Abstract
The generation of disease-specific induced pluripotent stem cell (iPSC) lines from patients with incurable diseases is a promising approach for studying disease mechanisms and drug screening. Such innovation enables to obtain autologous cell sources in regenerative medicine. Herein, we report the generation and characterization of iPSCs from fibroblasts of patients with sporadic or familial diseases, including Parkinson's disease (PD), Alzheimer's disease (AD), juvenile-onset, type I diabetes mellitus (JDM), and Duchenne type muscular dystrophy (DMD), as well as from normal human fibroblasts (WT). As an example to modeling disease using disease-specific iPSCs, we also discuss the previously established childhood cerebral adrenoleukodystrophy (CCALD)- and adrenomyeloneuropathy (AMN)-iPSCs by our group. Through DNA fingerprinting analysis, the origins of generated disease-specific iPSC lines were identified. Each iPSC line exhibited an intense alkaline phosphatase activity, expression of pluripotent markers, and the potential to differentiate into all three embryonic germ layers: the ectoderm, endoderm, and mesoderm. Expression of endogenous pluripotent markers and downregulation of retrovirus-delivered transgenes [OCT4 (POU5F1), SOX2, KLF4, and c-MYC] were observed in the generated iPSCs. Collectively, our results demonstrated that disease-specific iPSC lines characteristically resembled hESC lines. Furthermore, we were able to differentiate PD-iPSCs, one of the disease-specific-iPSC lines we generated, into dopaminergic (DA) neurons, the cell type mostly affected by PD. These PD-specific DA neurons along with other examples of cell models derived from disease-specific iPSCs would provide a powerful platform for examining the pathophysiology of relevant diseases at the cellular and molecular levels and for developing new drugs and therapeutic regimens.
Keywords: drug evaluation, preclinical; drug screening; induced pluripotent stem cells; models, biological; tissue therapy
Introduction
Human embryonic stem cells (hESCs) are isolated from the inner cell mass of the developing blastocyst, indefinitely maintaining their capability for self-renewal and pluripotent nature (Thomson et al., 1998). By taking advantage of hESC characteristics, studies on early human development, drug development, and cell-based therapies for incurable diseases have become possible (Hattori and Fukuda, 2010). However, ethical issues have arisen, since hESCs are derived from pre-implantation human embryos (Robertson, 2001). Moreover, challenging obstacles associated with immune rejection after transplantation for cell-replacement therapy remained unresolved.
As a breakthrough discovery in regenerative medicine, adult human fibroblasts have been successfully reprogrammed via overexpression of pluripotency-related transcription factors, which include OCT4 (POU5F1), SOX2, KLF4, and c-MYC, to establish human induced pluripotent stem cells (hiPSCs) (Takahashi et al., 2007). Many studies have demonstrated close similarities between hiPSCs and hESCs at the molecular and genetic level, and in terms of their morphologies and functions (Lowry et al., 2008; Chin et al., 2010; Hawkins et al., 2010). Disease-specific iPSC lines also are accessible since they can be generated from patients with sporadic or genetically inherited diseases (Dimos et al., 2008; Park et al., 2008; Maehr et al., 2009; Soldner et al., 2009; Chamberlain et al., 2010; Urbach et al., 2010). The hESC-like characteristics allow them to be differentiated into various cell types in response to developmental cues. Recent reports have described that cell types related to the respective disorder are differentiated from disease-specific iPSCs and have been able to faithfully recapitulate disease phenotypes (Ebert et al., 2009; Ku et al., 2010; Itzhaki et al., 2011; Liu et al., 2011; Zhang et al., 2011). This landmark discovery of hiPSCs has opened a new window for developing human disease models in vitro in order to investigate pathophysiology and drug development (Marchetto et al., 2010; Rashid et al., 2010; Jang et al., 2011; Nguyen et al., 2011; Yazawa et al., 2011). Although the recent studies have shown substantial differences between hiPSCs and hESCs at the transcriptional, epigenetic, genetic and functional levels (Hanna et al., 2009; Ohi et al., 2011), the value of hiPSCs has been proven as a useful tool for in vitro disease modeling.
