In vitro work in trabecular meshwork reveals molecular mechanisms behind the differential sensitivities for glucocorticoids among normal and glaucoma populations.
Abstract
Purpose.
Glaucoma is a leading cause of visual impairment and blindness, with elevated intraocular pressure (IOP) as a major causative risk factor. Glucocorticoid (GC) therapy causes morphologic and biochemical changes in the trabecular meshwork (TM), an ocular tissue involved in regulating IOP, which can lead to the development of glaucoma in susceptible individuals (steroid responders). Steroid responders comprise 40% of the general population and are at higher risk of developing glaucoma. In addition, a majority of glaucoma patients are steroid responders. Differential distribution of various isoforms of GC receptor (GR) may be responsible for this heterogeneity in the steroid response. The alternatively spliced GRβ isoform acts as dominant negative regulator of classical GRα transcriptional activity. mRNA splicing is mediated by spliceosomes, which include serine-arginine rich proteins (SRps). The purpose of this study was to determine whether specific SRps regulate levels of these isoforms and thereby GC response in TM cells.
Methods.
Quantitative RT-PCR, Western blot analysis, and immunocytochemistry were used to determine the differential expression of different SRps (SRp20, 30c, and 40) in human normal and glaucomatous TM cell strains. Bioinformatics was used to find putative binding sites for SRp20 and SRp40 on exon 9 of the GR gene. A peptide modulator of splicing (bombesin) and SRp expression vectors were used to modulate SRp levels and determine their effects on GRα/GRβ ratios as well as dexamethasone (DEX) responsiveness via GRE- luciferase reporter activity, fibronectin, and myocilin induction in TM cells.
Results.
SRp20, SRp30c, and SRp40 regulate GR splicing and the GC response in TM cells. Modulation of SRp levels altered the GRβ/α ratio that correlated with DEX responsiveness. Bombesin decreased SRp20; increased SRp30c, SRp40 levels, and GRβ/α ratio, and suppressed DEX response in TM cells.
Conclusions.
Relative levels of SRp20, SRp30c, and SRp40 in TM cells control differential expression of the two alternatively spliced isoforms of the GR and thereby regulate GC responsiveness. Different levels and/or activities of these SRps may account for differential GC sensitivity among the normal and glaucoma populations.
Glaucoma represents a group of optic neuropathies that result in retinal ganglion cell death leading to a slow and progressive loss of vision and blindness. Glaucoma involves multifactorial etiologies and occurs in many forms, with primary open angle glaucoma (POAG) being the most prevalent form in the United States. Glucocorticoids (GCs) have been suggested to play an important role in pathophysiology of POAG. Elevated levels of cortisol have been reported in plasma1–3 and aqueous humor2 of POAG patients. In addition, altered cortisol metabolism has been reported in the trabecular meshwork (TM)4–6 and peripheral blood cells7 in POAG patients.
Steroid-induced glaucoma shares many physiological and clinical symptoms with POAG. GCs cause various morphologic and biochemical changes in the TM that are associated with increased aqueous humor outflow resistance leading to ocular hypertension (OHT). GCs inhibit TM cell functions, including phagocytosis8,9 and cell migration.10 GCs also reorganize the cytoskeleton to form cross-linked actin networks (CLANs) in cultured human TM cells10 and tissues11 that mimic CLANs observed in cultured glaucoma (GTM) cells12 as well as TM tissue from glaucoma eyes.13 A potent and well-established GC, dexamethasone (DEX) is known to induce the glaucoma gene myocilin (MYOC) in TM cells.14,15
Physiologically characterized as potent regulators of metabolism, GCs are also used as potent anti-inflammatory, antiallergic and immune-suppressant agents. However, systemic and ocular side effects, including GC-induced OHT, limit their use. In addition to GC potency, duration and route of administration, GC-induced OHT depends on the inherent GC sensitivity of the patient.16 Approximately 4% to 6% of the general population develop a large increase (>15 mm Hg) in intraocular pressure (IOP) after topical ocular GC administration for 4 to 6 weeks, while one-third of the population develops moderate pressure rise (6–15 mm Hg).17,18 These individuals are categorized as steroid responders. The rest of the population are considered nonresponders. It has been reported that majority of POAG patients are moderate to high steroid responders.17 In addition, it has also been observed that steroid-responsive nonglaucomatous individuals are at much higher risk of developing POAG compared with steroid nonresponders.19,20 The molecular mechanisms underlying this differential sensitivity between normal and POAG patients toward GCs are poorly understood.
Alternative splicing of NR3C1, human glucocorticoid (GC) receptor (GR) gene, generates two isoforms: GRα and GRβ.21 GRα is the physiological and pharmacological receptor for GCs and is the classical active isoform through which most GCs work. The GRβ isoform is a truncated form of GRα that lacks the ligand-binding domain and does not transactivate GC-responsive genes. In fact, GRβ functions as a dominant negative regulator of GRα transcriptional activity.21 GRβ levels are elevated in a variety of GC resistant diseases, including asthma,22 rheumatoid arthritis,23 and inflammatory bowel disease,24 among others.25–30 We have previously reported that most normal TM cells express relatively higher amounts of GRβ compared with glaucomatous TM cells, making GTM cells more sensitive to GCs.31
Alternative splicing is one of the many mechanisms that regulate gene expression in eukaryotes, and this process itself must also be regulated. An imbalance or mutation in either trans-acting splicing factors or cis-acting DNA elements might disturb this RNA processing event. A specialized assembly of proteins, known as the spliceosome, mediates this mRNA splicing process. A spliceosome is composed of approximately 100 proteins, collectively known as splicing factors, in addition to the five small nuclear ribonucleoprotein particles (snRNPs).32 The critical balance of these serine/arginine-rich proteins (SRp) and snRNPs is necessary for controlling the level of exon inclusion/exclusion in the mRNA transcript.
There have been reports that different SRps such as SRp30c33,34 and SRp4035 could be involved in alternative splicing of GR, thus altering relative levels of GRα and GRβ. The potential role of these splicing factors in TM and GC responsiveness has not been previously explored. Studying the role of these SRps in the TM will directly answer the heterogeneous response to GCs in terms of elevated IOP and glaucoma pathogenesis. Our laboratory recently examined single nucleotide polymorphisms (SNPs) in some of these splicing factors in clinically well-characterized individuals (normal, POAG patients, and steroid responders).36 There was no significant differences in SNP allele frequencies between the 3 cohorts, suggesting that there are no heritable differences in these examined SRp genes associated with steroid responsiveness or POAG. Therefore, differences in the levels and/or activities of these different SRps may determine the GRβ/α ratio and GC response in steroid responders and POAG patients.
