Abstract
We aimed to identify whether there is any correlation between chromosomal/genetic changes, nuclear morphology and the histological grade of urothelial carcinomas of the urinary bladder. Morphometry and multicolour fluorescence in situ hybridisation (FISH) techniques were applied to 250 cells in five low-grade cases and 350 cells in seven high-grade cases of urothelial carcinoma. Compared with low-grade carcinomas, most high-grade cases showed larger and more variable nuclear size, more frequent polysomy of centromere enumeration probes (CEPs) 3, 7 and 17, and the loss of the 9p21 locus. The number of CEP signals in cells was increased as the nuclear area of the cells became larger. Cells with gains in two or more types of CEP had significantly larger nuclei than cells with normal FISH signal patterns. In conclusion, the present study indicates that there was a correlation between nuclear morphology and chromosomal/genetic changes which were related to histological grading. Thus, we show that differences in the chromosomal/genetic aberrations present in low- and high-grade tumours can affect not only nuclear morphology but also the histopathological and clinical behaviour of urothelial carcinomas.
Keywords: urothelial carcinoma, urinary bladder, chromosome, nuclear morphology, UroVysion
I. Introduction
Urothelial carcinoma is the most common malignancy of the urinary bladder [30]. The relative frequency of urothelial carcinoma in Japan is lower than in Western countries. However, in recent years, the incidence rate has increased and the disease accounts for more than 6,000 deaths per year in Japan [17]. According to the 2004 World Health Organization classification of tumours of the urinary system, papillary urothelial carcinomas are classified into four subgroups (low-grade or high-grade, and infiltrating or non-infiltrating) based on structural and cellular atypia or stromal invasion [4, 38]. These subtypes of urothelial carcinomas generally show divergent clinical behaviours. The low-grade type usually shows frequent recurrence but rare invasion with good prognosis, whereas the high-grade type is typified by stromal invasion with poor prognosis or recurrence with stromal invasion [25]. These different clinical behaviours necessitate different clinical treatments for each subgroup of urothelial carcinoma. Transurethral resection (TUR) is applied in low-grade cases, but in the high-grade type TUR is followed by Bacillus Calmette-Guerin (BCG) and/or chemotherapy. Moreover, in cases presenting muscular invasion (which tend to be high-grade tumours) total cystectomy is often also required [36].
Various molecular and immunohistochemical analyses for urothelial carcinomas have been documented [3, 16, 33, 44]. Studies using comparative genomic hybridisation (CGH) and loss-of-heterozygosity (LOH) analyses reported both chromosomal gains (1q, 5p, 8q, and 17q) and losses (2q, 5q, 8p, 9p, 9q, 10q, 11p, 18q and Y) [1, 5, 6, 10, 18, 19, 35, 45]. Furthermore, genetic anomalies or aberrant expression of genes such as Her2/neu, p53, p16INK4, cytokeratins and fibroblast growth factor receptor 3 (FGFR3) have been found to exist in urothelial carcinoma [2, 3, 6, 26, 42, 43]. Fluorescence in situ hybridisation (FISH) is a useful technique to detect abnormalities of chromosomes and genes in various tumours [29, 32]. UroVysion, a modification of the FISH technique for urothelial carcinomas, has been used successfully to assess aberrations in chromosomes 3, 7, and 17 by centromere enumeration probes (CEPs), and to detect deletions of p16 at the 9p21 locus [8]. UroVysion has been revealed as useful cytological diagnostic tool [13, 24, 40]. Halling et al. suggested that there was a strong correlation between nuclear size and the number of abnormal chromosomes present in the nucleus of urothelial carcinoma cells using UroVysion study [7]. However, combined analysis of FISH signals and nuclear morphology on a cell by cell basis has not been done.
Biological characteristics of carcinomas (e.g. invasiveness or potential for recurrence, prognosis and/or histopathological findings) are thought to be related to chromosomal or genetic aberrations, but a clear correlation has yet to be defined in carcinoma of other organs as well as urothelial carcinoma [22, 23, 28]. From a clinical viewpoint, accurate evaluation of cellular atypia is important for distinguishing low- and high-grade urothelial carcinomas [4, 36]. Some previous reports indicated chromosomal and genetic difference between low- and high-grade cases. However, there is no previous study which clarifies the relationship between nuclear morphology and chromosomal/genetic aberrations of urothelial carcinoma in each cell level [14]. In this study, we analysed chromosomal/genetic aberrations and nuclear morphology of the same urothelial carcinoma cells and clarified the correlation between chromosomal/genetic changes and cell nuclear atypia on a cell by cell basis.
II. Materials and Methods
Case selection and histological diagnosis
TUR or surgically-resected samples containing papillary urothelial carcinomas from 12 patients (10 male, two female; mean age at diagnosis was 67.5 years, range 46–79 years) were selected from the files of Saitama Medical University International Medical Center (Saitama, Japan). Histologic classification, grading and staging were performed in accordance with the 2004 World Health Organization classifications and TNM classification of Malignant Tumours seventh edition. Twelve papillary urothelial carcinomas were diagnosed (by histological grading and staging) as follows: five non-invasive low-grade types; two non-invasive high-grade types; and five invasive high-grade types, including four cases with lamina propria invasion (pT1) and one with muscular invasion (pT2). The four cases with stage of pT1 were divided two with focal invasion and two with diffuse invasion. Patient case summaries can be seen in Table 1. This study was carried on approval by IRB committee of International Medical Center Saitama Medical University (No. 08-070).