Herein, we report the generation and characterization of hiPSCs from fibroblasts of patients who have been previously diagnosed with sporadic or familial diseases including Parkinson's disease (PD), Alzheimer's disease (AD), juvenile-onset, type I diabetes mellitus (JDM), and Duchenne type muscular dystrophy (DMD), as well as from normal human fibroblasts (WT). The disease-specific hiPSCs can be differentiated into cell-types of their relevant diseases. Therefore, these disease-specific hiPSCs hold great potential for research and clinical purposes by providing a useful platform for disease modeling and drug screening. As recently reported, we have established childhood cerebral adrenoleukodystrophy (CCALD)- and adrenomyeloneuropathy (AMN)-iPSCs, and, as a proof-of-principle, modeled its diseases (Jang et al., 2011). In this study, we generated PD-specific DA neurons from the hiPSCs derived from fibroblasts of PD patients. The possibility of applying the PD-specific cell models to research and clinical purposes is currently under investigation.
Results and Discussion
Establishment and characterization of disease-specific hiPSC lines
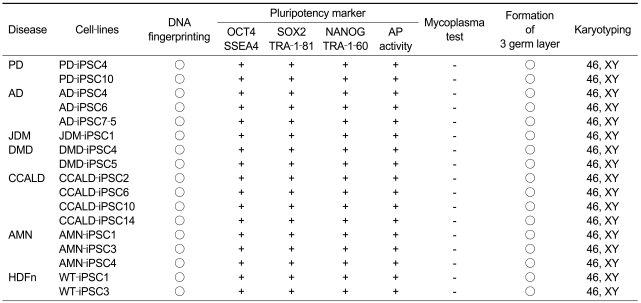
Patient-derived disease-specific fibroblasts and normal neonatal human dermal fibroblasts were reprogrammed by retroviral-mediated delivery of four defined factors (OCT4, SOX2, KLF4, and c-MYC) (Table 1). After transduction, the fibroblasts were cultured for about 4-5 weeks on a feeder layer until hESC-like colonies appeared. The colonies were picked mechanically and cultured on feeder cells for expansion. Since the early-passage hiPSCs showed a tendency of losing their stemness or spontaneous differentiation, we selected colonies that were passage 10 or more to establish stable hiPSC lines. These bona fide iPSC lines might be derived from either the fully reprogrammed cells or epigenetic adjustment of the early-passage reprogrammed cells during continued serial passages (Mikkelsen et al., 2008; Polo et al., 2010). Consistently, the previous report showed that extended passaging enabled the global gene expression pattern of hiPSCs similar to that of hESCs (Chin et al., 2009; Ohi et al., 2011).
Table 1.
Fibroblasts of patients with sporadic or familial diseases were used to generate iPSC lines
Generation of CCALD- and AMN-iPSCs was previously reported (Jang et al., 2011), and these cell lines are being stored with other cell lines and open to researchers as shown in Table 3.