Methods
TM Cell Culture
Human TM cells were isolated from carefully dissected human TM tissue explants derived from patients with glaucoma or from normal donors and were characterized as previously described.10,37–39 All donor tissues were obtained from regional eye banks and managed according to the guidelines in the Declaration of Helsinki for research involving human tissue. Isolated TM cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen-Gibco, Grand Island, NY) containing l-glutamine (0.292 mg/mL; Gibco BRL Life Technologies, Grand Island, NY), penicillin (100 U/mL)/streptomycin (0.1 mg/mL; Gibco BRL Life Technologies), amphotericin B (250 μg/μL; Thermo Scientific Ltd., Rockford, IL), and 10% fetal bovine serum (FBS; Gibco BRL Life Technologies). Stably transformed human TM cell lines GTM3 and NTM540 were also used and cultured in the same medium.
TM Cell Treatment
TM cells were grown to 100% confluence in serum containing medium. TM cells were incubated with fresh medium containing different doses (0–10 μM) of bombesin acetate hydrate (Cat # B4272; Sigma Aldrich, St. Louis, MO) for 0.5–48 hours. In some studies, bombesin (1 μM) treatment was followed by treatment with or without DEX (100 nM) for 6 to 12 hours for mRNA isolation and 24 to 72 hours for protein isolation.
Bioinformatics Approach
Bioinformatics software (Splicing Rainbow; Morais and Valcarcel EMBL-EBI; available at http://www.ebi.ac.uk/asd-srv/wb.cgi?method=8) was used to find putative binding sites for various spliceosome proteins in exon 9α and 9β sequences. From the list of binding sites received after the exon 9 sequence analysis, we only selected putative binding sites for SRp20 and SRp40 (two splicing factors that have been shown to regulate GR isoforms expression in various cell lines and GC-sensitive diseases). Information on potential SRp30c binding sites was not available in this software package. A sequence was considered a putative binding site for SRp20 if the score (S) was >6 and for SRp40 if S >5. We reduced the stringency for SRp40 to include more sites for analysis.
Overexpression Plasmids and Transfection
Expression plasmids for human SFRS3, SFRS5 and SFRS9 (encoding SRp20, SRp40, and SRp30c, respectively; see Table 1) were purchased from Origene (Rockville, MD). Transfection of plasmids (pCMV6-XL5) was performed as described in the Origene protocol for transient transfection of mammalian cells. In one tube, transfection reagent (Surefect; SA Biosciences, Frederick, MD) was mixed gently with 250 μL of medium (Opti-MEM; Invitrogen) and incubated for 5 minutes at room temperature. In separate tubes, 1 μg (100 ng/μL) of overexpression plasmid or control empty vector plasmid were mixed gently with 250 μL of medium (Opti-MEM; Invitrogen, Grand Island, NY). These two tubes were combined, gently mixed, and incubated for 20 minutes at room temperature. After incubation, medium (Opti-MEM; Invitrogen) without FBS and antibiotics was added to obtain a final volume of 1 mL for each well. One milliliter of GTM3 cells (dissolved in Opti-MEM [Invitrogen] medium containing 5% FBS) were plated in 6-well plates (3 × 105 cells per mL) and incubated with transfection mixture for 24 hours at 37°C. Cells were washed with sterile PBS and incubated with 10% FBS containing DMEM for 24 hours (for mRNA) or 36 to 48 hours (for protein) at 37°C. In experiments involving DEX treatment, cells were kept for an additional 24 hours in 10% FBS containing DMEM with or without DEX (100 nM). Cell lysates were analyzed for various proteins by Western blot analysis (see Table 3 for antibodies used).
Table 1.
Summary of SFRS Genes and SR Proteins and Their Effects on GR Splicing and DEX Effects in TM Cells
| Gene Name | Protein Name | Effect on GRβ/GRα Ratio | Effect on DEX-Mediated GRE-Luciferase Activity |
|---|---|---|---|
| SFRS3 | SRp20 | No significant change in GTM3 | No significant change in GTM3; increase in HTM5 |
| SFRS5 (SFRS5.1 and SFRS5.2 differs in 5′ UTR) | Both isoforms encode SRp40 | Increase in GTM3 | Decrease in GTM3; no change in HTM5 |
| SFRS9 | SRp30c | Increase in GTM3 | Decrease in GTM3; no change in HTM5 |
UTR, untranslated region.
Table 3.
List of Antibodies
| Protein Name | Primary Antibody | Dilution |
|---|---|---|
| SRp20 | Santa Cruz Biotech, Santa Cruz, CA; SC-73059; mouse monoclonal | 1:200 for WB; 1:50 for ICC |
| SRp30c | Santa Cruz Biotech, Santa Cruz, CA; SC134036; rabbit polyclonal | 1:200 for WB; 1:50 for ICC |
| SRp40 | Novus Biologicals, Littleton, CO; H00006430-B01P; mouse polyclonal | 1:500 for WB; 1:100 for ICC |
| GRα | Santa Cruz Biotech, Santa Cruz, CA; SC-1002; rabbit polyclonal | 1:300 for WB; 1:50 for ICC |
| GRβ | Custom-made; rabbit polyclonal | 1:2000 for WB; 1:500 for ICC |
| Fibronectin | Millipore, Billerica, MA; AB1945; rabbit polyclonal | 1:500 for WB; 1:500 for ELISA |
| Myoclin | Santa Cruz Biotech, Santa Cruz, CA; SC-20976; goat polyclonal | 1:500 for WB |
| GAPDH | Cell Signaling, Boston, MA; 14C10; rabbit monoclonal | 1:1000 for WB |
| β-Actin | Millipore, Billerica, MA; MAB 1501; mouse monoclonal | 1:500 for WB |
RNA Isolation, RT-PCR, and Agarose Gel Electrophoresis
Total cellular RNA was extracted from cultured TM cells (TRI Reagent RT, Cat. # RL-311; MRC Inc., Cincinnati, OH). A kit (SuperScript VILO cDNA Synthesis, Cat. # 11754; Invitrogen) was used for first strand cDNA synthesis. Primers for the various SR proteins were designed using software (Primer3; http://frodo.wi.mit.edu/primer3/). The primer pairs are listed in Table 2. The PCR products were loaded and electrophoresis performed on a 1.5% agarose gel containing ethidium bromide to detect DNA bands under UV light.
Table 2.
