Abstract
Many DNA polymerases (Pol) have an intrinsic 3′→5′ exonuclease (Exo) activity which corrects polymerase errors and prevents mutations. We describe a role of the 3′→5′ Exo of Pol δ as a supplement or backup for the Rad27/Fen1 5′ flap endonuclease. A yeast rad27 null allele was lethal in combination with Pol δ mutations in Exo I, Exo II, and Exo III motifs that inactivate its exonuclease, but it was viable with mutations in other parts of Pol δ. The rad27-p allele, which has little phenotypic effect by itself, was also lethal in combination with mutations in the Pol δ Exo I and Exo II motifs. However, rad27-p Pol δ Exo III double mutants were viable. They exhibited strong synergistic increases in CAN1 duplication mutations, intrachromosomal and interchromosomal recombination, and required the wild-type double-strand break repair genes RAD50, RAD51, and RAD52 for viability. Observed effects were similar to those of the rad27-null mutant deficient in the removal of 5′ flaps in the lagging strand. These results suggest that the 3′→5′ Exo activity of Pol δ is redundant with Rad27/Fen1 for creating ligatable nicks between adjacent Okazaki fragments, possibly by reducing the amount of strand-displacement in the lagging strand.
DNA polymerases (Pol) play essential roles in DNA replication, repair, and recombination. Several of the prokaryotic and eukaryotic DNA polymerases have an intrinsic 3′→5′ exonuclease (Exo) activity. Structural studies (1–3) indicate that the Exo catalytic site is located in an N-terminal domain that is separated from the polymerase domain. The movement of the 3′ end of the nascent DNA strand from the polymerase to the Exo site requires denaturation of 3–4 terminal base pairs. If the 3′ end of the nascent DNA strand is delayed in the polymerase site, for example when Pol generates misincorporation or misalignment, terminal nucleotides are likely to be partitioned to Exo site and hydrolyzed, resulting in proofreading of an error (reviewed in refs. 4–6). The structure of the Exo active site is highly conserved. Two divalent metal ions are present, and they are coordinated by the amino acids of three signature Exo motifs conserved throughout all families of DNA polymerases (ref. 7 and Fig. 1).
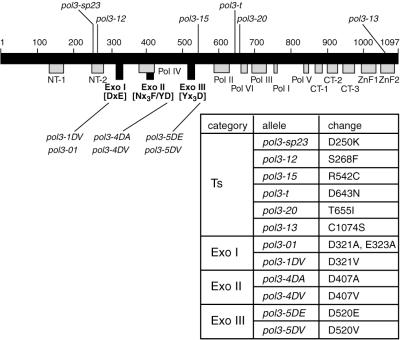
Figure 1.
Mutations in the yeast POL3 gene. The 1097-aa yeast POL3 ORF is shown schematically as a black bar. Names and positions of conserved regions are designated as in ref. 53. Gray boxes, conserved regions, other than nuclease; black boxes, exonuclease conserved regions. Consensus sequences of the 3′→5′ exonuclease signature motifs (7) are shown in brackets. Ts mutations are shown above and Exo mutations below the POL3 gene. Amino acid changes in different mutant alleles are shown in the table.
The proofreading function of Pol Exo is consistent with the observation that deficiency in a polymerase-associated exonuclease can lead to a strong mutator phenotype and a high error rate during in vitro replication (reviewed in refs. 1 and 8). Replication errors that escape Pol Exo proofreading can be corrected by postreplication mismatch repair. Cells that are deficient in both proofreading and mismatch repair are hypermutable and in some cases experience catastrophic rates of recessive lethal mutations (9–11). The biological importance of Pol Exo is emphasized by the observation of high incidence of tumors in mutant mice homozygous for a Pol δ Exo-deficient allele (B. Preston and R. Goldsby, personal communication).
A function for the 3′→5′ exonuclease of Pol δ distinct from correcting replication errors was proposed to explain why yeast double mutants carrying pol3-01 and rad27 (homolog of human FEN1, encoding for 5′ flap endonuclease) are inviable (12, 13). Synthetic lethality was observed not only with a rad27-Δ allele, but also with the subtle rad27-p defect (12). This mutation results from two amino acid changes F346A/F347A that eliminate the interaction between Rad27 and PCNA and has few phenotypic effects on its own. Unlike the pattern of synthetic lethality of pol3-01 in combination with mutations in mismatch repair genes (9, 14), pol3-01 rad27 was lethal not only in haploid, but also in diploid strains, even in pol3-01/POL3+ heterozygotes. This is inconsistent with a catastrophic rate of recessive lethal mutations leading to synthetic lethality.
There were several possible explanations for the synthetic lethality of pol3-01 with the rad27 defect. For example, it could have resulted not from the Pol δ exonuclease defect, but from some other change in function of this polymerase. Because both pol3-01 and rad27 defects interact with checkpoint mechanisms (15, 16), synthetic lethality could also be the consequence of cell cycle arrest caused by simultaneous activation of different branches of checkpoint control.