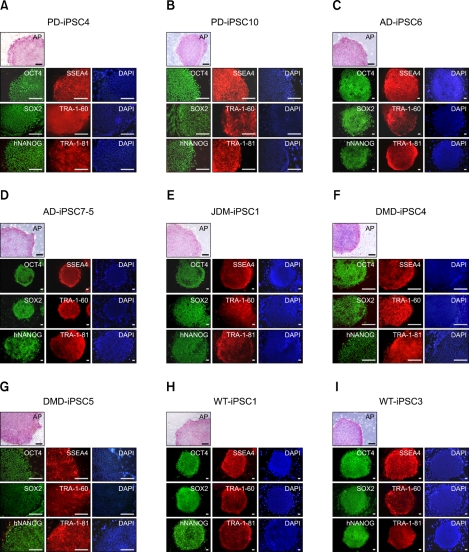
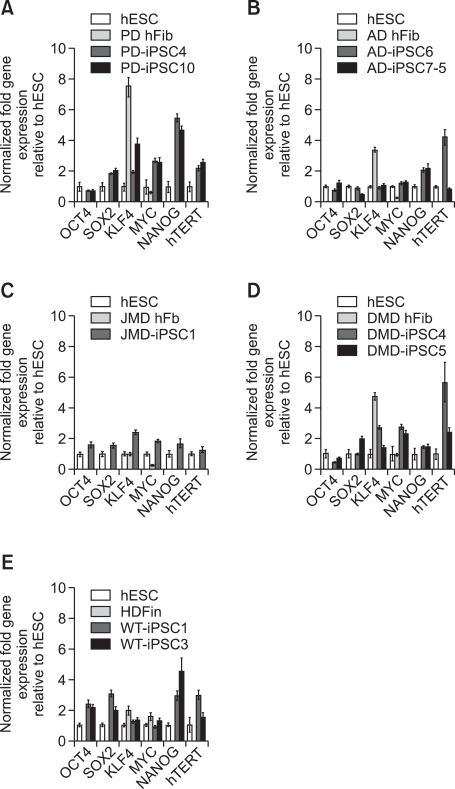
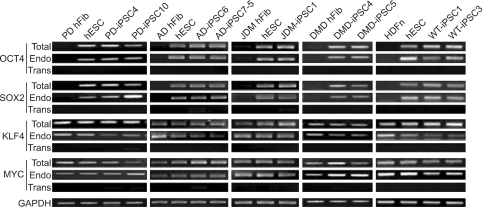
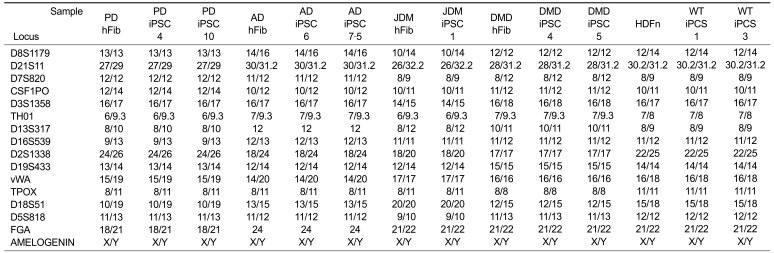
To confirm the presence of hESC-like characteristics, the expanded iPSC colonies were characterized at the molecular and genetic level; the colonies showed significant expression of multiple undifferentiation markers such as alkaline phosphatase (AP), OCT4, SOX2, NANOG, SSEA4, TRA-1-81, and TRA-1-60 (Figure 1). In addition, quantitative real-time PCR (qRT-PCR) analysis also demonstrated comparable level of expression of the well-known undifferentiation marker genes including OCT4, SOX2, KLF4, c-MYC, NANOG, and hTERT, as in H9-hESCs (Figure 2). In harmony with the previous observations, the exogenously introduced transgenes were silenced in iPSC lines while high expressions of pluripotency genes (OCT4, SOX2, KLF4, and c-MYC) were found from endogenous loci (Figure 3). Bisulfite genomic sequencing analyses showed that OCT4 promoter regions were hypomethylated in disease-specific hiPSCs relative to the parental fibroblasts (data not shown). DNA fingerprint analysis of short tandem repeats (STRs) revealed that each established hiPSC line was identical to its parental somatic cell line (Table 2). No aneuploidy was observed in all of the hiPSC lines (Figure 4). Although each hiPSC line is morphologically indistinguishable and meets the gold standard requirements for hiPSCs, the colonies vary in the extent of pluripotent marker expression and transgene silencing of hiPSCs, which was potentially caused by differential viral integration, culture conditions, passage number, and batch effects (Ohi et al., 2011). In addition, differences in origin, the epigenetic signatures and the genetic background of the patients' somatic cells used for reprogramming might also influence the variability in reprogramming status of the hiPSCs obtained (Mikkelsen et al., 2008; Kim et al., 2010a). For example, Raya et al. showed that somatic cells from patients with Franconi's anemia (FA) were refractory to reprogramming, making it difficult to model such condition in vitro, due to the genetic instability and apoptotic predisposition of FA cells. They successfully generated hiPSCs from patients with FA only after correction of FA-related gene (Raya et al., 2009).
Figure 1.