List of PCR Primers
| Gene Name | Left Primer Sequence (5′ to 3′) | Right Primer Sequence (5′ to 3′) |
|---|---|---|
| SFRS1.1 | GAAGACGCGGTGTATGGTC | GATCTGCTATGACGGGGAGA |
| SFRS2 | GACCGCTACACCAAGGAGTC | TTGGATTCCCTCTTGGACAC |
| SFRS3 | ATGCATCGTGATTCCTGTCC | ACCCTTAAACTGGCAGGACA |
| SFRS4 | CTCACAAGGGACGCAAAAAT | CTCACACTCCCTCGCTTCTC |
| SFRS5.1 | AGTGCGTCAGTTGTGGAGTG | CCACGTTTCTGGCTCTTCTC |
| SFRS5.2 | GCTGCTAAGTGCGTCAGTTG | CCACGTTTCTGGCTCTTCTC |
| SFRS6 | TATGCGACAAGCAGGTGAAG | TTGAGGGTGGAACAGGTAGC |
| SFRS7 | GTTGGTAACCTGGGAACTGG | CCATTCTTTCAGGACTTGCAC |
| SFRS8 | GGCGGATCTCACTACAGCTC | AGACCGAGGAGGACTTGGAT |
| SFRS9 | ACAATGCAGTGCGGCTGA | TTCTGAGCACAAAGCAGCTC |
| SFRS12 | ACGGGAAAAAGAGCATGAGA | TCCAACTTCTCCTTCCCATC |
| SFRS17A | GCTTCTCCGACATCCTGAAG | CCTTCTCTCTGCGCTTTTGT |
| NR3C1 (GRα) | TACCCTGCATGTACGACCAA | TCATCCAGCCAACTGTGAAA |
| NR3C1 (GRβ) | CTTCCAGAACCATGGTAGCC | TACGAAACTCCACCCAAAGG |
| FN1 (fibronectin) | ACCAACCTACGGATGACTCG | GCTCATCATCTGGCCATTTT |
| MYOC (myocilin) | CTGGAAACCCAAACCAGAGA | CATTGGCGACTGACTGCTTA |
| GAPDH | GGGAGCCAAAAGGGTCAT | TTCTAGACGGCAGGTCAGGT |
| ACTA1 (β-actin) | GGCATGGAGTCCTGTGG | GAAGCATTTGCGGTGG |
Quantitative Real Time PCR
Real-time PCR was performed as described previously.41 Briefly, 2.5 μL of cDNA was used in a reaction consisting of 1.5 units per reaction of antibody-bound Taq enzyme (Jump Start; Sigma-Aldrich, St. Louis, MO), 10x PCR buffer, 1.5 mM MgCl2, 200 nM dNTP mix, 100 nM respective primers (Table 2), 2.5 μL green nucleic acid dye (EvaGreen; Biotium, Hayward, CA), as well as 30 nM passive reference dye (Rox; USB, Cleveland, OH) per 50-μL reaction. PCR was performed on a real-time thermal cycler (model Mx3000p; Stratagene, La Jolla, CA), with cycling parameters of initial denaturation at 95°C; 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 60 seconds, and a denaturation cycle for creation of dissociation curves. Reactions for each sample and gene of interest were run in duplicate, cycle thresholds (Ct) were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression as a housekeeping gene, and comparative quantitation was performed using software (MxPro 4.0; Stratagene). Only individual PCR samples with single-peak dissociation curves were selected for data analysis.
Protein Extraction and Western Blot (WB) Analysis
Total cellular protein was extracted from the TM cells using mammalian protein extraction buffer (MPER, Cat # 78,501; Pierce Biotechnology, Rockford, IL) containing protease inhibitor (Cat. # 78415; Pierce Biotechnology) and phosphatase inhibitor (Cat. # 78420; Pierce Biotechnology) cocktails. Protein concentration was determined using a protein assay system (Bio-Rad Dc, Cat. # 500-0111; Bio-Rad Laboratories, Richmond, CA). The cellular proteins were separated on denaturing polyacrylamide gels and then transferred to polyvinylidene fluoride (PVDF) membranes by electrophoresis. Blots were blocked with 10% fat-free dry milk in tris-buffered saline Tween buffer (TBST) for 1 hour and then incubated overnight with primary antibodies (Table 3). The membranes were washed with TBST and processed with corresponding horseradish peroxidase-conjugated secondary antibodies. The proteins were then visualized in an imager (Fluor ChemTM 8900; Alpha Innotech, San Leandro, CA) using enhanced chemiluminescence (ECL) detection reagent (SuperSignal West Femto Maximum Sensitivity Substrate, Cat. # 34096; Pierce Biotechnology). To ensure equal protein loading, the same blot was subsequently reprobed for β-actin or GAPDH expression.
Immunocytochemistry (ICC) of TM Cells
TM cells from 5 different primary cell strains were grown on glass coverslips in 24-well plates. At 100% confluency, cells were fixed with 3.5% formaldehyde (Fisher Scientific, Pittsburgh, PA) in PBS for 20 minutes. Cells were treated with 0.2% Triton X-100 in PBS for 20 minutes, and then incubated for 1 hour in a blocking solution (SuperBlock blocking buffer; Cat. # 375717; Thermo Scientific). Cells were then incubated with primary antibodies (Table 3) overnight at 4°C and secondary antibodies at 1:200 in 1x PBS for 1 hour at room temperature. Negative controls consisted of omission of primary antibody. To visualize nuclei, cells were treated with 300 nM DAPI nuclear stain and mounted (Aqua-Mount; Lerner Laboratories, Pittsburgh, PA). Slides were stored in the dark at 4°C until visualized on a confocal imaging system (Zeiss 410; Carl Zeiss, Thornwood, NY).
ELISA Immunoassay for FN
Conditioned medium was obtained from three TM cell lines after 24 to 72 hours of 1 μM bombesin treatment with or without 100 nM DEX and centrifuged at 2000 rpm to remove cellular debris. Fifty microliters of conditioned medium was diluted to 150 μL with dilution buffer, and soluble FN was quantified using a commercially available ELISA kit (cat no. ECM 300; Chemicon International, Temecula, CA). Amounts of soluble FN (ng/mL) were plotted for each treatment using software (Graph-Pad Prism 5, La Jolla, CA).
GRE-Luciferase Reporter Assays
In a 96-well opaque plate (BD, Falcon, NJ), 2 × 104 GTM3 cells were transfected with 100 ng GRE reporter plasmid (Cignal CCS-006L; SA Biosciences) and 0.6 μL transfection reagent (Surefect; SA Biosciences) with or without 100 ng empty vector/pCMV6-XL5, SFRS3/pCMV6-XL5, SFRS5/pCMV6-XL5, or SFRS9/pCMV6-XL5 vectors or 100 nM siRNA against SFRS 3, 5, or 9 (Dharmacon, Lafayette, CO). For experiments to study the effect of bombesin on DEX activity, cells transfected with GRE reporter plasmid were treated with or without different doses of bombesin for 24 hours. Forty-eight hours after transfection, cells were treated with or without 100 nM DEX (in ethanol) in DMEM (Invitrogen) containing 10% fetal bovine serum (Invitrogen), 1% penicillin + streptomycin and 2 mM glutamine (Thermoscientific, Rockford, IL). Six hours later, substrate (Dual-Glow; Promega, San Luis Obispo, CA) was added to each well, and the signal was detected with a plate reader (M200; Tecan, Durham, NC). Firefly luciferase activity was normalized to renilla luciferase activity. Experiments were performed in quintuplets (n = 5).