Alternatively, synthetic lethality could be the result of synergy between Pol δ Exo and 5′ flap endonuclease Rad27/Fen1 participating as redundant functions in an important DNA metabolic process such as DNA replication. Based on in vitro studies, Rad27/Fen1 is required for processing Okazaki fragments into ligatable nicks because of its capability to remove 5′ ribonucleotide and 5′ flaps caused by strand displacement by DNA polymerase. Flap removal is essential for maturation of shorter Okazaki fragments into long continuous DNA strand (17, 18). Yeast rad27-Δ null mutants are prone to an unusual category of mutations, extended duplications of up to 108 bp flanked by short direct repeats (19). It was suggested that the duplications result from incorporation of unremoved 5′ flaps into replicated DNA. Increased recombination and chromosome loss in the rad27-Δ mutant indicated that unremoved 5′ flaps can lead to double-strand breaks (DSB). In support of this, the rad27-Δ mutant required the RAD52 group of recombinational DSB repair genes for viability (19, 20). We speculated that both the 5′ flap endonuclease Rad27/Fen1 and the 3′→5′ exonuclease of Pol δ could act together to process adjacent Okazaki fragments to create ligatable nicks and that toxic intermediates could occur in replication forks when mutant pol3-01 enzyme is included in the replication complex. We proposed that the accumulation of DSBs was the cause of inviability in the double mutant.
In this study we show that the synthetic lethality with a rad27 defect is a general feature of nuclease-deficient Pol δ Exo-motif mutants and is not observed with other Pol δ mutations. A specific prediction of the hypothesis of functional redundancy was that the combination of partial Pol δ Exo and Rad27/Fen1 defects could cause genetic changes typical of mutants lacking the Rad27/Fen1 function. Some Pol δ Exo mutations were viable in combination with a partial Rad27 defect, and the viability required a functional DSB recombinational repair system. Surprisingly, the double mutants did not increase the rate of point mutations that might result from DNA polymerase errors, but instead caused synergistic increases in the rates of duplications and mitotic recombination. This suggests that the Pol δ 3′→5′ exonuclease is important not only for correcting replication errors, but also is redundant with Rad27/Fen1 in processing Okazaki fragments and preventing genome instability.
Materials and Methods
Yeast Strains and Plasmids.
The wild type, rad27-p, rad27-Δ, and pol3-01 isogenic strains used in this study have been described (12, 13, 21). Each strain was MATα ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52. Strains also carried mutant lys2 alleles used as mutation or recombination reporters. Interchromosomal recombination strains ALE100 and ALE101 were described in refs. 12 and 22. Strains ALE1000 and ALE1001 (a gift from K. Lobachev, National Institute of Environmental Health Sciences, Research Triangle Park, NC) with an intrachromosomal recombination reporter carried the 5′ truncated lys2 sequence, and the LEU2 gene has been integrated into chromosome II as a direct repeat with the lys2∷HS-D allele.
The plasmids used to make two-step gene replacements of genomic POL3 by pol3-t (p171; ref. 21) and by pol3-01 (YIpAM26; ref. 9) mutations have been described. Other pol3 mutations were created either in the plasmid YIpKhr5 or in p170. The YIpKhr5 plasmid with the POL3 N-terminal region was a gift from A. Sugino and Y. Pavlov (Osaka University, Osaka). pol3 mutations in the plasmids were made with the Quick Change Site Directed Mutagenesis Kit (Stratagene). Mutated plasmids were sequenced to verify that there were no additional mutations. The LC80B–ARS-CEN-TRP1 plasmid with wild-type RAD27 was described in ref. 12. Construction of deletions was as described for rad50–pNKY83 (23), rad51–pΔRAD51 (24), rad52–pΔ52Blast (25), and pep4∷KanMX (26). Sequences of mutant plasmids and oligonucleotides are available on request.
Yeast Genetic Methods.
Yeast genetic methods were as described in ref. 12. pol3-Ts (temperature-sensitive) mutants were propagated at 23°C. Temperatures used in specific experiments are given in footnotes to Tables 1–4. CAN1 mutants were obtained from independent single colony isolates. The CAN1 gene was PCR amplified and sequenced as described in ref. 19.
Table 1.