Both disease-specific and wild-type iPSC lines expressing pluripotent markers similar to hESCs. PD-iPSC4, PD-iPSC10, AD-iPSC6, AD-iPSC7-5, JDM-iPSC1, DMD-iPSC4, DMD-iPSC5, WT-iPSC1, and WT-iPSC3 were derived from patient-derived fibroblasts as well as normal neonatal human dermal fibroblasts as shown in Table 1. The derivation of CCALD- and AMN-iPSC lines has been previously reported (Jang et al., 2011). All established iPSC colonies exhibited hESC-like morphology. Moreover, the established iPSC lines showed alkaline phosphatase (AP) activity and expressed pluripotency markers. 4,6-Diamidino-2-phenylindole (DAPI) staining was used to determine the total cell content in the image. Scale bars = 200 µm.
Figure 2.
Pluripotency-associated genes in disease-specific cell lines show expressions similar to the levels of hESCs. Quantitative real-time PCR showed the total expression levels of reprogramming factors, OCT4, SOX2, KLF4, and c-MYC as well as the pluripotency-related factors NANOG and hTERT in hESCs (H9), the indicated parent fibroblasts (hFib), and iPSC lines. Distinct expressions similar to those of hESCs, but different from those of the parent fibroblasts were observed in most of the genes. However, expression levels of KLF4 were high even in most of the fibroblasts when compared to those of hESCs. H9 is a human ES cell line used as a positive control. PCR reactions were normalized against internal controls (β-actin) and plotted relative to expression levels in the hESC control. HDFn, normal neonatal human dermal fibroblasts.
Figure 3.
Analysis for the reactivation of endogenous pluripotency-related genes such as OCT4, SOX2, KLF4, and c-MYC in iPSC lines. The semi-quantitative RT-PCR analysis showed the relative expression levels of four pluripotency-promoting genes: OCT4, SOX2, KLF4, and c-MYC in PD-, AD-, JDM-, DMD-, WT-iPSCs, and their fibroblasts. Specific primers were used to determine the expression levels of endogenous (Endo), virally-delivered transgenes (Trans) and the total expression (Total) for each cell line. All the disease-specific iPSC lines showed reactivation of endogenous reprogramming transcription factors while the expressions of retrovirus-delivered transgenes were reduced to low or undetectable levels in the iPSC lines. hESCs and fibroblasts were used as positive and negative controls. GAPDH shown at the bottom was used as a loading control for each lane.
Table 2.
The genetic identity of disease-specific and wild-type iPSCs to their parental fibroblasts was confirmed by DNA fingerprinting analysis
DNA fingerprinting analysis performed at 16 independent loci of STR (short tandem repeats) confirmed that iPSC lines have identical genetic information to their parental fibroblasts.
Figure 4.
Disease-specific and wild-type iPSC lines have normal karyotypes. When chromosomal contents were analyzed with high resolution G-banding karyotypes, the cell lines, which included PD-iPSC4, PD-iPSC10, AD-iPSC6, AD-iPSC7-5, JDM-iPSC1, DMD-iPSC4, DMD-iPSC5, WT-iPSC1, and WT-iPSC3 at over 20 passages, indicated normal diploid chromosomal contents.
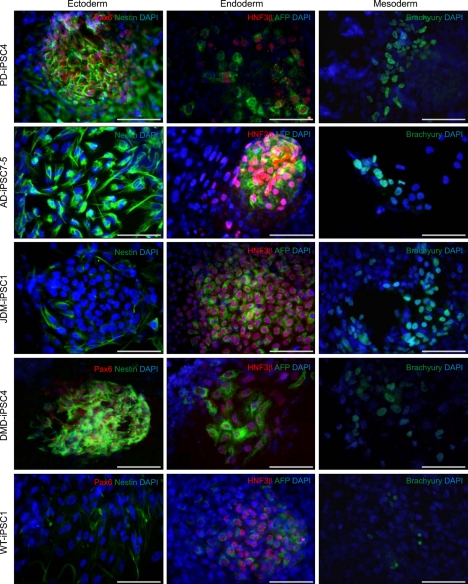
Human pluripotent stem cells including hiPSCs have the ability to differentiate into all three embryonic germ layers: ectoderm, endoderm, and mesoderm. It was reported that early-passage iPSC lines have the tendency of differentiate preferentially into the cell lineage of origin and continuous passaging abrogates these preferences in the differentiation potentials to established iPSCs (Chin et al., 2009; Polo et al., 2010). Using the previously described method (Kim et al., 2010b), embryoid bodies (EBs) were formed from the iPSC lines at over 10 passages in order to avoid preferential differentiation potentials, and then allowed to spontaneously differentiate into three germ layers. Ectodermal lineages were confirmed by expressions of Nestin and Pax6, while endodermal lineages were determined through expressions of HNF3β and AFP. Expressions of brachyury, on the other hand, confirmed mesodermal lineages of spontaneously differentiated iPSC lines (Figure 5).