Table 1.
Case summary and results of Urovysion analysis
| Case # | Age (years) | Gender | Grade | pT category (Invasiveness) | CEP3,7,17 | 9p21 | 9p21/CEP9 | |
|---|---|---|---|---|---|---|---|---|
| Single signal | No signal | |||||||
| 1 | 63 | M | Low | pTa (non-invasive) | 11 | 4 | 0 | 12 |
| 2 | 60 | F | 1 | 0 | 0 | 0 | ||
| 3 | 77 | M | 2 | 8 | 0 | 3 | ||
| 4 | 64 | M | 0 | 5 | 0 | 4 | ||
| 5 | 79 | M | 2 | 14 | 32 | 46 | ||
| 6 | 60 | M | High | pTa (non-invasive) | 49 | 19 | 12 | 39 |
| 7 | 77 | M | 3 | 2 | 0 | 2 | ||
| 8 | 64 | M | pT1 (focally invasive) | 34 | 10 | 0 | 7 | |
| 9 | 78 | M | 15 | 19 | 22 | 25 | ||
| 10 | 67 | F | pT1 (diffusely invasive) | 46 | 30 | 8 | 37 | |
| 11 | 46 | M | 40 | 2 | 43 | 48 | ||
| 12 | 75 | M | pT2 (diffusely invasive) | 42 | 1 | 44 | 46 | |
In total, samples were used from 12 patients. Details of patient age and gender, and the grade and invasiveness of their carcinoma are shown. Cells were analysed from each case sample for a number of FISH staining characteristics, including the expression of chromosome enumeration probes (CEPs) 3, 7 and 17; loss of one or both 9p21 loci, and the ratio of cells expressing 9p21 and CEP9. Shown are the numbers of cells (out of 50 cells per sample) showing each type of aberration. Boxes with light and dark grey background indicate aberrant results based on criteria of Vysis. Boxes with dark grey backgrounds exhibit aberrant results with complete loss of 9p21.
Cell isolation
For our FISH study, we obtained single cells (with whole nuclei) isolated from formalin-fixed paraffin-embedded (FFPE) blocks of the tumour tissue. We took 50 µm-thick sections from FFPE blocks of each sample. After dewaxing and hydration in PBS (60°C, 24 hr), the thick sections were minced and incubated with proteinase K (Sigma, St. Louis, MO, USA), (38 µg/ml in PBS, 37°C, 10 min). After repeated pipetting to disperse cells, the samples were filtered and fixed with Carnoy’s fluid (methanol: acetic acid, 3:1). The isolated single cells in the samples were smeared on slide glass using by Hanabi Metaphase Spreader (Adstec, Chiba, Japan). The same procedures for fixation, preparation of FFPE block and cell isolation were also used to study nuclear morphology in each case.
Staining for multicolour FISH
For multicolour FISH, we used centromere enumeration probe (CEP) 9 (GSP, Kanagawa, Japan) together with the UroVysion (Abbott, Des Moines, IA, USA) containing VysisTM CEP probes for chromosomes 3, 7, 17 and a unique probe for the 9p21 locus of p16 gene (LSI p16 (9p21)). These probes are labelled with the Vysis fluorophores SpectrumOrange, SpectrumRed, SpectrumGreen, SpectrumAqua and SpectrumGold, respectively. CEP9 was used to detect cases in which the concurrent gain of chromosome 9 and loss of 9p21 resulted in an apparently unchanged appearance of 9p21. Multicolour FISH was performed according to the manufacturer’s instructions and recommendations. Briefly, the slides were incubated in 2× saline sodium citrate (SCC) at room temperature for 5 min, 0.2 N HCl for 20 min, then treated with Vysis pre-treatment solution kit (Abbott, Des Moines, IA, USA) following the manufacturer’s instructions. Finally, cells were fixed in buffered formalin for 5 min at room temperature. The slides were placed in 70%, 85% and 100% ethanol for 1 min each, and then dried by air. They were denatured in 70% formamide/2× SCC for 5 min at 72°C. The FISH probe mixture was denatured for 5 min at 72°C. Denatured slides were hybridised overnight at 37°C. The slides were washed in 2× SCC/0.3% NP40 for 2 min at 72°C. After nuclear counterstaining with 4,6-diamino-2-phenylindole dihydrochloride (DAPI), the numbers of FISH signals were counted under a Zeiss microscope (Carl Zeiss Japan, Tokyo, Japan).
Assessment of FISH signals and nuclear morphology
We analysed 50 tumour cells on each smear slide. Morphological parameters and FISH signals of the five probes were quantitatively measured for each cell using the Isis FISH Imaging system v5.1 (MetaSystems GmbH, Altlussheim, Germany). As morphological parameters, we measured nuclear area, maximum nuclear length and nuclear shape factor (NSF), which was calculated by the formula (major axis/2)2×3.14/area [21]. The NSF value of a perfectly round nucleus should be 1.0. To assess gain of chromosomes and loss of the 9p21 locus, FISH signals from the five probes were evaluated in the same 50 cells used for measurement of the nuclear morphological parameters. Normal cells have two FISH signals per probe. Gain of chromosomes and loss of 9p21 locus were defined based on criteria defined by Vysis [39]. Briefly, the cases including more than eight nuclei with two or more CEPs for chromosomes 3, 7, and 17 were defined as polysomy. Loss of 9p21 was defined as the cases exhibiting more than 24 cell nuclei with only one or no 9p21 signal. Ratio of 9p21 and CEP9 was calculated to detect those cases where gain of chromosome (CEP) 9 masked the concurrent loss of 9p21.