Statistical Analysis
For comparing results between two groups, Student's t-test was performed. For comparison of results between more than two groups, one-way ANOVA was employed. Statistical tests used for each individual experiment are listed in the respective figure legends.
Results
Profiling of SRps in TM cells
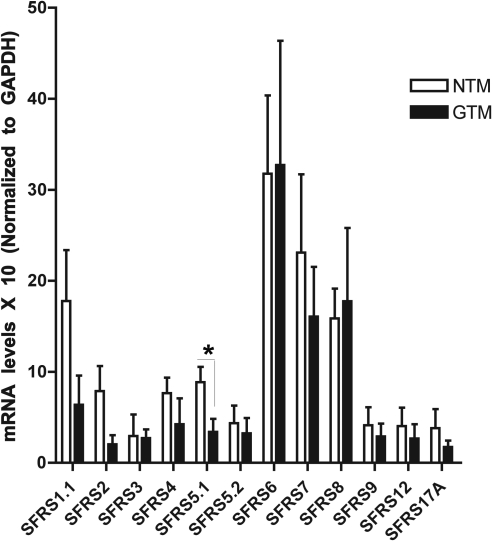
The expression of different members of the SR protein family in the TM has not been previously studied. Therefore, we determined whether these SRps are expressed in cultured human TM cells. Using quantitative RT-PCR (qRT-PCR), we profiled the cDNA samples obtained from four normal and four glaucomatous TM cell strains (Fig. 1). Numerous SRps were expressed in multiple TM cell strains. Although there appeared to be differences in their basal expression among the TM cell strains, SFRS5.1 (SRp40) expression was higher in NTM cells (P < 0.05). Both GRα and GRβ were expressed in all strains, and GRβ was higher in NTM cell strains, confirming our previous findings (data not shown).
Figure 1.
Expression of SRp genes in TM cells. qRT-PCR was done on RNA samples from four normal (NTM) and four glaucomatous TM (GTM) cell strains for 12 different SRp genes that are involved in constitutive and alternative splicing. GAPDH was used as a housekeeping gene. SFRS5.1 (SRp40) mRNA levels are significantly higher in NTM compared with GTM cells. Mean ± SEM; *P < 0.05 using unpaired t-test.
Bioinformatics Identification of Putative SRp Binding Sites
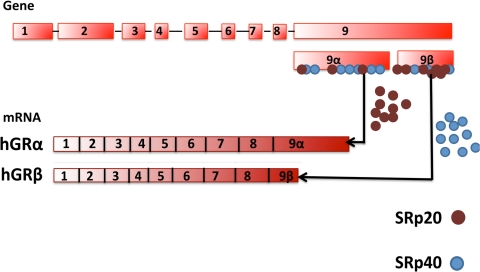
The bioinformatics software (Splicing Rainbow; Morais and Valcarcel EMBL-EBI; available at http://www.ebi.ac.uk/asd-srv/wb.cgi?method=8) predicted more binding sites for SRp20 on exon 9β compared with exon 9α. In contrast, SRp40 was found to have more binding sites on exon 9α compared with exon 9β (Fig. 2). Altered levels or function of these SRps might therefore affect GR mRNA slicing and thereby alter levels GRα and GRβ.
Figure 2.
Bioinformatics prediction of SRp binding. Exon 9α was predicted to have more binding sites for SRp40 (blue dots), while exon 9β had more binding sites for SRp20 (brown dots). More binding sites on exon 9α predicts elevated activity of SRp40 that would increase exon 9β/GRβ mRNA splicing. Similarly, more SRp20 binding sites on exon 9β predicted increased chances of exon 9α/GRα mRNA splicing.
Expression of SRp20, 30c, and 40 in TM Cells
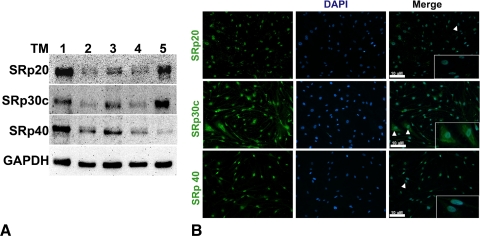
Given the mRNA expression of SRp20, 30c, and 40 (Fig. 1), we wanted to determine whether these SRps are expressed at the protein level in five TM cell strains. Our Western blot analysis results showed that SRp20, 40, and 30c proteins were expressed in the three normal and two glaucomatous TM cells tested (Fig. 3A). Immunocytochemistry analysis showed these SRps mainly present in nucleus colocalized with DAPI staining, although there also was some staining in cytoplasm, particularly for SRp30c (Fig. 3B). These results are in agreement with previous studies implicating SRp involvement in post mRNA processing events such as nuclear export of mRNA and translation process.42–45
Figure 3.
Protein expression of SRp20, 30c and 40 in TM cells. (A) Western blot analysis shows SRp20, SRp30c, and SRp40 protein expression in five TM cell strains with GAPDH as a loading control. (B) Immunocytochemistry data showing levels and localization of SRp20, SRp30c, and SRp40 in NTM-210–04 at magnification × 20. Insets show magnification × 40 images of cells marked by arrowheads. These are representative data from five different TM cell strains.
Effect of Bombesin on SRps Expression and the GRβ/GRα Ratio
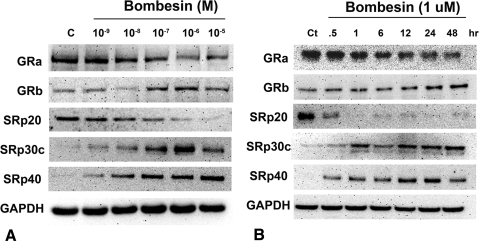
Bombesin has been shown to alter SRp30c levels and affect GR splicing in PC-3 cells.33 To study the effect of SRps on GR splicing in TM, we treated TM cell lines (n = 5) with increasing concentrations of bombesin (0–10 μM) for 24 hours. The protein expression of SRp20, SRp30c, SRp40, GRα, and GRβ was determined by Western blot analysis. Bombesin induced SRp30c and SRp40, decreased SRp20, and increased the GRβ/GRα ratio in a concentration-dependent manner (Fig. 4A). TM cell strains (n = 3) were treated with 1 μM bombesin for 0.5–48 hours to examine the time dependent effects on SRp expression and GR splicing (Fig. 4B). Bombesin induced SRp30c and SRp40 proteins maximally at 12 to 24 hours. The decrease in SRp20 was seen as soon as 30 minutes after bombesin treatment. Also, the maximum effect on GR splicing (i.e., highest GRβ/GRα ratio) was observed at approximately 24 hours. Results obtained were comparable among the normal and glaucomatous cell lines tested.
Figure 4.