Mutation rates of pol3 mutants
| POL3 allele | lys2∷InsLD | his7-2 | CAN1 |
|---|---|---|---|
| WT* | |||
| POL3+ | 1.0 | 1.0 | 1.0 |
| Ts | |||
| pol3-t | 120 | 1.2 | 5.9 |
| pol3-12 | 67 | 1.1 | 2.9 |
| pol3-13 | 69 | 2.7 | 4.8 |
| pol3-15 | 8.7 | 0.9 | 1.4 |
| pol3-20 | 23 | 1.1 | 2.2 |
| pol3-sp23 | 250 | 3.6 | 15 |
| Exo | |||
| pol3-01 | 0.6† | 74 | 110 |
| pol3-1DV | 3.3 | 37 | 86 |
| pol3-4DA | 1.3 | 90 | 55 |
| pol3-4DV | 1.0 | 68 | 76 |
| pol3-5DE | 3.5 | 39 | 79 |
| pol3-5DV | 2.1 | 7.4 | 20 |
Rates are relative to wild type: lys2∷InsLD reversion, 0.1 × 10−8; his7-2 reversion, 1.9 × 10−8; CAN1 forward mutation, 29 × 10−8. Yeast were grown at 25°C.
Wild type.
Data from ref. 21.
Table 4.
Effect DSB recombinational repair defects on the loss from yeast pol3-Exo III and rad27-p mutants of the TRP1 marker associated with the RAD27 plasmid
| Genotype | TRP1 loss, %* | Viability |
|---|---|---|
| pol3-5DE rad27-p | 12–48 | + |
| pol3-5DV rad27-p | 66–69 | + |
| rad27-p rad51Δ | 33–43 | + |
| pol3-5DE rad51Δ | 45–78 | + |
| pol3-5DV rad51Δ | 53–76 | + |
| pol3-5DE rad27-p rad51Δ | 0 | − |
| pol3-5DV rad27-p rad51Δ | 0–1.1† | − |
| rad27-p rad50Δ | 8.9–30 | + |
| pol3-5DE rad27-p rad50Δ | 0–1.6‡ | − |
| rad27-p rad52Δ | 15–24 | + |
| pol3-5DE rad27-p rad52Δ | 0–2.1§ | − |
Yeast were grown at 30°C.
Presented are the minimal and maximal values of the TRP1 marker loss obtained in two to eight independent experiments for viable and seven to eight experiments for nonviable combinations.
There was no plasmid loss in seven of eight isolates.
There was no plasmid loss in six of eight isolates.
There was no plasmid loss in six of seven isolates.
Purification of Pol δ.
The protease-deficient pep4∷KanMX strains were grown as described in (27). The cells (about 25 g wet weight) were resuspended in 10 ml water and frozen as kernels in liquid nitrogen. They were disrupted in a blender with dry ice in lysis buffer as described (28). The lysate was fractionated with polymin P and ammonium sulfate, and the dialyzed crude fraction was subjected to phosphocellulose chromatography and MonoQ FPLC (27).
The peak MonoQ fractions containing Pol δ activity were diluted with an equal volume of buffer A0 (30 mM Hepes-NaOH, pH 7.4/1 mM EDTA/0.5 mM EGTA/10% glycerol/0.01% Nonidet P-40/5 mM DTT/2 μM pepstatin A/2 μM leupeptin; and NaCl as indicated by a subscript in mM) and injected onto a 0.5-ml MonoS column equilibrated in buffer A50. The column was washed with 1 ml of buffer A50 and eluted with a 10-ml linear gradient from 30–500 mM NaCl in buffer A. Pol δ eluted at ≈A350. The elution positions of the mutant enzymes were identical to wild type. The total yield of partially purified DNA polymerase, measured as units of polymerase activity on activated DNA, from the pol3-5DE and pol3-5DV mutants was comparable to that of wild type. However, the yield from the pol3-01 mutant polymerase was about half that of wild type.
DNA Polymerase and Exonuclease Assays.
DNA polymerase assays were carried out on DNase I-activated salmon sperm DNA (29).
The 50-μl nuclease assay contained 20 mM Tris⋅HCl, pH 7.8, 8 mM MgAc2, 0.2 mg/ml BSA, 4% glycerol, 1 mM DTT, and 100 fmol (as 3′ termini) of 3′-endlabeled pUC19 DNA plus enzyme. This DNA concentration was saturating for both the wild-type and mutant polymerases. The DNA substrate was prepared by linearizing pUC19 DNA with EcoRI and filling in with dATP and [3H]dTTP, followed by purification of the DNA. Assays were assembled on ice in microfuge tubes and incubated at 37°C for 15 min. They were stopped by addition of 100 μl 25-mM EDTA, 25 mM sodium pyrophosphate, and 50 μg/ml carrier DNA, followed by 125 μl 10% trichloroacetic acid. After 10 min on ice, the tubes were spun in a microfuge for 10 min. Supernatant (200 μl) was added to a water miscible scintillation fluid and counted in a liquid scintillation counter.
Results
pol3 Mutations.
A series of isogenic Saccharomyces cerevisiae strains was constructed that carry POL3 alleles with mutations in different parts of the large Pol δ subunit (Fig. 1). In addition to the previously studied pol3-01 mutation in the Exo I motif, five alleles were created that contained mutations in conserved residues of the Exo I, Exo II, or Exo III motifs (Exo-mutations; Fig. 1). Four of these mutations, pol3-01 (9, 10), pol3-1DV, pol3-4DA (30), and pol3-5DV (B. Preston, personal communication) were previously characterized as strong mutators.