Combining FISH and morphometric analyses in low-grade and high-grade groups
The results of combined FISH and morphometric analyses in 250 cells in the low-grade group and 350 cells in the high-grade group were compared. The cells in both groups were divided to subgroups based on median values of nuclear size (113.6 µm2) and NSF (1.44). Namely, the cells in low- and high-grade groups were classified into ‘small’ (27.8 to 113.6 µm2) and ‘large’ (113.7 to 375.2 µm2) nuclear subgroups, and into ‘round’ (1 to 1.44) and ‘non-round’ (>1.44) nuclear subgroups. Finally, the mean number of FISH signals was compared between small versus large and round versus non-round nuclear subgroups in both low- and high-grade carcinoma cell samples.
Correlation between nuclear morphology and number of FISH signals
The 600 cells analysed as above were taken together and grouped into six groups according to their FISH signal characteristics. Normal (‘NM’) cells showed two signals for all probes. Cells exhibiting gain of CEPs were grouped into either ‘GoneCEP’ or ‘GmultiCEP’ groups if they contained a single or multiple CEP gain, respectively. ‘L9p21’ group cells were those missing the 9p21 locus but without gain of CEPs, while cells in the ‘GoneCEP & L9p21’ group had lost the 9p21 locus but with concurrent gain of a single CEP. Finally, the ‘GmultiCEPs & L9p21’ group contained those cells with missing 9p21 and concurrent gain of multiple CEPs. Nuclear morphometric parameters were statistically compared between NM and the other five groups, and the correlation between nuclear morphology and the number of FISH signals was analysed.
Statistical analysis
Statistical analysis was performed using two-sided Mann-Whitney U-test and Steel-Dwass test. A p value of less than 0.05 was considered statistically significant.
III. Results
Assessment of FISH signals in low-grade and high-grade cases
Representative images of normal and aberrant patterns of FISH signals are shown in Figure 1. Results of FISH signal analysis of each case are summarised in Table 1. Among five non-invasive low-grade urothelial carcinomas, one showed polysomy and another indicated loss of 9p21. In seven high-grade urothelial carcinomas, six revealed polysomy, and five indicated loss of 9p21. All five invasive high-grade cases showed polysomy. Complete loss of the 9p21 locus (manifested as the absence of any FISH signal for 9p21) was found in the cases with diffuse invasion. The aberrant 9p21/CEP9 ratios were consistent with the loss of 9p21.
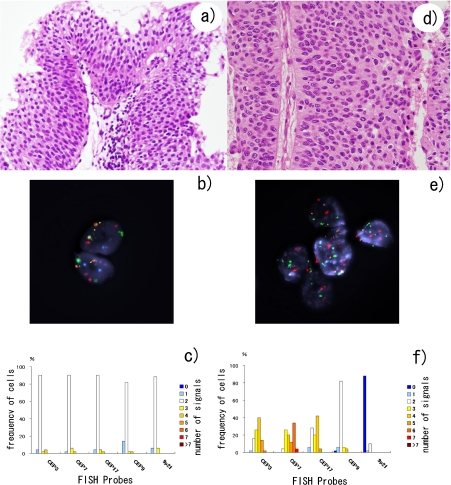
Fig. 1.
Representative results of multicolour FISH analyses of low- and high-grade urothelial carcinomas. a–c) Low-grade case (case no. 3); d–f) High-grade case (case no. 12); a, d) HE stain (original magnification ×40); b, e) Multicolour FISH stain (original magnification ×63); c, f) Frequency of cells with each number of FISH signals in that sample. Normal (two FISH signals from all five probes (b, c)) and abnormal (more than two CEP signals and loss of 9p21 signals (e, f)) patterns can be seen in these Figures.
Assessment of FISH signals and nuclear morphology in low-grade and high-grade groups
Table 2 shows the averages and standard deviation of number of CEPs and 9p21 signals, nuclear area, and nuclear shape factor based on our assessment of FISH signals and nuclear morphology of 250 and 350 cells from low- and high-grade cases, respectively. Figure 2 graphically indicates the results shown in Table 2. Figure 2a shows distribution of nuclear area in the cells in low- and high-grade groups. Nuclear area of the cells in the high-grade group (mean±SD: 136.9±65.1 µm2) was significantly larger than that in the low-grade group (mean±SD: 108.9±51.0 µm2) (Table 2, p<0.001). The cells in the high-grade group also had wider variation in nuclear size. Figure 2c reveals the distribution of NSF in the cells in low- and high-grade groups. There was no significant difference in the NSF of cells in low-grade (mean±SD: 1.67±0.74) and high-grade (mean±SD: 1.55±0.50) groups. The number of CEP signals was significantly greater in cells from the high-grade group (mean±SD: 9.8±3.1) than the low-grade group (mean±SD: 6.4±1.4) (Table 2, p<0.001), and the number of 9p21 signals was significantly lower (high-grade: mean±SD: 1.2±1.2; low-grade: mean±SD: 1.7±0.8) (Table 2, p<0.001).