Bombesin concentration- and time-dependent increases in SRp30c, SRp40, and GRβ/GRα ratio with decreases in SRp20. (A) Representative data showing dose-dependent induction of SRp30c, SRp40, and GRβ as well as decreased expression of SRp20 and GRα proteins by 0 to 10 μM bombesin in cultured TM cells (n = 5 TM cell strains). (B) Time course induction (0.5–48 hours) of SRp30c, SRp40, and GRβ proteins by 1 μM bombesin in cultured TM cells (n = 3 TM cell strains). Maximum induction was seen with 1 μM bombesin at 12 to 24 hours for SRp30c and SRp40 and 24 hours for GRβ.
Effect of Bombesin on DEX Responsiveness in TM Cells
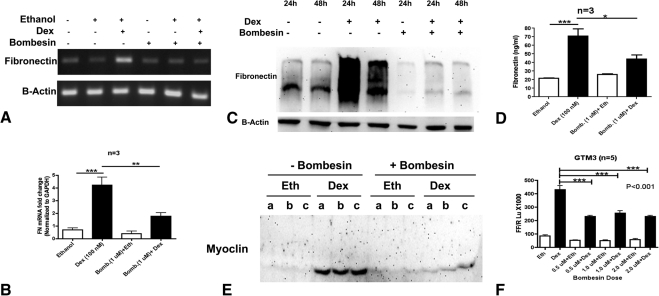
GTM3 cells were pretreated with or without bombesin (1 μM) for 24 hours before treatment with or without DEX (100 nM) for 12 hours. Total RNA was isolated for RT-PCR analysis. DEX treatment elevated fibronectin expression compared with untreated or bombesin only-treated samples. Pretreatment with bombesin (Fig. 5A for regular PCR and Fig. 5B for real time PCR) completely blocked the DEX- mediated fibronectin induction.
Figure 5.
Bombesin decreases DEX responsiveness in TM cells. An increased GRβ/GRα ratio by bombesin is associated with decreased DEX activity in TM cells. One- to 7-day treatment with DEX (100 nM) increased fibronectin mRNA (A) and (B), (n = 3), cell-associated fibronectin protein (C), (n = 8), and soluble fibronectin in conditioned medium (D), (n = 8) of cultured TM cells. Seven-day treatment of DEX (100 nM) also increased myocilin in conditioned medium of cultured TM cells (E) (n = 5); a, b, and c denote biological replicates. Bombesin pretreatment (1 μM) for 24 hours decreased DEX- mediated induction of FN mRNA (A, B), FN protein (C, D), and myocilin protein (E) levels. Bombesin pretreatment (0.5–2 μM) for 24 hours significantly decreased DEX-mediated GRE-luciferase reporter activity in GTM3 (F) (n = 5). Mean ± SEM, *P < 0.05, **P < 0.01, and ***P < 0.001 (one- way ANOVA).
We also treated TM cell strains (n = 8) with or without DEX (100 nM) for 1 to 7 days (1 day for transformed cell lines; 3 and 7 days for primary cell strains to check for DEX induction of fibronectin and myocilin, respectively), with or without 24 hours pretreatment with 1 μM bombesin. DEX elevated fibronectin and myocilin protein expression compared with untreated or vehicle-treated samples. Bombesin completely inhibited the DEX-mediated fibronectin induction in whole cell lysates (Fig. 5C, Western blot analysis) and in conditioned medium (Fig. 5D, ELISA). Bombesin alone appeared to affect cell-associated fibronectin (Fig. 5C), but not soluble fibronectin in conditioned medium (Fig. 5D). This suggests a role for some of these splicing factors, which are affected by bombesin treatment, in alternatively spliced and differentially distributed isoforms of fibronectin. Bombesin also inhibited the DEX induction of myocilin in TM cells (Fig. 5E).
In addition to these bombesin effects on the DEX induction of fibronectin and myocilin, we used a GRE-reporter assay to examine the effects of bombesin on DEX-induced GRE activity. GTM3 cells were transfected with a GRE-luciferase vector followed by pretreatment with 0, 0.5, 1, or 2 μM bombesin for 24 hours The cells were then treated with or without DEX (100 nM) and luciferase activity was determined 6 hours later. Consistent with the fibronectin and myocilin results, all three doses of bombesin significantly reduced DEX mediated GRE-luciferase activity compared with the untreated control (Fig. 5F). These results strongly suggest that bombesin affected SRp20, SRp30c, and SRp40 levels, which altered GR splicing. The increased GRβ/GRα ratio is associated with decreased DEX responsiveness in TM cells.
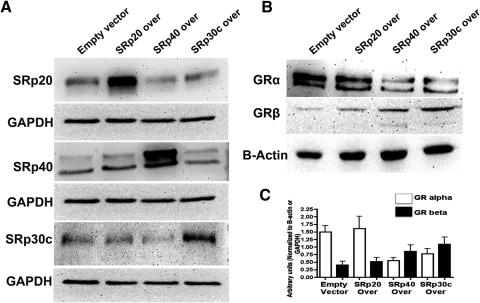
Effect of SRp Overexpression on GR Splicing
To study the direct role of SRps in alternative splicing of GR, we overexpressed different SRps in GTM3 cells using expression plasmids. The three different SRp expression plasmids increased the expression of their respective SRps (Fig. 6A), without affecting levels of the other two SRps tested (e.g., SRp20 overexpression did not affect SRp30c and SRp40 levels). SRp20 did not significantly change GRα and GRβ levels, therefore not changing the overall GRβ/GRα ratio (Figs. 6B and 6C). SRp30c and SRp40 overexpression resulted in higher GRβ and lower GRα levels compared with empty vector control (Figs. 6B and 6C), thereby increasing the GRβ/GRα ratio.
Figure 6.
SRp30c and SRp40 overexpression increases GRβ/GRα ratio in GTM3. (A) Western blot analysis confirming overexpression of SRp20, SRp30c, and SRp40 after transfection. (B) Representative Western immunoblot showing that SRp30c and SRp40 overexpression decreased GRα and increased GRβ levels whereas SRp20 overexpression did not significantly change GRα and GRβ levels in GTM3 cells (n = 3). (C) Densitometric analysis of data shown in (B); mean ± SEM.
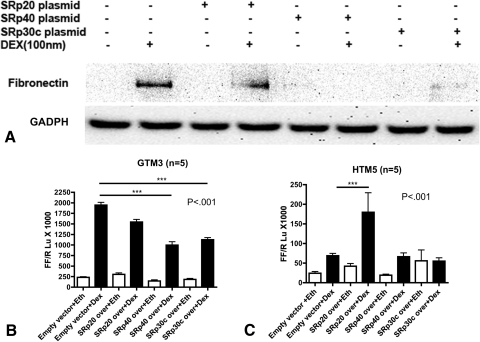
Effect of SRp30c and SRp40 Overexpression on DEX Activity
We examined the effect of SRp overexpression on GC activity by determining DEX induction of fibronectin protein levels in TM cells. GTM3 cells were transfected with SRp20, 30c, or 40 expression vectors followed by treatment with or without 100 nM DEX for 24 hours. DEX significantly increased fibronectin expression in the empty vector control compared with untreated samples. In contrast, SRp30 and SRp40 overexpression significantly blocked this DEX induction (P < 0.001) (Fig. 7A).
