Abstract
The SecY/Sec61α family of membrane proteins are the central subunits of the putative protein translocation channel. We introduced random mutations into a segment of Escherichia coli SecY within its cytoplasmic domain 5, which was shown previously to be important for the SecA-dependent translocation activity. Mutations were classified into those retaining function and those gaining a dominant-interfering ability caused by a loss of function. These analyses showed that Arg-357, Pro-358, Gly-359, and Thr-362 are functionally important; Arg-357, conserved in almost all organisms, was identified as an indispensable residue.
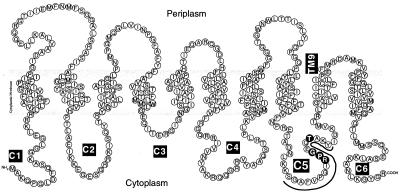
For proteins to make up a living cell, some of them must be translocated out of the cytosol. To allow transport of selected proteins, the cytoplasmic membrane in prokaryotic cells and the endoplasmic reticulum membrane in eukaryotic cells contain conserved heterotrimeric membrane protein complexes termed SecYEG and Sec61αβγ, respectively (1). They are thought to constitute a protein-translocating channel (2). SecY is the central subunit of the SecYEG complex (3) and is in close proximity of the translocating polypeptide chain (4). It interacts specifically with SecA to activate the translocation ATPase activity (5–7). SecY has 10 transmembrane segments with both termini facing the cytoplasm (Fig. 1). Among its six cytoplasmic domains, the fourth domain (C4) is important for the interaction with SecE (8), whereas the fifth and the most C-terminal cytosolic domains (C5 and C6) are important for translocation facilitation (9, 10). Thus, a number of cold-sensitive and export-defective mutations including secY39 (R357H) are known to affect the C5 domain (11). Consistent with the importance of C5, we isolated a dominant-negative mutation, secY−d1, with an internal deletion of 3 amino acids at the C5-transmembrane segment 9 junction, which produces an inactive but SecE-sequestering product (9). In vitro assays showed that inverted membrane vesicles from the C5 and C6 cold-sensitive mutants are defective in supporting the SecA translocation ATPase activities (10). Also known are a cold-sensitive mutation in C6 that does not allow the productive mode of SecA insertion reaction and suppressor mutations in secA against the defects of the C5/C6 mutations (refs. 7 and 12; unpublished data). Thus, the C5 and C6 regions are important for functional SecY-SecA interaction.
Figure 1.
Topology of SecY in the membrane and the target of mutagenesis. The cytoplasmic domains are indicated as C1–C6. The Ser-349 to Tyr-365 segment, shown by the wavy line, was subjected to the targeted random mutagenesis.
The preferential occurrence of sec mutations in C5 prompted us to investigate the importance of this region more systematically at the amino acid residue level. We thus undertook extensive random mutagenesis of a C5 segment and found that Arg-357 is particularly important for SecY activity.
Materials and Methods
Escherichi coli Strains.
Strains AD202 (MC4100, ompT∷kan), GN40 (AD202, leu82∷Tn10) and KI438 [CSH26, Δlac-pro secA-lacZ-f181 (λPR9)] were described previously (7, 9, 13). MR1 (AD202, secY39 leu82∷Tn10), NH178 (AD202, secA173 leu82∷Tn10), and NH193 (AD202, secA329 leu82∷Tn10) were constructed by appropriate P1 transductions. The secA173 (H545L) and the secA329 (I502S) mutations had been isolated as a suppressor against the secY205 defect (12) and the secY39 defect (unpublished data), respectively.
Plasmid Libraries with Targeted Random Mutations for a Ser-349/Tyr-365 Segment of SecY C5 Domain.
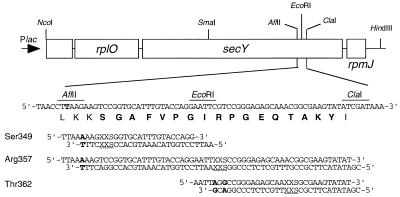
A secY plasmid named pHMC5A was constructed by introducing a unique AflII site at the position corresponding to Leu-346/Lys-347 without changing the coding (see Fig. 2) by site-directed mutagenesis. The parental plasmid had a NcoI–HindIII fragment (Fig. 2) from pCM10 (14) that was cloned into a variant of pTWV228 (11) having a NcoI site at the lacZα initiation codon. Thus, pHMC5A carried unique AflII, EcoRI, and ClaI sites in a segment of secY corresponding to Leu-346/Ile-366 (Fig. 2). To introduce directed random mutations, a series of degenerate oligonucleotides were synthesized chemically by using a mixture of four nucleotide precursors for the first and the second positions and a mixture of G and C precursors for the third position of each target codon of the Ser-349/Tyr-365 interval (Figs. 1 and 2). Two complementary chains were phosphorylated and annealed. They covered the AflII–EcoRI, AflII–ClaI, or EcoRI–ClaI segment, and they were used to substitute the corresponding portions of pHMC5A. The oligonucleotides additionally were designed to have their AflII ends or the EcoRI ends (in the case of the EcoRI–ClaI fragments) ligatable but not cleavable after ligation with the vector (Fig. 2). This design enabled us to eliminate, by a restriction enzyme digestion, the original plasmid molecules that escaped the restriction or reincorporated the original fragment. We thus constructed a series of plasmid mutant libraries in which the secY gene had random mutations at each specified codon of Ser-349 to Tyr-365. Random sampling of transformants and sequencing of the plasmids indicated that almost all of the plasmids had a nucleotide alteration(s) at the target sites.