Table 2.
Averages of numbers of FISH signals, nuclear area and nuclear shape factor
| Number of cells | Number of CEP signals | Number of 9p21 signals | Nuclear area (µm2) | NSF | |
|---|---|---|---|---|---|
| Low grade | 250 | 6.4±1.4 (6)* | 1.7±0.8 (2)* | 108.9±51.0 (101.2)* | 1.67±0.74 (1.48) |
| High grade | 350 | 9.8±3.1 (10)* | 1.2±1.2 (1)* | 136.9±65.1 (122.1)* | 1.55±0.50 (1.41) |
| Total | 600 | 8.3±3.0 (7) | 1.4±1.1 (2) | 125.2±61.2 (113.6) | 1.60±0.61 (1.44) |
Cells from low- and high-grade samples of urothelial carcinoma were analysed for the number of CEPs and 9p21 signals, their average nuclear area, and their average nuclear shape factor (NSF). Data are mean±SD (median in parentheses). Differences in these parameters between low- and high-grade samples were assessed for statistical significance using a two-sided Mann-Whitney U-test and Steel-Dwass test. * denotes p<0.001, which was considered statistically significant.
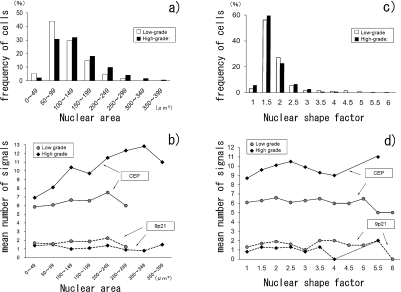
Fig. 2.
Nuclear area, nuclear shape factor (NSF) and mean number of FISH signals in 250 low-grade and 350 high-grade carcinoma cells. a) Distribution of nuclear area in low- and high-grade groups, b) Correlation of nuclear area and number of FISH signals (CEPs 3, 7, 17 and 9p21) per cell, c) Distribution of NSF in low- and high-grade groups, d) Correlation of NFS and number of FISH signals (CEPs 3, 7, 17 and 9p21) per cell. For comparison, mean numbers of CEP and 9p21 signals in normal cells are 6 and 2 per cell, respectively.
Combined analysis of FISH signals and nuclear morphology on a cell by cell basis in low-grade and high-grade groups
Figure 2b indicates the correlation between the mean number of FISH signals and the nuclear area in the cells from low- and high-grade carcinoma samples (total 250 and 350 cells, respectively). The number of CEP signals per cell was positively correlated to the nuclear area of the cells, and this tendency was more prominent in the high-grade group, as shown in Figure 2b. There was no apparent correlation between the number of CEP signals and the NSF (Fig. 2d).
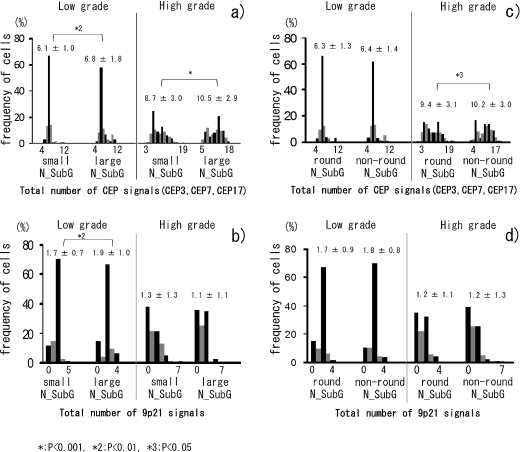
Figures 3a and 3b show comparisons of the numbers of FISH signals between the ‘small nucleus’ and ‘large nucleus’ subgroups of the low- and high-grade groups. In the low-grade group, there were 151 cells with small nuclei and 99 cells with large nuclei. In the high-grade group, 149 cells had small nuclei and 201 cells had large nuclei, indicating that high-grade tumours generally have larger nuclei. In both low- and high-grade groups, the number of CEP signals in the large nuclear subgroups (mean±SD: 6.8±1.8 (low grade) and 10.5±2.9 (high grade)) was significantly greater than in the small nuclear subgroups (mean±SD: 6.1±1.0 (low grade) and 8.7±3.0 (high grade)) (Fig. 3a). The number of 9p21 signals in the small nuclear subgroup (mean±SD: 1.7±0.7) was significantly less than that in large nuclear subgroup (mean±SD: 1.9±1.0) in the low-grade group, but was no different when comparing small and large nuclear subgroups in the high-grade group (Fig. 3b). Figures 3c and 3d show comparisons of the numbers of FISH signals in the round (112 (low grade); 188 (high grade)) and non-round (138 (low grade); 162 (high grade)) nuclear subgroups. The number of CEP and 9p21 signals were not significantly different between round and non-round nuclear subgroups, with the exception of CEPs in the round (mean±SD: 9.4±3.1) and non-round (mean±SD: 10.2±3.0) nuclear subgroups of the high-grade group (p<0.05, Fig. 3c and 3d).