We also studied six Ts mutations of Pol δ. Five of these cause genome instability and/or repair defects in S. cerevisiae: pol3-t (13, 21, 22, 31); pol3-13 (32); pol3-12, pol3-15, and pol3-20 (33). The sixth mutation, pol3-sp23 results in a mutant protein with the same amino acid change as a Ts mutant characterized in the fission yeast Schizosaccharomyces pombe (cdc6-23; ref. 34). None of the Ts mutants grew at 37°C, whereas all Exo mutants grew normally at all temperatures tested (23°C, 25°C, 30°C, and 37°C).
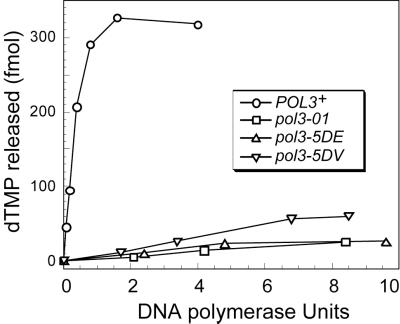
The pol3-4DA mutation in the Exo II motif eliminates the 3′→5′ exonuclease activity of Pol δ in vitro (30). To confirm the loss of exonuclease activity for mutants in the Exo I and Exo III motifs, Pol δ was purified to ≈30% purity from pol3-01, pol3-5DE, pol3-5DV, and wild-type strains (data not shown). Relative to wild-type Pol δ, the exonuclease activities of pol3-01, pol3-5DE, and pol3-5DV mutants were estimated to be < 0.9%, 1.3%, and 1.7%, respectively (Fig. 2).
Figure 2.
Exonuclease activity of mutant Pol δ. Assays were as described in Materials and Methods.
pol3-Exo and pol3-Ts Mutations Cause Different Types of Genetic Instability.
The effects of twelve pol3 alleles on genetic stability (Table 1) were studied with several reporter systems previously used to characterize the effects of the pol3-t (Ts) and pol3-01 (Exo) mutations: (i) lys2∷InsLD is a 31-bp insertion mutation that only reverts by deletions between 7-bp direct repeats, most likely via replication slippage (21, 31); (ii) the his7-2 allele contains a −1 frameshift in a run of eight A-T base pairs and reverts via +1 or −2 frameshifts (35); and (iii) canavanine-resistant forward mutants of the CAN1 gene can occur via a broad spectrum of point mutations and rearrangements (ref. 15 and references therein).
The exonuclease-deficient Pol δ Exo-motif mutations had different effects on genetic stability than the Pol δ alleles that are Ts (Table 1). The Exo-motif alleles did not cause a statistically significant increase in lys2∷InsLD deletions, but the Ts mutations elevated the rate of these events from 9- to 250-fold. In marked contrast to Ts alleles, Pol δ Exo-motif alleles were stronger mutators in the his7-2 frameshift reversion or can1 forward mutation detection systems (7.4- to 110-fold).
Synthetic Lethality of pol3-Exo with rad27 Defects.
The genetic interaction between POL3 and RAD27 was examined by determining the viability of strains carrying different alleles of these two genes (Table 2). The viability of a double mutant was evaluated by the ability to lose a plasmid carrying a wild-type copy of the RAD27 gene and a TRP1 marker (12). Double mutants of rad27 mutations with pol3-Ts showed high frequencies of TRP1 marker loss. In contrast, all of the double mutants carrying rad27-Δ and pol3-Exo mutations showed little or no loss of the TRP1 plasmid marker, indicating that the combination of Exo-deficient pol3 alleles with a deficiency of RAD27 function is lethal. (Rare TRP1 marker loss could be due to intraplasmid rearrangement or to reversion of the rad27 or pol3-Exo mutation.) Mutations in the Exo I and Exo II motifs were lethal even when combined with a subtle rad27-p defect. However, mutations in the Exo III motif (pol3-5DE and pol3-5DV) were viable with rad27-p. A reduced frequency of cells without the plasmid was observed in several cultures of the pol3-5DE rad27-p double mutant, but this is likely due to the double mutant growing at a reduced rate in the absence of the plasmid (data not shown).
Table 2.
Loss from pol3 rad27 strains of the TRP1 marker associated with the plasmid containing a wild-type RAD27 gene
| POL3 allele |
rad27
Δ
|
rad27-p
|
||
|---|---|---|---|---|
| TRP1 loss, %* | Viability | TRP1 loss, % | Viability | |
| Ts | ||||
| pol3-sp23 | 11–38 | + | 43–58 | + |
| pol3-12 | 7.3–33 | + | 21–43 | + |
| pol3-15 | 14–31 | + | 21–51 | + |
| pol3-t | 7.2–29 | + | 19–42 | + |
| pol3-20 | 15–31 | + | 33–61 | + |
| pol3-13 | 19–27 | + | 55–66 | + |
| Exo | ||||
| pol3-01† | 0–0.06 | − | 0–1.0 | − |
| pol3-1DV | 0 | − | 0 | − |
| pol3-4DA | 0 | − | 0 | − |
| pol3-4DV | 0 | − | 0 | − |
| pol3-5DE | 0 | − | 2.6–46‡ | + |
| pol3-5DV | 0 | − | 20–59 | + |
Yeast were grown at 23°C.