Figure 2.
Plasmid and oligonucleotides used for targeted random mutagenesis of a C5 segment of SecY. (Upper) The chromosomal segment cloned into pHMC5A, which contains an engineered AflII site created by a site-directed nucleotide change, is shown. (Lower) Either the AflII–EcoRI, EcoRI–ClaI, or the AflII–ClaI region was replaced by oligonucleotides that had mutations at a particular secY codon. Three representative oligonucleotides (mutation targets for Ser-349, Arg-357, and Thr-362, respectively) are shown. X, either A, C, G, or T; S, G or C. The AflII and EcoRI recognition sequences at the upstream end of the oligonucleotides had been modified as shown in bold face.
Reconstitution of Translocase.
Wild-type SecA protein and SecA329 mutant protein were overproduced and purified as described (refs. 7 and 15; unpublished data). Plasmid pCM66 carried lacIq as well as a region encoding an N-terminally (His)6-tagged SecY, which was fused in-frame to the lacZα initiation codon of a pUC-based lac promoter vector (E. Matsuo, personal communication). The R357E mutation was introduced into it to yield pCM66(R357E). One of these plasmids and a pACYC184-based pTYE100 (T. Yoshihisa, personal communication) encoding (His)6-tagged SecE and SecG (expression level of SecG was low for unknown reasons) were introduced into strain AD202 lacking OmpT (13). Overproduction of SecYE and SecY(R357E)E were achieved by induction with 1 mM isopropyl-thio-β-D-thiogalactoside for 1 h. The SecYE complex was solubilized from a membrane fraction with 2.5% β-octylglucoside and purified by DEAE2 column chromatography. (His)6-tagged SecYE complexes were eluted earlier than the chromosomal product without the tag. The SecYE preparations were mixed with separately purified SecG and E. coli phospholipids for reconstitution of proteoliposomes essentially as described by van der Does et al. (16). The proteoliposomes were energized by valinomycin/K-acetate treatments (17) and subjected to proOmpA translocation reactions essentially as described (7).
Results
Functional Substitutions Identified Among SecY Mutant Libraries for the Ser-349/Tyr-365 Interval in Domain C5.
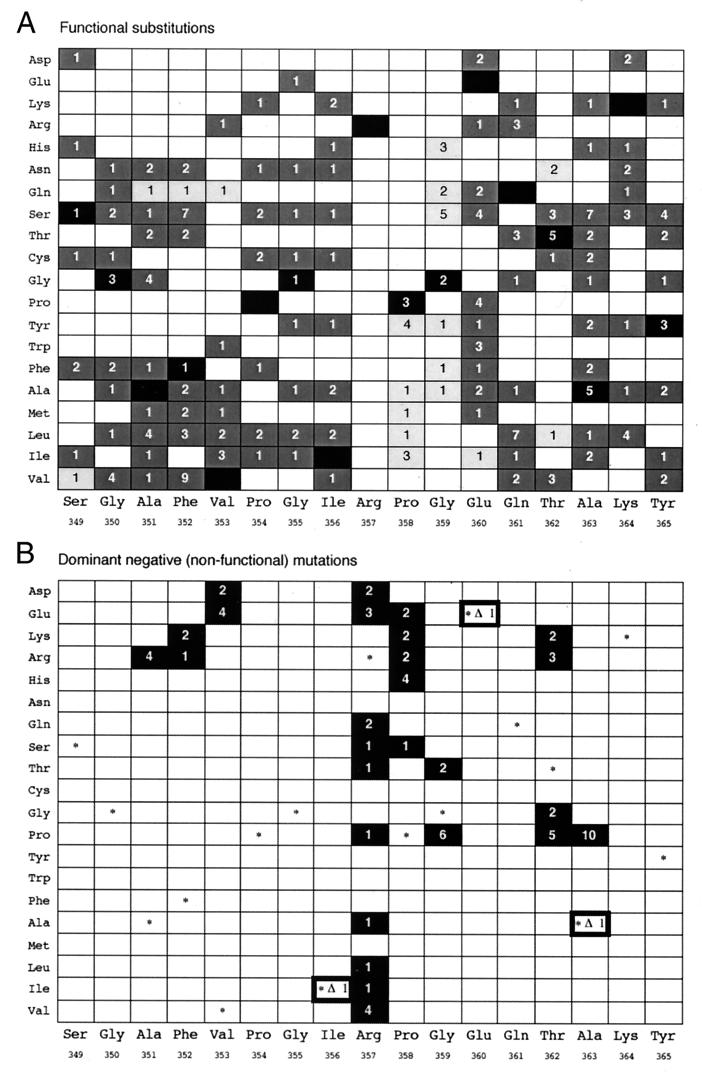
We constructed a series of plasmid mutant libraries in which secY had random mutations at each specified codon of a Ser-349/Tyr-365 segment in domain C5. To assess functional importance of each residue, mutations that did not impair the functionality of SecY significantly were screened first; each mutant library was introduced into the secY39(Cs) mutant (18), and transformants (20–239 colonies) were examined for their ability to grow at 20°C (Table 1). The frequencies of Cs+ transformants were 30% or more for most positions. Although a majority of the Cs+ cells showed nearly full growth (Fig. 3A, dark gray), some of them, especially those with alterations at residues 358, 359, and 362, exhibited only slow growth (Fig. 3A, light gray). It was noticed that complementation-positive clones were less than 10% at three consecutive residues of Arg-357, Pro-358, and Gly-359. Functional mutations were infrequent also (23%) at the Thr-362 position. Most notably, no complementing mutation was obtained at the Arg-357 position. Nucleotide sequencing identified 132 amino acid changes in SecY, among which 123 single amino acid changes are summarized in Fig. 3A. Some changes were obtained repeatedly (Fig. 3A) among the initial 600 clones that had been sequenced.
Table 1.
Targeted random mutagenesis of the Ser-349/Tyr-365 segment of SecY
| Target position | Screening for functional
mutations
|
Screening for dominant-negative mutations
|
||||
|---|---|---|---|---|---|---|
| Number examined | Number of Cs+ | Percentage of Cs+ | Number examined | Number of Sec− | Percentage of Sec− | |
| Ser-349 | 58 | 45 | 78 | 282 | 3 | 1 |
| Gly-350 | 42 | 16 | 40 | 340 | 3 | 1 |
| Ala-351 | 28 | 20 | 73 | 298 | 20 | 7 |
| Phe-352 | 64 | 30 | 48 | 358 | 17 | 5 |
| Val-353 | 50 | 19 | 38 | 266 | 7 | 3 |
| Pro-354 | 36 | 16 | 44 | 288 | 0 | 0 |
| Gly-355 | 20 | 19 | 95 | 863 | 42 | 5 |
| Ile-356 | 26 | 23 | 88 | 494 | 35 | 7 |
| Arg-357 | 22 | 0 | 0 | 612 | 415 | 68 |
| Pro-358 | 64 | 3 | 5 | 836 | 152 | 18 |
| Gly-359 | 34 | 2 | 6 | 1,018 | 111 | 11 |
| Glu-360 | 92 | 57 | 62 | 811 | 5 | 1 |
| Gln-361 | 80 | 24 | 30 | 791 | 7 | 1 |
| Thr-362 | 138 | 31 | 22 | 1,171 | 221 | 19 |
| Ala-363 | 239 | 174 | 73 | 355 | 17 | 5 |
| Lys-364 | 33 | 31 | 94 | 802 | 9 | 1 |
| Tyr-365 | 23 | 22 | 96 | 729 | 12 | 2 |
Figure 3.
Summary of the mutations obtained. (A) Functional substitutions. Shown at the bottom are the amino acid sequence of the target region (Ser-349 to Tyr-365) of SecY. Amino acid changes, specified at Left, are shown above each residue by dark gray boxes (which fully complemented secY39) and light gray boxes (which partially complemented secY39). Black boxes indicate the original amino acid. The numbers of the clones identified are shown in the boxes. (B) Dominant-negative (nonfunctional) mutations. Mutations that were obtained after screening for dominant-negative Sec phenotypes are summarized. Black boxes, substitutions; white boxes with thick frames and Δ, single residue deletions; *, the wild-type residue. The numbers of the clones identified are shown in the boxes.