Figure 7.
SRp30c and SRp40 overexpression decreases DEX activity in TM cells. (A) SRp30c and SRp40 overexpression decreased DEX-mediated FN induction compared with empty vector control in GTM3 cells, corresponding to an increased GRβ/GR ratio. The Western immunoblot is representative of three experiments. (B, C) SRp40 and SRp30c overexpression significantly decreased DEX mediated GRE-luciferase activity compared with empty vector control (CT) in GTM3 cells (n = 5), whereas SRp20 overexpression significantly increased GRE-luciferase activity compared with empty vector CT in HTM5 (C, n = 5). Mean ± SEM, ***P < 0.001 (one-way ANOVA).
We also employed the GRE-luciferase reporter assay as an independent way to assess GC activity in TM cells. GTM3 and HTM5 cell lines (n = 5) overexpressing individual SRps were also transfected with the GRE promotor constructs followed by DEX (100 nM) or vehicle (0.1% ethanol) treatment for 6 hours. DEX increased luciferase activity four-fold in the empty vector control. SRp30c and SRp40 overexpression significantly reduced DEX-mediated luciferase activity compared with empty vector control in GTM3 cells (Fig. 7B). SRp20 overexpression modestly reduced the DEX activity, but this effect was not statistically significant. As expected, the basal induction of luciferase activity by DEX in HTM5 cells, which already have a high GRβ/GRα ratio, was not as high compared with GTM3 cells. However, SRp20 overexpression significantly increased DEX-mediated luciferase activity, whereas SRp30c and SRp40 did not have significant effects (Fig. 7C).
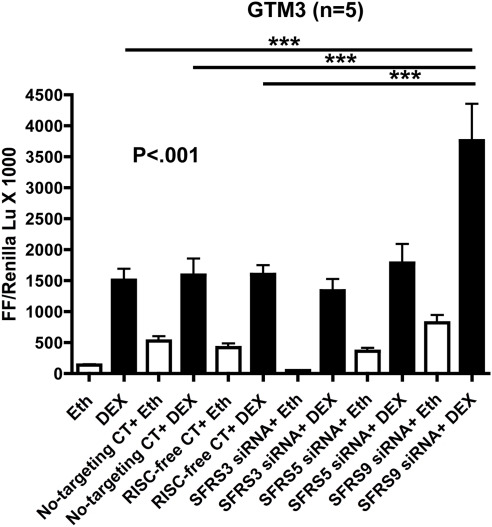
Effect of SRp Knockdown on DEX Activity
To complement our SRp overexpression results on DEX activity, we also knocked down expression of different SRps in GTM3 cells using specific SRp siRNAs. These siRNA-treated cells were cotransfected with GRE-promotor plasmids for 48 hours followed by 6 hours of treatment with or without DEX (100 nM). DEX increased luciferase activity in untransfected as well as in the nontargeting siRNA and RNAi-mediated silencing complex (RISC)-free siRNA controls (negative controls used to test for possible nonspecific siRNA effects). This increased activity was further enhanced with SRp30c knockdown (Fig. 8), which would alter the splicing of GR, favoring GRα expression. These results along with the overexpression data strongly suggest that SRp20, SRp30c, and SRp40 are involved in GR alternative splicing in TM cells, with the latter two increasing the GRβ/GRα ratio and thus increasing GC resistance.
Figure 8.
SRp30c knockdown increases DEX-activity in GTM3 cells. Six hours of 100 nM DEX treatment increases GRE-luciferase activity in untreated, nontargeting siRNA treated and RISC-free siRNA treated GTM3 cells compared with the corresponding vehicle (ethanol) controls (n = 5). SRp30c knockdown significantly augmented DEX activity when compared with no siRNA, no-targeting siRNA, and RISC-free siRNA-treated samples. Mean ± SEM; ***P < 0.001 (one-way ANOVA).
Discussion
The association between GC therapy, OHT, steroid glaucoma, and POAG has been known for >60 years.16,18 A number of laboratories have studied the effects of GCs on the TM to better understand steroid glaucoma and POAG because of the similarities in clinical phenotypes as well as the similar morphologic and biochemical changes in the TM.46 There is considerable heterogeneity in an individual's response to GCs, and approximately a third of the population develop GC-induced OHT compared with most POAG patients, who are steroid responders. We previously suggested that the molecular mechanism responsible for these differences in steroid responsiveness is due to differential expression of the GRβ isoform between normal and glaucoma TM cells.9,31 However, the mechanisms responsible for this differential expression of GRβ in TM cells had not been previously explored.
The relative levels of the two alternatively spliced isoforms of the GR, GRα and GRβ, regulate GC responsiveness. Increased expression of the dominant negative GRβ isoform has been associated with numerous steroid-resistant diseases16,22,24–27,30. We previously demonstrated lower levels of GRβ in GTM cells, making these cells more sensitive to GCs. Increased expression of GRβ made the TM cells more resistant to GCs.9,31
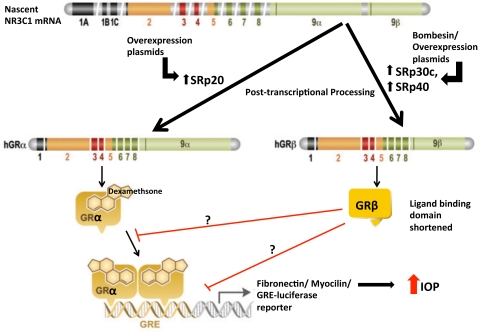
We have now shown that TM cells express a number of different SRp splicing proteins and that the three SRps tested are involved in the alternative splicing of GR in TM cells. It would be interesting to explore whether additional SRps expressed in the TM would also modify GR splicing. The peptide bombesin increased the expression of SRp30c and SRp40 as well as decreased SRp20 levels. Both SRp30c and SRp40 increased the expression of GRβ, whereas SRp20 did not alter the GRβ/GRα ratio. Increased expression of either SRp30c or SRp40 with SRp expression vectors decreased the DEX induction of FN and myocilin as well as blocked the DEX induction of a GRE-luciferase reporter gene. These results demonstrate one important mechanism regulating GR isoform expression and GC response in TM cells (Fig. 9).
Figure 9.