Presented are the minimal and maximal values of the TRP1 marker loss obtained in 4–12 independent experiments for viable and 9–12 experiments for nonviable combinations. About 100–300 colonies were counted in each experiment.
Data from ref. 12.
Median value is 11%.
It was previously shown (12) that heterozygous pol3-01/POL3+ diploids were also inviable with either rad27-Δ or rad27-p. Therefore, the viability of diploid heterozygotes for other mutations in Exo I, Exo II, and Exo III motifs was examined in this study. All pol3-Exo/POL3+ were synthetic lethal with rad27-Δ, and heterozygotes for pol3-1DV and pol3-4DV were synthetic lethal even with rad27-p (data not shown).
Genetic Instability in pol3-Exo rad27-p Double Mutants.
Synthetic lethality in pol3-Exo rad27 could result from an accumulation of DNA lesions in the double mutant cells that might be detectable as genome instability in the viable double mutants, pol3-5DE or -5DV with rad27-p. Double mutants were created in the presence of the plasmid LC80B carrying wild-type RAD27 and TRP1. The rates of mutation and recombination were determined by using fresh Trp− isolates that lost the plasmid (Table 3). This approach was taken to avoid accumulation of suppressor mutants that might confer growth advantage and possibly alter the genetic instability phenotype. All strains carried the reporters for +1 frameshift mutation (his7-2), forward mutation (CAN1), and either interchromosomal or intrachromosomal recombination.
Table 3.
Mutation and recombination rates in pol3 rad27 double mutants
| Genotype | his7-2 |
CAN1
|
Recombination
|
||
|---|---|---|---|---|---|
| Total | DUPL* | INTRA | INTER | ||
| wild type | 1.0 | 1.0 | 0 (0/20) | 1.0 | 1.0 |
| rad27-p | 1.8 | 2.7 | 1.3 (9/19) | 1.9 | 1.9 |
| rad27-Δ | 15 | 24 | 18 (15/20)§ | 18 | 15 |
| pol3-5DE | 51 | 26 | ND | 1.1 | 1.1 |
| pol3-5DE rad27-p | 62† | 100‡ | ND | 33‡ | 14‡ |
| pol3-5DV | 14 | 5.9 | 0 (0/20) | 1.3 | 0.9 |
| pol3-5DV rad27-p | 27† | 49‡ | 36 (19/26)§ | 14‡ | 3.9‡ |
Rates are relative to wild type: his7-2 reversion, 1.2 × 10−8; CAN1 forward mutation rate, 51 × 10−8; INTRA (intrachromosomal recombination), 1400 × 10−8; INTER (interchromosomal recombination), 9.9 × 10−8. Yeast were grown at 30°C.
Rates of extended duplications (DUPL) in CAN1 relative to the total rate of can1 mutations in the wild type were calculated based on the fraction of duplications among all can1 mutants sequenced (numbers of duplications and the total numbers of sequenced can1 mutants are given in parenthesis as numerator and denominator, respectively). Mutation spectrum is presented in the supplemental data.
No statistically significant increase over rates in the corresponding single pol3-Exo III mutant based on overlapping 95% confidence intervals.
Statistically significant increase over the rates in single mutants based on nonoverlapping 95% confidence intervals.
No statistically significant difference between fractions of duplications, P = 0.88.
Unlike the synergistic interaction between mutations inactivating mismatch repair and defects of either Pol δ or Pol ɛ Exo (9, 10, 14), there was no synergistic increase in the rate of +1 frameshift reversions of his7-2 in the pol3-Exo III rad27-p double mutants. This supports the conclusion that the negative interaction of Pol δ 3′→5′ exonuclease and rad27 defects resulting in lethality and genetic instability is not due simply to high levels of unrepaired mismatches generated by replication.