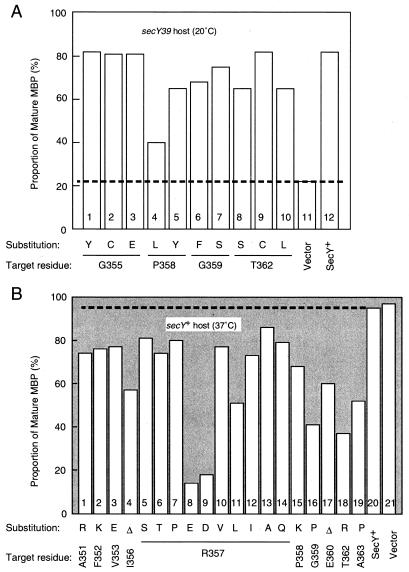
Export of maltose-binding protein (MBP) was examined by pulse-labeling the secY39 mutant cells carrying one of the mutant secY plasmids. Although only ≈20% of labeled MBP in this mutant was in the mature form at 20°C (Fig. 4A, column 11 and broken line), the secY mutant plasmids that fully complemented the growth defect allowed ≈80% processing (columns 1, 2, 3, and 9) as the wild-type secY plasmid did (column 12). Thus, the amino acid substitutions shown in dark gray in Fig. 3A have been assigned as compatible with the functioning of SecY. Partially complementing mutations (shown in light gray), which were common among mutations in residues 358, 359, and 362, gave lower processing efficiencies (40–70%; columns 4–8 and 10). The latter mutations were weakly dominant-negative when overexpressed in secY+ cells (data not shown). Generally, good correlation was observed between protein export proficiency and growth of the complemented secY39 cells.
Figure 4.
Complementing and interfering phenotypes of the secY mutations. (A) Complementation of the secY39 protein export defect. Strain MR1 (secY39) was transformed with a SecY mutant plasmid indicated at the bottom. Cells were grown with glycerol and maltose as carbon sources, first at 37°C and then at 20°C for 30 min, and pulse-labeled with [35S]methionine for 1 min (7). Labeled MBP was immunoprecipitated and proportions of radioactivities associated with the mature (presumably exported) form are quantitated by a PhosphorImager (7). The broken line shows the noncomplemented value. (B) Dominant interference. Strain AD202 (secY+) was transformed with two plasmids, pSTD343 (lacIq; Y. Akiyama, personal communication) and one of the dominant-negative SecY plasmids as specified at the bottom. Cells were grown at 37°C in the presence of 1 mM isopropyl-thio-β-D-thiogalactoside and 5 mM cAMP and pulse-labeled with [35S]methionine for 30 sec. The extents of MBP processing are shown as in A. The broken line shows the noninterfered value.
Nonfunctional and Dominant-Negative Substitutions.
A lack of function can result from any nonsense, deletion, and frame-shift mutations, which are not of our primary interests. Nonfunctionality caused by an extreme instability of the product is not informative either. We showed that loss-of-function mutations at the C-terminal region of SecY would be dominant-negative because they will still bind to SecE and sequester this partner molecule (8, 9, 11). To screen nonfunctional mutants, we introduced each plasmid mixture into a strain carrying a secA-lacZ fusion, which can report the protein export status of the cell by the β-galactosidase activity (19). We saved transformant colonies that developed blue color because of increased β-galactosidase-mediated hydrolysis of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (9). Thus we were able to avoid simple knocking-out and destabilizing mutations.
Blue-colored colonies were obtained at frequencies that were correlated inversely with those of the functional mutations (Table 1; compare Fig. 2 A and B). Thus, the dominant-negative mutations were frequent for the positions Arg-357, Pro-358, Gly-359, and Thr-362, at which functional mutations were rare. In particular, as many as 68% of the clones after mutagenesis of the Arg-357 position were of the nonfunctional/dominant-negative class. There were some variations in the extents of color development. We identified 27 single amino acid substitutions and three one-residue deletions (Fig. 2B) as well as 41 mutations (data not shown) with deletions, insertions, or substitutions of more than one amino acid residue in this region. These mutations may have been introduced by some aberrant products of oligonucleotides. None of the dominant-negative class of mutations complemented the secY39 mutant. Strikingly, 10 different dominant-negative substitutions were identified at the Arg-357 position, at which no functional substitution was obtained. Five and four different mutations were found at Pro-358 and Thr-362, respectively.