Modulation of levels of splicing factors either by overexpression plasmids or bombesin peptide alters GRβ/α ratio in TM cells. Increased GRβ/GRα ratio decreases GRα-mediated responses in presence of DEX. Increased GRβ/GRα ratio also decreases DEX-mediated extracellular matrix (ECM) changes in TM and elevated intraocular pressure in the eye. Adapted with permission from Revollo JR, Cidlowski JA. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann N Y Acad Sci. 2009;1179:167–178. © John Wiley & Sons, Inc.
There are emerging data that establish an association between altered expression of normal or mutant SR proteins and a number of human diseases.47–53 Involvement of SRp30c in alternative splicing of GR in neutrophils34 and of SRp40 in HeLa cells35 show the cell/tissue-specific effect of SR proteins. There was no significant difference of protein expression of SRp20 and SRp30c between normal and glaucomatous cell strains tested. However, expression of SFRS5.1 (SRp40) was significantly higher in NTM compared with GTM cells. This would favor the alternative splicing and expression of GRβ, making NTM cells more resistant to GCs, as we previously demonstrated.31 To confirm this, it would be necessary to perform large population-based studies and measure the mRNA levels of these SRps in normal, glaucomatous, and steroid responders. Alternatively, it might be the stoichiometry of SR proteins and/or their relative activities that regulate GR splicing in particular cell types.
There appeared to be a discrepancy on the effects of over- or underexpression of SRp40 in our GRE-luciferase assays. It is possible that SRp40 knockdown results in compensatory effects on other SR proteins. This possibility needs to be considered in our other experiments with bombesin or SRp overexpression, which are indirect measures of the DEX response executed by changes in GR levels. It is possible that changes associated with other components of GR signaling are involved in regulating the GC response.
In conclusion, this is the first report demonstrating the role of SR proteins in GR splicing in human TM cells. To best of our knowledge, no study had previously correlated the effect of GR splicing with functional GC response assays. In the future, translation of our in vitro results to an ex vivo anterior segment perfusion organ culture (POC) model and/or an in vivo model will help better understand the role of GR splicing in GC regulation of IOP.
Acknowledgments
The authors thank Raghu Krishnamoorthy for providing custom-made GR beta antibody, and John Fuller (presently at Johns Hopkins University) and Weiming Mao for help in various molecular biological techniques.
Footnotes
Supported by NEI Grant RO1 EY016242 (AFC, TY) and Alcon Research Limited (AFC).
Disclosure: A. Jain, None; R.J. Wordinger, None; T. Yorio, None; A.F. Clark, Alcon Research Limited (F)
References
- 1. Ray S, Mehra KS, Misra S, Singh R. Plasma cortisol in glaucoma. annals of ophthalmology. 1977;9:1151–1154 [PubMed] [Google Scholar]
- 2. Rozsival P, Hampl R, Obenberger J, Starka L, Rehak S. Aqueous humour and plasma cortisol levels in glaucoma and cataract patients. Curr Eye Res. 1981;1:391–396 [DOI] [PubMed] [Google Scholar]
- 3. Schwartz B, McCarty G, Rosner B. Increased plasma free cortisol in ocular hypertension and open angle glaucoma. Arch Ophthalmol. 1987;105:1060–1065 [DOI] [PubMed] [Google Scholar]
- 4. Southren AL, Gordon GG, Munnangi PR, et al. Altered cortisol metabolism in cells cultured from trabecular meshwork specimens obtained from patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 1983;24:1413–1417 [PubMed] [Google Scholar]
- 5. Southren AL, Gordon GG, Weinstein BI. Genetic defect in cortisol metabolism in primary open angle glaucoma. Trans Assoc Am Physicians. 1985;98:361–369 [PubMed] [Google Scholar]
- 6. Weinstein BI, Munnangi P, Gordon GG, Southren AL. Defects in cortisol-metabolizing enzymes in primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 1985;26:890–893 [PubMed] [Google Scholar]
- 7. Weinstein BI, Iyer RB, Binstock JM, et al. Decreased 3 alpha-hydroxysteroid dehydrogenase activity in peripheral blood lymphocytes from patients with primary open angle glaucoma. Exp Eye Res. 1996;62:39–45 [DOI] [PubMed] [Google Scholar]
- 8. Matsumoto Y, Johnson DH. Dexamethasone decreases phagocytosis by human trabecular meshwork cells in situ. Invest Ophthalmol Vis Sci. 1997;38:1902–1907 [PubMed] [Google Scholar]
- 9. Zhang X, Ognibene CM, Clark AF, Yorio T. Dexamethasone inhibition of trabecular meshwork cell phagocytosis and its modulation by glucocorticoid receptor beta. Exp Eye Res. 2007;84:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark AF, Wilson K, McCartney MD, Miggans ST, Kunkle M, Howe W. Glucocorticoid-induced formation of cross-linked actin networks in cultured human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 1994;35:281–294 [PubMed] [Google Scholar]
- 11. Clark AF, Brotchie D, Read AT, et al. Dexamethasone alters F-actin architecture and promotes cross-linked actin network formation in human trabecular meshwork tissue. Cell Motil Cytoskeleton. 2005;60:83–95 [DOI] [PubMed] [Google Scholar]
- 12. Clark AF, Miggans ST, Wilson K, Browder S, McCartney MD. Cytoskeletal changes in cultured human glaucoma trabecular meshwork cells. J Glaucoma. 1995;4:183–188 [PubMed] [Google Scholar]
- 13. Hoare MJ, Grierson I, Brotchie D, Pollock N, Cracknell K, Clark AF. Cross-linked actin networks (CLANs) in the trabecular meshwork of the normal and glaucomatous human eye in situ. Invest Ophthalmol Vis Sci. 2009;50:1255–1263 [DOI] [PubMed] [Google Scholar]
- 14. Nguyen TD, Chen P, Huang WD, Chen H, Johnson D, Polansky JR. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem. 1998;273:6341–6350 [DOI] [PubMed] [Google Scholar]
- 15. Polansky JR, Fauss DJ, Chen P, et al. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–139 [DOI] [PubMed] [Google Scholar]
- 16. Clark AF. Basic sciences in clinical glaucoma: steroids, ocular hypertension, and glaucoma. J Glaucoma. 1995;4:354–369 [PubMed] [Google Scholar]
- 17. Armaly MF, Becker B. Intraocular pressure response to topical corticosteroids. Fed Proc. 1965;24:1274–1278 [PubMed] [Google Scholar]
- 18. Wordinger RJ, Clark AF. Effects of glucocorticoids on the trabecular meshwork: towards a better understanding of glaucoma. Prog Retin Eye Res. 1999;18:629–667 [DOI] [PubMed] [Google Scholar]