The rad27-Δ strains exhibit increased mutation and recombination rates (Table 3; ref. 12 and references therein). Similar to ref. 19, we also observed a large increase in duplications flanked by short direct repeats among forward mutations in the CAN1 gene. The rad27-p mutation has little or no effect on the overall rates of recombination and mutation (Table 3; ref. 12). Importantly, it caused a small but detectable increase in duplications (11–96 bp flanked by 5–13-bp direct repeats), indicating that the mutant protein may be only partially defective in flap removal. Single mutations in the Pol δ Exo III motif are mutators for both his7-2 and CAN1. Sequencing 20 CAN1 mutants in the pol3-5DV revealed only point mutations and the spectrum did not differ significantly from the wild type (see supplemental data, which is published on the PNAS web site, www.pnas.org). A 4- to 8-fold increase in the CAN1 forward mutation rate over a single pol3-Exo III mutants was observed in the double mutants. Importantly, the increased mutability of CAN1 in the pol3-5DV rad27-p double mutant is mainly due to extended duplications (7–64 bp flanked by 2–10-bp direct repeats). Thus, the addition of the pol3-5DV mutation to the rad27-p caused about 28-fold increase in the rate of duplications (Table 3, second and last lines).
The single mutations rad27-p, pol3-5DE, and pol3-5DV had little if any effect on recombination. However, the double mutants pol3-5DV rad27-p and pol3-5DE rad27-p exhibited elevated rates of interchromosomal and intrachromosomal recombination.
Viability of pol3-Exo III rad27-p Double Mutants Requires a DSB Recombinational Repair System.
One explanation for increased recombination in the viable double mutants carrying rad27-p and either pol3-5DE or pol3-5DV is that these cells have a higher than normal level of DSBs. To test this, mutations in DSB repair genes were introduced into these double mutants and their viability was determined (Table 4). Based on in vitro studies, RAD50, RAD51, and RAD52 play different roles in DSB repair (36). Each of these genes was deleted in single or double pol3-Exo III and rad27-p mutants carrying the RAD27-TRP1 plasmid LC80B.
The double mutants carrying rad27-p or pol3-Exo III and a DSB-repair defect were able to lose the RAD27-TRP1 plasmid. The lower frequency of cells lacking the RAD27-TRP1 plasmid in strains with rad27-p and a DSB repair defect is in agreement with the reduced growth rate of such double mutants (ref. 12 and data not shown). However, there was no loss or a drastic reduction in the frequency of loss of the RAD27-TRP1 plasmid in the triple mutants carrying rad27-p pol3-Exo III and any of the DSB repair defects. Therefore, the viability of pol3-Exo III rad27-p double mutants depends on the presence of a functional DSB recombinational repair system.
Discussion
Our study demonstrates that a general feature of 3′→5′ exonuclease deficient is synthetic lethality with a rad27 defect. Synthetic lethality cannot be explained by a catastrophic error rate resulting from the lack of Pol δ proofreading (see the Introduction). The nature of the biological function of the Pol δ Exo activity, which is distinct from correcting replication errors, can be deduced from the phenotype of the rad27-p pol3-Exo III double mutant.
The 3′→5′ Exonuclease Activity of Pol δ Can Participate in Processing of Okazaki Fragments.
We propose that the 3′→5′ exonuclease activity of Pol δ can act in place of Rad27/Fen1 to process Okazaki replication intermediates. The strongest support for this is the large increase in duplication mutations caused by the pol3-5DV mutation when combined with the rad27-p partial defect of the 5′ flap endonuclease (Table 3). It is generally accepted (see the Introduction) that the Rad27/Fen1 5′ flap endonuclease is important for prevention and/or removal of displaced 5′ flaps and creating ligatable nicks at the border between Okazaki fragments in the lagging strand. Duplications in rad27-Δ mutants are considered to result from the decreased flap removal. The partial rad27-p defect that disrupts the interaction between Rad27/Fen1 and PCNA also increased the rate of duplications, but to a smaller extent than found with the rad27-Δ mutant. This is in agreement with in vitro results indicating that PCNA enhances Fen1 cleavage efficiency via stabilization of the endonuclease on the flap but is not required for flap binding and cleavage (ref. 37 and references therein). The absolute increase in duplication rate caused by addition of pol3-5DV to the rad27-p is not less than the rate of duplications in the rad27-Δ strains. This supports our proposal that, along with the Rad27/Fen1, the Pol δ Exo can process (or prevent) 5′ flap intermediates.
In the presence of rad27-p both pol3-Exo III mutations led to increased recombination (Table 3). The increase in the recombination rate in most cases was comparable with the hyperrec phenotype of the rad27-Δ. Because both rad27-p pol3-Exo III mutants required the DSB recombinational repair genes for viability, we suggest that the most likely cause of hyperrecombination is the appearance of DSBs. Similar levels of hyperrecombination and requirements for DSB repair genes were found for the rad27-Δ single mutants (19, 20). The DSBs that are proposed to result from retained flaps could be due to unligated Okazaki fragments continuing into the next round of replication. Alternatively, DSBs could be formed in association with a stalled replication fork (38).