Fig. 3.
a, b) Comparison (between small and large nuclear subgroups (N_SubG) in the low- and high-grade groups) of a) the distribution of the total number of CEP signals (CEPs 3, 7, 17) per cell, and b) the number of 9p21 signals per cell. c, d) Comparison (between round and non-round nuclear subgroups (N_SubG) in low- and high-grade groups) of c) the distribution of the total number of CEP signals (CEPs 3, 7, 17) per cell, and d) the number of 9p21 signals per cell. Value above columns is the mean±SD for each condition.
Correlation between nuclear morphology and number of FISH signals based on a cell by cell analysis
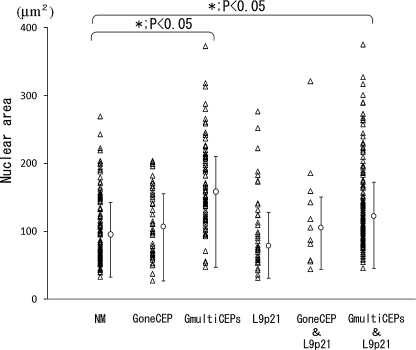
A total of 600 cells were divided into six groups according to the number of identifiable FISH signals, and these groups were analysed to determine whether there was any correlation between nuclear morphology and the number of FISH signals. Cells exhibiting gains in two or more kinds of CEPs (GmultiCEPs group, and GmultiCEPs & L9p21 group) had significantly larger nuclei than cells with normal FISH signal patterns (NM group) (p<0.05, Fig. 4). However, compared with NM group, cells having gain of only one kind of CEP or a loss of 9p21 showed no significant difference in nuclear area. There was no correlation between NFS and gain of chromosome/loss of 9p21.
Fig. 4.
Correlation between nuclear area and gain of chromosome or loss of 9p21. A total of 600 cells was grouped by the number of identifiable FISH signals. ‘NM’ denotes normal cells with two signals for each probe; ‘GoneCEP’ denotes cells with gain of one kind of chromosome; ‘GmultiCEPs’ group indicates cells with gains of two or more CEPs; ‘L9p21’ contains those cells exhibiting loss of 9p21 but without any gain of CEPs; ‘GoneCEP & L9p21’ denotes cells with both gain of one kind of CEP and loss of 9p21; and the ‘GmultiCEPs & L9p21’ group of cells exhibit a gain of two or more kinds of CEPs together with the loss of 9p21.
IV. Discussion
Urothelial carcinomas show diverse histological grading and stages associated with a variety of pathological and clinical behaviors [15]. Typically, low-grade cases with good prognosis rarely show invasion despite frequent recurrence, whereas high-grade cases with poor prognosis indicate stromal invasion or recurrence with invasion. These divergent behaviours may be explained by genetic alterations [19]. Previous studies have reported that urothelial carcinomas have diverse chromosomal and genetic abnormalities related with histopathological finding, grading and staging [1, 6, 10, 19, 24, 31, 43]. For instance, FGFR3 and p53 clearly play an important role in carcinogenesis of low-grade papillary urothelial carcinomas and high-grade urothelial carcinomas, respectively [35, 41, 43]. Thus, some pathologically and clinically interesting correlations between histopathological grading and chromosomal/genetic aberration have been suggested. However, there is no study which clarifies the relationship between nuclear morphology and chromosomal/genetic aberrations of urothelial carcinoma on a cell by cell basis.
The UroVysion multicolour FISH technique, which detects the most common chromosomal and genetic aberrations in urothelial carcinoma cells, has been a popular clinical application for detection of carcinoma cells in urine [4, 8, 40]. The present study indicates a strong correlation between histological grading and chromosomal/genetic aberration. Most high-grade cases showed both polysomy and loss of 9p21 locus, whereas most low-grade cases did not. The result is compatible with a previous study describing a positive correlation between FISH results and both tumour invasiveness and histological grade [40]. Moreover, two of three diffusely invasive high-grade cases indicated complete loss of the 9p21 locus. Since chromosomes 3, 7, and 17 include tumour growth factors such as RAS family, EGFR and c-erbB-2, and the 9p21 locus encodes p16 tumour suppressor factor, there is a possibility that the cell proliferation activity of urothelial carcinomas might affect their histological grading and invasive potential [26]. The findings of our study suggest that cytogenetic analysis by multicolour FISH can be a useful ancillary tool for diagnosis and surveillance of histological and clinical behaviours in urothelial carcinomas.