Figure 5.
EB-based differentiation of iPSC lines reveals three lineage-specific marker expressions. Disease-specific and wild-type iPSC lines were spontaneously differentiated in hESC medium without bFGF for the formation of EBs. Two weeks later, EBs were attached on a gelatin coated dish and allowed to differentiate into all three germ layers for 10 days. Immunostaining was performed using antibodies specific to ectoderm (Nestin, green; Pax6, red), endoderm [AFP (α-fetoprotein), green; HNF3β, red], and mesoderm (Brachyury, green) lineages. Scale bars = 100 µm.
Taken together, these data indicate that primary fibroblasts from patients with sporadic or familial diseases were successfully reprogrammed into hiPSCs.
Differentiation of disease-specific hiPSCs into disease-relevant cells: disease modeling and platforms for new drug discovery
One of the most important aspects of hiPSC technology is modeling human diseases using patient-derived hiPSCs. Recapitulation of phenotypes of relevant diseases using disease-specific hiPSCs would provide a unique opportunity to study etiopathophysiology of the diseases and to develop new therapeutic regimens or drugs. To generate cell models of diseases, differentiation of disease-specific iPSCs into cell types relevant to the diseases is absolutely required.
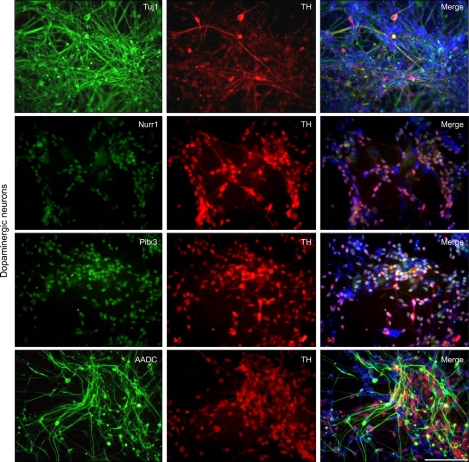
In an effort to establish cell models of PD, we sought to differentiate PD-iPSCs into DA neurons which are mostly affected in PD by introducing morphogens and neurotrophins (Cho et al., 2008). As previously reported (Kim et al., 2010b), BMP and Activin/Nodal signals were simultaneously inhibited by introduction of small molecules such as dorsomorphin (DM) and SB431542 (SB) during EB formation to obtain neural rosette structures. Using sonic hedgehog homolog (SHH) and fibroblast growth factor 8 (FGF 8), we regionalized PD-iPSC-derived neural rosette cells, and terminally differentiated them into DA neurons with GDNF, BDNF, TGF-β3, cAMP and ascorbic acid. After the terminal differentiation, Tuj1 (beta-tubulin class III; a microtubule element expressed exclusively in neurons), TH (tyrosine hydroxylase; a rate-limiting enzyme in the production of dopamine), Nurr1 (nuclear receptor related 1 protein; a key nuclear receptor protein implicated in the DA differentiation of the embryonic brain), Pitx3 (paired-like homeodomain 3, a essential transcription factor in the development and survival of A9 midbrain DA neurons), and AADC (aromatic-L-amino-acid decarboxylase; a key enzyme in the formation of biogenic trace amines such as DA and serotonin)-expressing cells were detected by immunostaining using antibodies against Tuj1, TH, Nurr1, Pitx3, and AADC (Figure 6). Our result indicated that the PD-iPSCs we generated retained potential to differentiate into DA neurons in vitro. The possibility of using the PD-DA neurons as a cell model for research and clinical purposes is under investigation.