DSBs are highly toxic and 20–30 DSBs can kill a repair proficient yeast cell, whereas it takes only one DSB to inactivate a repair deficient rad52 cell (39, 40). Thus, it is possible that the synthetic lethality observed in certain pol3-Exo rad27 double mutants (Table 2 and refs. 12 and 13) results from an accumulation of DSBs to a level that exceeds the capacity of the DSB-repair system and/or from unrepairable DSBs. Partially defective Rad27/Fen1 (rad27-p) is unable to prevent DSBs even when pol3-Exo is heterozygous (pol3-Exo/POL3+), suggesting that unremoved flaps leading to DSBs may arise via concerted action of the mutant proteins in some replication forks.
An Alternative Way to Reduce 5′ Flap Formation.
Presented in Fig. 3 is a model explaining how a deficiency in 3′→5′ exonuclease activity could lead to increased formation of 5′ flaps. We propose that Exo-deficient Pol δ has increased strand displacement capacity similar to Exo-deficient mutants of replicative DNA polymerases such as T7 Pol (41–43) and T4 Pol (43–46).
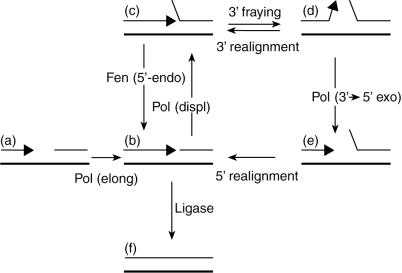
Figure 3.
Possible roles of the 3′→5′ exonuclease of Pol δ and the Rad27/Fen1 5′ flap endonuclease in creating a ligatable nick after strand displacement. The arrowhead shows the growing 3′ end of the nascent (Top) strand. Enzymatic reactions and conformational changes of DNA: Pol (elong), Pol δ elongation; Ligase, DNA ligase; Pol (displ), Pol δ displacement; Fen (5′-endo), Rad27/Fen1 5′ flap endonuclease; 3′ fraying, fraying at the 3′ end of the nascent strand; 5′ realignment, realignment of displaced 5′ end; Pol (3′→5′ exo), Pol δ 3′→5′ exonuclease.
Shown in Fig. 3 is the action of Pol δ, Rad27/Fen1, and DNA ligase at the junction between Okazaki fragments from which the RNA primer has already been removed by one of the 5′ nucleases, such as RNase H (47), Exo1 (48), or Dna2 (49). Strand displacement by DNA Pol (b→c) can create a 5′ flap intermediate that cannot be ligated. The 5′ flap can be removed by Fen1 (c→b). Alternatively, a ligatable nick could be created as a result of 3′ degradation by the 3′→5′ activity of DNA Pol (c→d→e→b). This could occur if strand separation (fraying) at the 3′ end (c→d) is followed by removal of terminal 3′ nucleotide(s) (d→e) similar to proofreading reaction (see the Introduction). Realignment of a 5′ end into the gap (e→b) could then create a ligatable nick.
The primary roles for Pol Exo activity is generally considered to be the removal of polymerization errors and prevention of mutations. Here we show that the Exo activity of Pol δ can also prevent extended duplications and recombination, and possibly DSBs, by suppressing excessive strand displacement and 5′ flap formation. The role of Pol δ Exo in suppressing genome instability and DSBs in yeast becomes evident only if the Rad27/Fen1 function is reduced. The Exo deficiency of yeast Pol δ on its own did not cause increased duplications. However, in partially defective Rad27/Fen1 (rad27-p) the rate of duplications caused by Exo defect is very high. pol3-Exo mutations on their own did not cause an increase in recombination or a requirement for the DSB recombinational repair genes including RAD52. These results suggests that in yeast the Rad27/Fen1 protein efficiently removes 5′ flaps in Pol δ Exo-deficient yeast mutants, although it might be working under stress. Delayed cell cycle progression in the pol3-01 Exo-mutant could be due to increased flap formation activating S-phase checkpoint (15). In higher eukaryotes, the relative roles of Pol δ Exo and Rad27/Fen1 in reducing 5′ flap formation and preventing genome instability remain to be determined. The contribution of Pol δ Exo may vary depending on the activity of Fen1, whose expression (50, 51) and nuclear localization (52) can be altered by proliferation status and DNA damage.
Supplementary Material
Acknowledgments
We thank Dr. K. Lobachev for yeast strains, Drs. A. Sugino and Y. Pavlov for the plasmid YIpKhr5, Drs. K. Bebenek, R. Schaaper, M. Longley, F. Kadyrov, K. Lewis, and E. Johansson for discussions and advice on the manuscript, B. Preston for communicating unpublished results, and H. Westmoreland for help in conducting experiments. The work in the P.M.J.B. laboratory was supported by National Institutes of Health Grant GM58534.