- 19. Lewis JM, Priddy T, Judd J, et al. Intraocular pressure response to topical dexamethasone as a predictor for the development of primary open-angle glaucoma. Am J Ophthalmol. 1988;106:607–612 [DOI] [PubMed] [Google Scholar]
- 20. Kitazawa Y, Horie T. The prognosis of corticosteroid-responsive individuals. Arch Ophthalmol. 1981;99:819–823 [DOI] [PubMed] [Google Scholar]
- 21. Oakley RH, Sar M, Cidlowski JA. The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J Biol Chem. 1996;271:9550–9559 [DOI] [PubMed] [Google Scholar]
- 22. Hamid QA, Wenzel SE, Hauk PJ, et al. Increased glucocorticoid receptor beta in airway cells of glucocorticoid-insensitive asthma. Am J Respir Crit Care Med. 1999;159:1600–1604 [DOI] [PubMed] [Google Scholar]
- 23. Derijk RH, Schaaf MJ, Turner G, et al. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. J Rheumatol. 2001;28:2383–2388 [PubMed] [Google Scholar]
- 24. Honda M, Orii F, Ayabe T, et al. Expression of glucocorticoid receptor beta in lymphocytes of patients with glucocorticoid-resistant ulcerative colitis. Gastroenterology. 2000;118:859–866 [DOI] [PubMed] [Google Scholar]
- 25. Longui CA, Vottero A, Adamson PC, et al. Low glucocorticoid receptor alpha/beta ratio in T-cell lymphoblastic leukemia. Horm Metab Res. 2000;32:401–406 [DOI] [PubMed] [Google Scholar]
- 26. Hamilos DL, Leung DY, Muro S, et al. GRbeta expression in nasal polyp inflammatory cells and its relationship to the anti-inflammatory effects of intranasal fluticasone. J Allergy Clin Immunol. 2001;108:59–68 [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, Song L, Li B. [The expression of glucocorticoid receptor beta messenger RNA in peripheral white blood cells of hormone-resistant nephrotic syndrome patients]. Zhonghua Nei Ke Za Zhi. 2001;40:725–728 [PubMed] [Google Scholar]
- 28. Zhao YH, Zhou J, Li XX. [A study on the relationship between alpha- and beta-isoform of glucocorticoid receptors and glucocorticoid-resistant idiopathic thrombocytopenia purpura]. Zhonghua Nei Ke Za Zhi. 2005;44:363–365 [PubMed] [Google Scholar]
- 29. Xie YC, Jiao YJ, Xu XH, Wang H, Yin J. [The expression and clinical implications of glucocorticoid receptor isoforms in peripheral blood mononuclear cells in myasthenia gravis]. Zhonghua Nei Ke Za Zhi. 2007;46:56–59 [PubMed] [Google Scholar]
- 30. Hagg PM, Hurskainen T, Palatsi R, Ilves M, Oikarinen A. Increased expression of glucocorticoid receptor beta in lymphocytes of patients with severe atopic dermatitis unresponsive to topical corticosteroid. Br J Dermatol. 2010;162:318–324 [DOI] [PubMed] [Google Scholar]
- 31. Zhang X, Clark AF, Yorio T. Regulation of glucocorticoid responsiveness in glaucomatous trabecular meshwork cells by glucocorticoid receptor-beta. Invest Ophthalmol Vis Sci. 2005;46:4607–4616 [DOI] [PubMed] [Google Scholar]
- 32. Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu J, Gong JY, Goodman OB, Jr., Cartegni L, Nanus DM, Shen R. Bombesin attenuates pre-mRNA splicing of glucocorticoid receptor by regulating the expression of serine-arginine protein p30c (SRp30c) in prostate cancer cells. Biochim Biophys Acta. 2007;1773:1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Q, Leung DY, Kisich KO. Serine-arginine-rich protein p30 directs alternative splicing of glucocorticoid receptor pre-mRNA to glucocorticoid receptor beta in neutrophils. J Biol Chem. 2003;278:27112–27118 [DOI] [PubMed] [Google Scholar]
- 35. Yan XB, Tang CH, Huang Y, et al. Alternative splicing in exon 9 of glucocorticoid receptor pre-mRNA is regulated by SRp40. Mol Biol Rep. 2010;37:1427–1433 [DOI] [PubMed] [Google Scholar]
- 36. Fingert JH, Alward WL, Wang K, Yorio T, Clark AF. Assessment of SNPs associated with the human glucocorticoid receptor in primary open-angle glaucoma and steroid responders. Mol Vis. 2010;16:596–601 [PMC free article] [PubMed] [Google Scholar]
- 37. Wordinger RJ, Clark AF, Agarwal R, et al. Cultured human trabecular meshwork cells express functional growth factor receptors. Invest Ophthalmol Vis Sci. 1998;39:1575–1589 [PubMed] [Google Scholar]
- 38. Steely HT, Browder SL, Julian MB, Miggans ST, Wilson KL, Clark AF. The effects of dexamethasone on fibronectin expression in cultured human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 1992;33:2242–2250 [PubMed] [Google Scholar]
- 39. Wilson K, McCartney MD, Miggans ST, Clark AF. Dexamethasone induced ultrastructural changes in cultured human trabecular meshwork cells. Curr Eye Res. 1993;12:783–793 [DOI] [PubMed] [Google Scholar]
- 40. Pang IH, Shade DL, Clark AF, Steely HT, DeSantis L. Preliminary characterization of a transformed cell strain derived from human trabecular meshwork. Curr Eye Res. 1994;13:51–63 [DOI] [PubMed] [Google Scholar]
- 41. Wordinger RJ, Fleenor DL, Hellberg PE, et al. Effects of TGF-beta2, BMP-4, and gremlin in the trabecular meshwork: implications for glaucoma. Invest Ophthalmol Vis Sci. 2007;48:1191–1200 [DOI] [PubMed] [Google Scholar]
- 42. Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189 [DOI] [PubMed] [Google Scholar]
- 45. Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. Embo J. 2007;26:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clark AF, Wordinger RJ. The role of steroids in outflow resistance. Exp Eye Res. 2009;88:752–759 [DOI] [PubMed] [Google Scholar]
- 47. Watanuki T, Funato H, Uchida S, et al. Increased expression of splicing factor SRp20 mRNA in bipolar disorder patients. J Affect Disord. 2008;110:62–69 [DOI] [PubMed] [Google Scholar]
- 48. Ghigna C, Moroni M, Porta C, Riva S, Biamonti G. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 1998;58:5818–5824 [PubMed] [Google Scholar]
- 49. Fischer DC, Noack K, Runnebaum IB, et al. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–1090 [PubMed] [Google Scholar]
- 50. Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582 [DOI] [PubMed] [Google Scholar]
- 51. Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384 [DOI] [PubMed] [Google Scholar]
- 52. Dowling D, Nasr-Esfahani S, Tan CH, et al. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neugebauer KM, Merrill JT, Wener MH, Lahita RG, Roth MB. SR proteins are autoantigens in patients with systemic lupus erythematosus. Importance of phosphoepitopes. Arthritis Rheum. 2000;43:1768–1778 [DOI] [PubMed] [Google Scholar]