Figure 6.
Differentiation of PD-iPSCs into DA neurons. The identity of DA neurons was confirmed via TH positive neurons co-expressing Tuj1, Nurr1, Pitx3, AADC. The total cell content in the merged image was represented via DAPI (blue) staining. Scale bar = 100 µm.
Many studies have recently demonstrated patient-specific iPSCs as an important source for cell models to study etiopathophysiology of several diseases. For example, a progressive degeneration of motor neurons was observed during differentiation from spinal muscular atrophy (SMA)-iPSCs (Ebert et al., 2009). Cardiomyocytes derived from LEOPARD syndrome iPSCs were found to reflect the hypertrophic features related to this disease (Carvajal-Vergara et al., 2010). Similarly, cardiomyocytes produced from Long QT and Timothy syndrome patient iPSCs had the prolonged duration of action potentials, as is seen in these patients (Moretti et al., 2010; Yazawa et al., 2011). RETT syndrome patient iPSCs reproduced the decreased number of glutamatergic synapses and calcium transients, as might be relevant to the clinical features of RETT (Marchetto et al., 2010). Familial dysautonomia iPSC-derived neural crest precursor cells exhibited reduced neurogenic differentiation and migration associated with these disease phenotypes (Lee et al., 2009). Smooth muscle cells differentiated from Hutchinson-Gilford progeria patient iPSCs gave rise to premature senescence, vascular aging defects seen in patients (Liu et al., 2011; Zhang et al., 2011). Importantly, the defects of neurons derived from SMA-, familial-dysautonomia-, and RETT-iPSCs were partially ameliorated by the treatment with small-molecule candidates or previously reported candidate drugs, further supporting the idea that disease specific-iPSCs would provide cell models for research and clinical purposes (Ebert et al., 2009; Lee et al., 2009; Marchetto et al., 2010).
Our group has recently established iPSCs from fibroblasts of patients with CCALD and AMN and used the cells to generate cell models to study X-linked Adrenoleukodystrophy (X-ALD) (Jang et al., 2011). Oligodendrocytes differentiated from CCALD-iPSCs displayed a significant accumulation of intracellular very long chain fatty acids (VLCFAs), recapitulating pathophenotypical characteristics observed in the patients with this disease. Furthermore, we also observed a decrease in the accumulated VLCFA by chemical treatment to induce expression of the closest homolog of the ABCD1 transporter, ABCD2. Therefore, the cell model derived from CCALD-iPSCs would be potentially of great use in screening or testing the efficacy of new drugs to treat patients suffering from CCALD.
Herein, we have shown the establishment of iPSCs from patients who were previously diagnosed with sporadic or familial diseases and the potential of iPSCs to differentiate into disease-relevant cells. The generated disease-specific iPSC lines were exclusively stored and maintained by the Stem Cell Bank in Korea and are available to all researchers who study the pathophysiologies of incurable diseases and drug discovery (Table 3). Recent studies regarding disease models developed from patient-derived iPSCs have shown the effectiveness of disease-specific iPSCs in terms of potentials in specialized cell differentiation, understanding human disease mechanisms and new drug screening. Although the field of iPSCs has developed rapidly in the past several years, the field remains relatively young and holds many questions to be answered.
Table 3.
List of iPSC lines in Korea Stem Cell Bank (http://koreastemcellbank.org/)
17 cell lines are being distributed for research purposes.
Methods
Culture of fibroblasts
Primary fibroblast of patients with Parkinson's disease (PD, AG20442), Alzheimer's disease (AD, AG07889), juvenile-onset, type I diabetes mellitus (JDM, GM01911), Duchene type muscular dystrophy (DMD, GM05128), and normal control fibroblasts (HDFn, SKU#C-004-5C) were purchased from Coriell and Invitrogen. Purchased primary fibroblasts were cultured under culture conditions recommended by the manufacturers.