Abbreviations
- Pol
DNA polymerase
- Exo
exonuclease
- DSB
double-strand break
- Ts
temperature-sensitive
References
- 1.Derbyshire V, Pinsonneault J K, Joyce C M. Methods Enzymol. 1995;262:363–385. doi: 10.1016/0076-6879(95)62030-3. [DOI] [PubMed] [Google Scholar]
- 2.Steitz T A. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 3.Shamoo Y, Steitz T A. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 4.Johnson K A. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 5.Goodman M F, Fygenson K D. Genetics. 1998;148:1475–1482. doi: 10.1093/genetics/148.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 7.Blanco L, Bernad A, Salas M. Gene. 1992;112:139–144. doi: 10.1016/0378-1119(92)90316-h. [DOI] [PubMed] [Google Scholar]
- 8.Reha-Krantz L J. Genetics. 1998;148:1551–1557. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison A, Johnston A L, Johnston L H, Sugino A. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison A, Sugino A. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 11.Fijalkowska I J, Schaaper R M. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gary R, Park M S, Nolan J P, Cornelius H L, Kozyreva O G, Tran H T, Lobachev K S, Resnick M A, Gordenin D A. Mol Cell Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H T, Gordenin D A, Resnick M A. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta A, Schmeits J L, Neelam S A, Lau P J, Myung K, Kolodner R K. Mol Cell. 2000;6:593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 16.Vallen E A, Cross F R. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 18.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 19.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 20.Symington L S. Nucleic Acids Res. 1998;26:5589–5595. doi: 10.1093/nar/26.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran H T, Degtyareva N P, Gordenin D A, Resnick M A. Genetics. 1999;152:47–59. doi: 10.1093/genetics/152.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobachev K S, Shor B M, Tran H T, Taylor W, Keen J D, Resnick M A, Gordenin D A. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alani E, Padmore R, Kleckner N. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 24.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 25.Lewis L K, Kirchner J M, Resnick M A. Mol Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 27.Burgers P M. Methods Enzymol. 1995;262:49–62. doi: 10.1016/0076-6879(95)62008-7. [DOI] [PubMed] [Google Scholar]
- 28.Burgers P M. Methods. 1999;18:349–355. doi: 10.1006/meth.1999.0796. [DOI] [PubMed] [Google Scholar]
- 29.Burgers P M, Gerik K J. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 30.Simon M, Giot L, Faye G. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran T H, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giot L, Chanet R, Simon M, Facca C, Faye G. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giot L, Simon M, Dubois C, Faye G. Mol Gen Genet. 1995;246:212–222. doi: 10.1007/BF00294684. [DOI] [PubMed] [Google Scholar]
- 34.Iino Y, Yamamoto M. Mol Gen Genet. 1997;254:93–97. doi: 10.1007/s004380050395. [DOI] [PubMed] [Google Scholar]
- 35.Shcherbakova P V, Kunkel T A. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung P, Trujillo K M, Van Komen S. Mutat Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- 37.Tom S, Henricksen L A, Bambara R A. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 38.Rothstein R, Michel B, Gangloff S. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 39.Resnick M A, Martin P. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 40.Resnick M A. J Theor Biol. 1978;71:339–346. doi: 10.1016/0022-5193(78)90164-9. [DOI] [PubMed] [Google Scholar]
- 41.Engler M J, Lechner R L, Richardson C C. J Biol Chem. 1983;258:11165–11173. [PubMed] [Google Scholar]
- 42.Lechner R L, Engler M J, Richardson C C. J Biol Chem. 1983;258:11174–11184. [PubMed] [Google Scholar]
- 43.Canceill D, Viguera E, Ehrlich S D. J Biol Chem. 1999;274:27481–27490. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 44.Reha-Krantz L J, Stocki S, Nonay R L, Dimayuga E, Goodrich L D, Konigberg W H, Spicer E K. Proc Natl Acad Sci USA. 1991;88:2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Silva E F, Reha-Krantz L J. J Biol Chem. 2000;275:31528–31535. doi: 10.1074/jbc.M004594200. [DOI] [PubMed] [Google Scholar]
- 46.Chastain P D, II, Mackhov A M, Nossal N G, Griffith J D. Mol Cell. 2000;6:803–814. doi: 10.1016/s1097-2765(05)00093-6. [DOI] [PubMed] [Google Scholar]
- 47.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu J, Qian Y, Chen V, Guan M X, Shen B. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 49.Bae S H, Seo Y S. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 50.Kimura S, Ueda T, Hatanaka M, Takenouchi M, Hashimoto J, Sakaguchi K. Plant Mol Biol. 2000;42:415–427. doi: 10.1023/a:1006349511964. [DOI] [PubMed] [Google Scholar]
- 51.Kim I S, Lee M Y, Lee I H, Shin S L, Lee S Y. Biochim Biophys Acta. 2000;1496:333–340. doi: 10.1016/s0167-4889(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 52.Qiu J, Li X, Frank G, Shen B. J Biol Chem. 2000;95:4901–4908. doi: 10.1074/jbc.M007825200. [DOI] [PubMed] [Google Scholar]
- 53.Hindges R, Hubscher U. Biol Chem Hoppe-Seyler. 1997;378:345–362. doi: 10.1515/bchm.1997.378.5.345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.