Generally, normal cells show round or non-round nuclei with smooth nuclear contours, whereas carcinoma cells have morphological abnormalities of the nucleus (i.e. nuclear atypia) represented by large nuclei and irregular nuclear contours [20]. This nuclear atypia is a characteristic feature of carcinoma cells and seems to be related to carcinogenesis [20, 22, 23, 37]. The general role of nuclear feature formation is not clear for either normal or carcinoma cells. Past reports have studied the nuclear morphogenesis of normal cells based mainly on nuclear envelope organisation and cytoskeletal features [11, 28, 34]. On the other hand, it is suggested that there is some correlation between nuclear atypia and chromosomal/genetic aberrations. However, few reports have studied the nuclear morphogenesis of carcinoma cells in relation to chromosomal or genomic aberrations on a cell by cell basis [9, 23]. The present study indicates several correlations in each cell level between chromosomal/genetic aberration and nuclear morphology. For instance, nuclear size in the high-grade group was significantly larger and more variable than that in low-grade group. Furthermore, the number of CEP signals was increased as the nuclear area of the cells grew larger (a trend that was more prominent in high-grade than low-grade samples), and the number of CEP signals was apparently related to nuclear size in both low- and high-grade cases. Namely a larger nucleus is linked to an increase in CEP signals. Finally, the number of 9p21 signals was also correlated to nuclear size, being lower in those cells with small nuclei than in those with larger nuclei, although only in low-grade cases. Interestingly, cells in the high-grade group not only had large nuclei but also many smaller nuclei and these small nuclei had prominent aberrations in CEP signals. Conversely, larger nuclei of cells in the low-grade group revealed less CEP aberrations, and thus the variation of nuclear size in the low-grade group may be due to cell cycle processes rather than chromosomal aberration [21]. There were no significant differences in NSF between low- and high-grade groups, and (with one exception) no correlation between the number of CEP or 9p21 signals and nuclear roundness. Thus, nuclear shape and chromosomal/genetic aberration appear not to be linked in the same way as nuclear area and chromosomal/genetic aberration. To analyse in more detail the correlation between nuclear shape and chromosomal/genetic aberration, further study using different NSFs may be required [21].
Interestingly, analysis based on a total of 600 tumour cells showed that those cells exhibiting gains of two or more kinds of CEPs had significantly larger nuclei than cells with normal FISH signal pattern. In contrast, cells gaining only one kind of CEP or losing 9p21 showed no significant difference in nuclear area. These results suggest that large changes in nuclear size depend on gross alterations at the chromosomal, rather than the genomic, level. This conclusion is supported by the observation that papillary thyroid carcinoma cells with only point mutation or transformation without any evident increase of chromosomes show negligible changes in nuclear morphology [12, 23, 27].
In conclusion, we show here that there is a correlation between chromosomal/genetic changes and nuclear morphology in each cell level. Differences in these chromosomal/genetic aberrations between low- and high-grade cases may affect the nuclear morphology as well as biological behaviour (invasivity or recurrence potential) of carcinomas and their prognosis.
V. Acknowledgments
This study was partly supported by SMU-FHMC Grant 09-01.
VI. References
- 1.Baffa R., Letko J., McClung C., LeNoir J., Vecchione A., Gomella L. G. Molecular genetics of bladder cancer: targets for diagnosis and therapy. J. Exp. Clin. Cancer Res. 2006;25:145–160. [PubMed] [Google Scholar]
- 2.Berggren de Verdier P. J., Kumar R., Adolfsson J., Larsson P., Norming U., Onelov E., Wijkstrom H., Steineck G., Hemminki K. Prognostic significance of homozygous deletions and multiple duplications at the CDKN2A (p16INK4a)/ARF (p14ARF) locus in urinary bladder cancer. Scand. J. Urol. Nephrol. 2006;40:363–369. doi: 10.1080/00365590600795396. [DOI] [PubMed] [Google Scholar]
- 3.Cina S. J., Lancaster-Weiss K. J., Lecksell K., Epstein J. I. Correlation of Ki-67 and p53 with the new World Health Organization/International Society of Urological Pathology Classification System for Urothelial Neoplasia. Arch. Pathol. Lab. Med. 2001;125:646–651. doi: 10.5858/2001-125-0646-COKAPW. [DOI] [PubMed] [Google Scholar]
- 4.Eble J. N., Sauter G., Epstein J. I., Sesterhenn I. A. ed. by S. L. Johansson, C. Busch and V. E. Reuter. IARC Press; Lyon: 2004. World-Health-Organization Tumours of the Urinary System and Male Genital Organs; pp. 115–118. [Google Scholar]