Retroviral transduction and maintenance of iPSCs
A pMSCV vector containing a c-MYC human sequence and pMIG vectors containing OCT4, SOX2, and KLF4 human sequences were obtained from Addgene. 293GPG cells were transfected using retroviral vectors. A day after transfection, viral supernatants were collected for a 9 day-long period. Obtained supernatants were concentrated by ultracentrifugation. Primary skin fibroblasts were transduced for a day and on the 5th day post infection, infected cells were re-plated onto STO feeder cells. Re-plated cells were cultured in hESC medium including basic fibroblast growth factor (bFGF; 10 ng/ml) for 3-4 weeks. Apparent colonies on STO feeder cells were mechanically picked and transferred once every week. After 10 passages, the concentration of bFGF was changed to 4 ng/ml.
Alkaline phosphatase staining and immunocytochemistry
Alkaline phosphatase staining was performed following the manufacturer's recommended procedure (Sigma-Aldrich). Cells were fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked in 2% BSA in PBS for 1 h. Cells were then incubated with the appropriate primary antibodies against NANOG (R&D systems), OCT4 (Santa Cruz), SOX2 (Chemicon), SSEA4 (Chemicon), TRA-1-81 (Chemicon), TRA-1-60 (Chemicon), HNF3β (Santa Cruz), Brachyury (Santa Cruz), PAX6 (DSHB), Tuj1 (COVANCE), Nestin (Chemicon), TH (Pelfreez), Nurr1 (Millipore), Pitx3 (Millipore), and AADC (Millipore). Appropriate Alexa Fluor 488- or 568-labeled secondary antibodies (Molecular Probes) were treated for 1 h.
Gene expression analysis
Total RNA preparation was conducted for iPSCs, control hESCs, and fibroblasts using the Total RNA extraction kit (Intron Biotechnology) and converted to complementary DNA using the iScript™ cDNA Synthesis Kit (BIO-RAD). Semi-quantitative RT-PCR was performed with EmeraldAmp GT PCR Master Mix (Takara Bio Inc.) and quantitative RT-PCR was carried out using specific primers (Park et al., 2008). Quantitative RT-PCR was performed with SYBR premix Ex taq (Takara Bio Inc.) in the C1000™ thermal cycler (BIO-RAD).
DNA fingerprinting analysis
PCR based DNA fingerprint analysis of iPSCs were performed by Humanpass Inc. (Seoul, Korea) using a standard protocol for short tandem repeats (STR).
Karyotype analysis
Chromosomal studies were performed at GenDix, Inc. (Seoul, Korea) using standard protocols for GTG banding.
Differentiation of iPSCs into three-germ layer cell types
Disease-specific iPSCs were washed with PBS and 2 mg/ml collagenase was introduced for an hour. Detached colonies were collected, washed with EB medium (80% DMEM/F12, 20% KO serum replacement, 1 mM L-glutamine, 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 100 U/ml penicillin and 100 µg/ml streptomycin) and gently resuspended in EB medium with 5% FBS. EBs were formed by culturing colonies in suspension and the medium was changed every other day. Suspension culture was continued for a week and EBs were then attached onto Matrigel (BD) coated culture dishes. They were maintained for an additional 7 days for differentiation and the medium was changed every other day.
Differentiation of iPSCs into DA neurons
Differentiation of DA neurons was performed according to our established protocols with a slight modification (Cho et al., 2008; Kim et al., 2010b). First, iPSCs were washed with PBS and 2 mg/ml collagenase was introduced for an hour. Following our previously established protocol, EBs were cultured for 4 days in the presence of DM and SB (Kim et al., 2010b).
For differentiation to DA neurons, EBs were attached onto Matrigel-coated culture dishes and cultured for 5 days to generate neural rosette structures expressing neural stem cell markers such as Nestin and Pax6. Neural rosettes were isolated mechanically to obtain a high population of neural precursor cells (NPCs), and they were cultured on Matrigel-coated culture dishes for expansion. In order to induce DA neurons, NPCs were cultured in medium with SHH and FGF8 for 8 days and terminally differentiated in the presence of GDNF, BDNF, TGF-β3, cAMP and ascorbic acid in the culture medium for about 2 weeks.
Acknowledgements
This work was supported by grants from Korean Health Technology R&D Project, Ministry for Health, Welfare and Family Affairs (A100694).
