marra M A, Kucaba T A, Dietrich N L, Green E D, Brownstein B, WIlson R K, McDonald K M, Hillier L W, McPherson J D, Waterson R H. Genet Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrol A M, Sweigard J A, Valent B. Fungal Genet Newslett. 1994;41:22. [Google Scholar]
- 27.Lampe D J, Grant T E, Robertson H M. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 30.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 31.Dear S, Durbin R, Hillier L, Marth G, Thierry-Mieg J, Mott R. Genome Res. 1998;8:260–267. doi: 10.1101/gr.8.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S C, Ham K S, Darvill A G, Albersheim P. Mol Plant-Microbe Interact. 1997;10:700–708. [Google Scholar]
- 34.Payne A C, Grosjean-Cournoyer M C, Hollomon D W. Curr Genet. 1998;34:100–104. doi: 10.1007/s002940050372. [DOI] [PubMed] [Google Scholar]
- 35.Fincham J R. Microbiol Rev. 1989;53:148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adachi K, Hamer J E. Plant Cell. 1998;10:1361–1373. doi: 10.1105/tpc.10.8.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban M, Bhargava T, Hamer J E. EMBO J. 1999;18:512–521. doi: 10.1093/emboj/18.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froeliger E H, Carpenter B E. Mol Gen Genet. 1996;251:647–656. doi: 10.1007/BF02174113. [DOI] [PubMed] [Google Scholar]
- 39.Law A, Kelton D. Simulation Modeling and Analysis. NY: McGraw-Hill; 1991. [Google Scholar]
- 40.Hewitt H G. Fungicides in Crop Protection. Oxfordshire, U.K.: Commonwealth Agricultural Bureaux; 1998. p. 221. [Google Scholar]
- 41.Fernandez-Canon J M, Penalva M A. Proc Natl Acad Sci USA. 1995;92:9132–9136. doi: 10.1073/pnas.92.20.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keon J, Hargreaves J. FEMS Microbiol Lett. 1998;15:337–343. doi: 10.1111/j.1574-6968.1998.tb12966.x. [DOI] [PubMed] [Google Scholar]
- 43.Norris S R, Shen X, DellaPenna D. Plant Physiol. 1998;117:1317–1323. doi: 10.1104/pp.117.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez-Canon J M, Penalva M A. J Biol Biochem. 1995;270:21199–21205. doi: 10.1074/jbc.270.36.21199. [DOI] [PubMed] [Google Scholar]
- 45.Orbach M J. Gene. 1994;150:159–162. doi: 10.1016/0378-1119(94)90877-x. [DOI] [PubMed] [Google Scholar]
- 46.Brody H, Griffith J, Cuticchia A J, Arnold J, Timberlake W E. Nucleic Acids Res. 1991;19:3105–3109. doi: 10.1093/nar/19.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz-Perez S V, Crouch V W, Orbach M J. Fungal Genet Biol. 1996;20:280–288. doi: 10.1006/fgbi.1996.0042. [DOI] [PubMed] [Google Scholar]