- 5.Florl A. R., Franke K. H., Niederacher D., Gerharz C. D., Seifert H. H., Schulz W. A. DNA methylation and the mechanisms of CDKN2A inactivation in transitional cell carcinoma of the urinary bladder. Lab. Invest. 2000;80:1513–1522. doi: 10.1038/labinvest.3780161. [DOI] [PubMed] [Google Scholar]
- 6.Florl A. R., Schulz W. A. Chromosomal instability in bladder cancer. Arch. Toxicol. 2008;82:173–182. doi: 10.1007/s00204-008-0280-3. [DOI] [PubMed] [Google Scholar]
- 7.Halling K. C., King W., Sokolova I. A., Meyer R. G., Burkhardt H. M., Halling A. C., Cheville J. C., Sebo T. J., Ramakumar S., Stewart C. S., Pankratz S., O’Kane D. J., Seelig S. A., Lieber M. M., Jenkins R. B. A comparison of cytology and fluorescence in situ hybridization for the detection of urothelial carcinoma. J. Urol. 2000;164:1768–1775. [PubMed] [Google Scholar]
- 8.Halling K. C., Kipp B. R. Bladder cancer detection using FISH (UroVysion assay) Adv. Anat. Pathol. 2008;15:279–286. doi: 10.1097/PAP.0b013e3181832320. [DOI] [PubMed] [Google Scholar]
- 9.Han J., Chang H., Giricz O., Lee G. Y., Baehner F. L., Gray J. W., Bissell M. J., Kenny P. A., Parvin B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput. Biol. 2010;6:e1000684 (1-12). doi: 10.1371/journal.pcbi.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim W. J., Bae S. C. Molecular biomarkers in urothelial bladder cancer. Cancer Sci. 2008;99:646–652. doi: 10.1111/j.1349-7006.2008.00735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King M. C., Drivas T. G., Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo T., Nakazawa T., Murata S., Kurebayashi J., Ezzat S., Asa S. L., Katoh R. Enhanced B-Raf protein expression is independent of V600E mutant status in thyroid carcinomas. Hum. Pathol. 2007;38:1810–1818. doi: 10.1016/j.humpath.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Krause F. S., Feil G., Zumbragel A., Bichler K. H. Molecular genetic methods in the diagnosis of invasive bladder cancer. Anticancer Res. 2000;20:5015–5021. [PubMed] [Google Scholar]
- 14.Lindgren D., Frigyesi A., Gudjonsson S., Sjodahl G., Hallden C., Chebil G., Veerla S., Ryden T., Mansson W., Liedberg F., Hoglund M. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Beltran A. Bladder cancer: clinical and pathological profile. Scand. J. Urol. Nephrol. Suppl. 2008:95–109. doi: 10.1080/03008880802325226. [DOI] [PubMed] [Google Scholar]
- 16.Mallofre C., Castillo M., Morente V., Sole M. Immunohistochemical expression of CK20, p53, and Ki-67 as objective markers of urothelial dysplasia. Mod. Pathol. 2003;16:187–191. doi: 10.1097/01.MP.0000056628.38714.5D. [DOI] [PubMed] [Google Scholar]
- 17.Ministry of Health L.a.W. ed. by Ministry of Health. L.a.W.; Tokyo: 2006. Vital Statistics of Japan; pp. 294–299. [Google Scholar]
- 18.Mitra A. P., Datar R. H., Cote R. J. Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J. Clin. Oncol. 2006;24:5552–5564. doi: 10.1200/JCO.2006.08.2073. [DOI] [PubMed] [Google Scholar]
- 19.Mitra A. P., Bartsch C. C., Cote R. J. Strategies for molecular expression profiling in bladder cancer. Cancer Metastasis Rev. 2009;28:317–326. doi: 10.1007/s10555-009-9196-5. [DOI] [PubMed] [Google Scholar]
- 20.Murata S., Mochizuki K., Nakazawa T., Kondo T., Nakamura N., Yamashita H., Urata Y., Ashihara T., Katoh R. Detection of underlying characteristics of nuclear chromatin patterns of thyroid tumor cells using texture and factor analyses. Cytometry. 2002;49:91–95. doi: 10.1002/cyto.10162. [DOI] [PubMed] [Google Scholar]
- 21.Murata S., Mochizuki K., Nakazawa T., Kondo T., Nakamura N., Yamashita H., Urata Y., Ashihara T., Katoh R. Morphological abstraction of thyroid tumor cell nuclei using morphometry with factor analysis. Microsc. Res. Tech. 2003;61:457–462. doi: 10.1002/jemt.10355. [DOI] [PubMed] [Google Scholar]
- 22.Murata S., Kishikawa T., Isojima Y., Tsuchihashi Y., Katoh R. Unusual cytologic findings in low grade papillary transitional cell carcinoma. Acta Cytol. 2004;48:492–496. doi: 10.1159/000326410. [DOI] [PubMed] [Google Scholar]
- 23.Murata S., Nakazawa T., Ohno N., Terada N., Iwashina M., Mochizuki K., Kondo T., Nakamura N., Yamane T., Iwasa S., Ohno S., Katoh R. Conservation and alteration of chromosome territory arrangements in thyroid carcinoma cell nuclei. Thyroid. 2007;17:489–496. doi: 10.1089/thy.2006.0328. [DOI] [PubMed] [Google Scholar]
- 24.Murata S., Iseki M., Kinjo M., Matsuzaki O., Moriuchi A., Ohtani H., Sakurai T., Satake T., Tsuzuki T. Molecular and immunohistologic analyses cannot reliably solve diagnostic variation of flat intraepithelial lesions of the urinary bladder. Am. J. Clin. Pathol. 2010;134:862–872. doi: 10.1309/AJCPACNUDWEN9GN4. [DOI] [PubMed] [Google Scholar]
- 25.Murphy W. M., Grignon D. J., Perlman E. J. In “AFIP Atlas of Tumor Pathology series 4. Tumors of the Kidney, Bladder, and Related Urinary Structures”, ed. by W. M. Murphy, D. J. Grignon and E. J. Perlman. AFIP; Washington DC: 2004. Tumors of the Urinary Bladder; pp. 255–275. [Google Scholar]