Abbreviations
- AADC
aromatic-L-amino-acid decarboxylase
- ABCD
ATP-binding cassette, sub-family D
- AD
Alzheimer disease
- AFP
alpha-fetoprotein
- AMN
adrenomyeloneuropathy
- AP
alkaline phosphatase
- BDNF
brain-derived neurotrophic factor
- cAMP
cathelicidin antimicrobial peptide
- CCALD
childhood cerebral adrenoleukodystrophy
- DA
dopaminergic
- DM
dorsomorphin
- DMD
duchenne type muscular dystrophy
- EBs
embryoid bodies
- FA
Franconi's anemia
- GDNF
glial cell derived neurotrophic factor
- HDFn
normal neonatal human dermal fibroblast
- hESCs
human embryonic stem cells
- HNF3β
hepatocyte nuclear factor 3 beta
- hTERT
human telomerase reverse transcriptase
- iPSCs
induced pluripotent stem cells
- JDM
juvenile-onset, type I diabetes mellitus
- KLF4
Kruppel-like factor 4
- MYC
myelocytomatosis oncogene
- NANOG
Tir Na Nog (Nanog homeobox)
- NPCs
neural precursor cells
- Nurr1
nuclear receptor related 1 protein
- OCT4
octamer-binding protein 4
- OPCs
oligodendrocyte precursor cells
- Pax6
paired box 6
- PD
Parkinson's disease
- Pitx3
paired-like homeodomain 3
- SB
SB431542
- SHH
sonic hedgehog
- SMA
spinal muscular atrophy
- SNM
spherical neural mass
- SOX2
SRY (sex determining region Y)-box 2
- SSEA4
stage-specific embryonic antigen-1
- STR
short tandem repeat
- TH
tyrosine hydroxylase
- TRA-1-60
tumor-related antigen-1-60
- TRA-1-81
tumor-related antigen-1-81
- Tuj1
beta-tubulin class III
- VLCFA
very long chain fatty acid
References
- 1.Carvajal-Vergara X, Sevilla A, D'Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho MS, Hwang DY, Kim DW. Efficient derivation of functional dopaminergic neurons from human embryonic stem cells on a large scale. Nat Protoc. 2008;3:1888–1894. doi: 10.1038/nprot.2008.188. [DOI] [PubMed] [Google Scholar]
- 6.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 7.Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori F, Fukuda K. Strategies for ensuring that regenerative cardiomyocytes function properly and in cooperation with the host myocardium. Exp Mol Med. 2010;42:155–165. doi: 10.3858/emm.2010.42.3.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 12.Jang J, Kang HC, Kim HS, Kim JY, Huh YJ, Kim DS, Yoo JE, Lee JA, Lim B, Lee J, Yoon TM, Park IH, Hwang DY, Daley GQ, Kim DW. Induced pluripotent stem cell models from X-linked adrenoleukodystrophy patients. Ann Neurol. 2011;70:402–409. doi: 10.1002/ana.22486. [DOI] [PubMed] [Google Scholar]
- 13.Kim DS, Lee JS, Leem JW, Huh YJ, Kim JY, Kim HS, Park IH, Daley GQ, Hwang DY, Kim DW. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 2010b;6:270–281. doi: 10.1007/s12015-010-9138-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010a;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAA·TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, Studer L. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C, Thompson J, Boue S, Fung HL, Sancho-Martinez I, Zhang K, Yates J, 3rd, Izpisua Belmonte JC. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W, Palmer TD, Pera RR. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, Yu J, Hebrok M, Hochedlinger K, Costello JF, Song JS, Ramalho-Santos M. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raya A, Rodríguez-Pizá I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellá M, Río P, Sleep E, González F, Tiscornia G, Garreta E, Aasen T, Veiga A, Verma IM, Surrallés J, Bueren J, Izpisúa Belmonte JC. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson JA. Human embryonic stem cell research: ethical and legal issues. Nat Rev Genet. 2001;2:74–78. doi: 10.1038/35047594. [DOI] [PubMed] [Google Scholar]
- 30.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 33.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF, Stewart CL, Colman A. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]