- 26.Nakazawa K., Murata S., Yuminamochi T., Ishii Y., Ohno S., Nakazawa T., Kondo T., Katoh R. p16(INK4a) expression analysis as an ancillary tool for cytologic diagnosis of urothelial carcinoma. Am. J. Clin. Pathol. 2009;132:776–784. doi: 10.1309/AJCP61KNVHJVHAFN. [DOI] [PubMed] [Google Scholar]
- 27.Nakazawa T., Murata S., Kondo T., Niu D., Mochizuki K., Kawasaki T., Yamane T., Nakamura N., Katoh R. RET/PTC rearrangements arising from a small population of papillary thyroid carcinoma cells, possible candidate for passenger mutation. Virchows Arch. 2009;455:35–41. doi: 10.1007/s00428-009-0789-8. [DOI] [PubMed] [Google Scholar]
- 28.Niu D., Murata S., Kondo T., Nakazawa T., Kawasaki T., Ma D., Yamane T., Nakamura N., Katoh R. Involvement of centrosomes in nuclear irregularity of thyroid carcinoma cells. Virchows Arch. 2009;455:149–157. doi: 10.1007/s00428-009-0802-2. [DOI] [PubMed] [Google Scholar]
- 29.Ooi A. Oncogene amplification detection by fluorescence in situ hybridization (FISH) Acta Histochem. Cytochem. 2001;34:391–397. [Google Scholar]
- 30.Parkin D. M. The global burden of urinary bladder cancer. Scand. J. Urol. Nephrol. Suppl. 2008:12–20. doi: 10.1080/03008880802285032. [DOI] [PubMed] [Google Scholar]
- 31.Pollard C., Smith S. C., Theodorescu D. Molecular genesis of non-muscle-invasive urothelial carcinoma (NMIUC) Expert Rev. Mol. Med. 2010;12:e10 (1-22). doi: 10.1017/S1462399410001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pothos A., Plastira K., Plastiras A., Vlachodimitropoulos D., Goutas N., Angelopoulou R. Comparison of chromogenic in situ hybridisation with fluorescence in situ hybridisation and immunohistochemistry for the assessment of her-2/neu oncogene in archival material of breast carcinoma. Acta Histochem. Cytochem. 2008;41:59–64. doi: 10.1267/ahc.07029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raica M., Zylis D., Cimpean A. M. Cytokeratin 20, 34betaE12 and overexpression of HER-2/neu in urine cytology as predictors of recurrences in superficial urothelial carcinoma. Rom. J. Morphol. Embryol. 2005;46:11–15. [PubMed] [Google Scholar]
- 34.Salpingidou G., Smertenko A., Hausmanowa-Petrucewicz I., Hussey P. J., Hutchison C. J. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulz W. A. Understanding urothelial carcinoma through cancer pathways. Int. J. Cancer. 2006;119:1513–1518. doi: 10.1002/ijc.21852. [DOI] [PubMed] [Google Scholar]
- 36.Sharma S., Ksheersagar P., Sharma P. Diagnosis and treatment of bladder cancer. Am. Fam. Physician. 2009;80:717–723. [PubMed] [Google Scholar]
- 37.Smith G. D., Bentz J. S. “FISHing” to detect urinary and other cancers: validation of an imaging system to aid in interpretation. Cancer Cytopathol. 2010;118:56–64. doi: 10.1002/cncy.20066. [DOI] [PubMed] [Google Scholar]
- 38.Sobin L. H., Gospodarowicz M. K., Wittekind C. In “TNM Classification of Malignant Tumours. 7th ed.”, ed. by M. K. Gospodarowicz. Wiley-Blackwell; London: 2009. Urinary Bladder; pp. 262–265. [Google Scholar]
- 39.Sokolova I. A., Halling K. C., Jenkins R. B., Burkhardt H. M., Meyer R. G., Seelig S. A., King W. The development of a multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J. Mol. Diagn. 2000;2:116–123. doi: 10.1016/S1525-1578(10)60625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song M. J., Lee H. M., Kim S. H. Clinical usefulness of fluorescence in situ hybridization for diagnosis and surveillance of bladder cancer. Cancer Genet. Cytogenet. 2010;198:144–150. doi: 10.1016/j.cancergencyto.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Sung M. T., Zhang S., Lopez-Beltran A., Montironi R., Wang M., Davidson D. D., Koch M. O., Cain M. P., Rink R. C., Cheng L. Urothelial carcinoma following augmentation cystoplasty: an aggressive variant with distinct clinicopathological characteristics and molecular genetic alterations. Histopathology. 2009;55:161–173. doi: 10.1111/j.1365-2559.2009.03363.x. [DOI] [PubMed] [Google Scholar]
- 42.Tomlinson D. C., Baldo O., Harnden P., Knowles M. A. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J. Pathol. 2007;213:91–98. doi: 10.1002/path.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Kwast T. H., Bapat B. Predicting favourable prognosis of urothelial carcinoma: gene expression and genome profiling. Curr. Opin. Urol. 2009;19:516–521. doi: 10.1097/MOU.0b013e32832eb45f. [DOI] [PubMed] [Google Scholar]
- 44.Varma M., Morgan M., Amin M. B., Wozniak S., Jasani B. High-molecular-weight cytokeratin antibody (clone 34betaE12) as a urothelial marker: a note of caution. Histopathology. 2004;44:189–190. doi: 10.1111/j.1365-2559.2004.01752.x. [DOI] [PubMed] [Google Scholar]
- 45.Yurakh A. O., Ramos D., Calabuig-Farinas S., Lopez-Guerrero J. A., Rubio J., Solsona E., Romanenko A. M., Vozianov A. F., Pellin A., Llombart-Bosch A. Molecular and immunohistochemical analysis of the prognostic value of cell-cycle regulators in urothelial neoplasms of the bladder. Eur. Urol. 2006;50:506–515. doi: 10.1016/j.eururo.2006.03.027. [DOI] [PubMed] [Google Scholar]