These mutant plasmids were expressed in a secY+ strain, and export of MBP was examined at 37°C. Although MBP processing was ≈95% in the presence of the vector plasmid or a secY+ plasmid (Fig. 4B, column 20 and broken line), it was retarded to below 80% in the presence of a mutant plasmid. Even this small reduction in the processing efficiency indicates a significant defect in the SecY mutant protein, because these assays were done in the presence of wild-type SecY molecules. The R357D and R357E substitutions conferred extremely strong translocation interference, giving only ≈20% processing of MBP (Fig. 4B, columns 8 and 9). These mutations had more adverse effects on cell growth than the other mutations, and they gave a deeper blue color in the colony assay. They seem to inactivate SecY almost completely, because their phenotypes were similar to those of deletions of as many as 12–15 residues in this region (data not shown). Mutations R357L, G359P, and T362R also inactivated SecY severely (Fig. 4B, columns 11, 16, and 18).
Although residue 357 was found to be particularly important, we did not obtain its substitution to a basic or aromatic amino acid. We thus constructed two substitutions, R357K and R357F, by site-directed mutagenesis. The former complemented weakly a secY defect and exhibited a weak dominant-negative character, whereas the latter was complementation-negative and strongly dominant-negative (data not shown). Thus, only a basic amino acid is allowed at this position, although Lys is still suboptimal.
Arg-357 Is Crucial for the Translocase Activity of SecY.
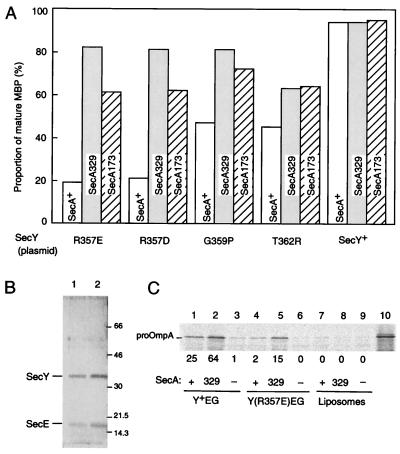
In our characterization of the secY205 (Cs) and the secY39 (Cs) mutants, a number of extragenic suppressor mutations were isolated in secA (ref. 12; unpublished data). Large fractions of them proved to suppress a number of different secY alleles. It was found that the altered SecA proteins, called “superactive SecA,” possess highly elevated intrinsic ATPase activity (ref. 15; unpublished data). We combined a chromosomal suppressor mutation, secA329 (I502S) or secA173 (H545L), with a dominant-negative SecY (R357D, R357E, G359P, or T362R) plasmid. The dominant interference with the MBP export was alleviated significantly in the presence of these suppressors (Fig. 5A). The efficiencies of suppression varied combination-specifically to certain extents; secA329 suppressed the severe SecY defects caused by R357E and R357D more effectively than the less severe T362R defect (Fig. 5A).
Figure 5.
Translocase inactivation by dominant-negative secY mutations and reactivation by suppressor secA mutations. (A) Effects of the suppressor secA mutations on the dominant interference of protein export by some SecY alterations. Strains GN40 (secA+ secY+; open columns), NH178 (secA329 secY+; shaded columns), and NH193 (secA173 secY+; striped columns) were transformed with two plasmids, pSTD343 (lacIq) and a mutant or a wild-type SecY plasmid, as specified at the bottom. The extents of MBP processing were assessed as described in Fig. 4B. (B) SDS/PAGE profiles of the purified preparations of (His)6-tagged SecYE (lane 1) and (His)6-tagged SecY(R357E)E (lane 2), each 1 μg of protein, as stained with Coomassie brilliant blue. (C) In vitro translocation activities of reconstituted proteoliposomes. Proteoliposomes were reconstituted with (His)6-tagged SecYE complex (0.2 μg, lanes 1–3) and SecG (0.2 μg), (His)6-tagged SecY(R357E)E complex (0.2 μg, lanes 4–6) and SecG (0.2 μg), or no added proteins (lanes 7–9). They were incubated at 37°C for 10 min with in vitro-translated and 35S-labeled proOmpA in the presence of either SecA+ (lanes 1, 4, and 7) or SecA329 (lanes 2, 5, and 8) or in the absence of SecA (lanes 3, 6, and 9). Lane 10 shows input proOmpA. ATP (1 mM) was present in all assays. Samples were treated with proteinase K (0.1 mg/ml) on ice for 20 min and subjected to SDS/PAGE and PhosphorImager exposure. The values below the gels represent the proportions (percentage) of translocated proOmpA.
We overproduced (His)6-tagged versions of SecY and SecY(R357E) together with (His)6-tagged SecE and purified the (His)6-tagged SecYE complexes (Fig. 5B). By using these preparations and separately purified SecG, proteoliposomes containing purified (His)6-SecYEG or (His)6-SecY(R357E)EG were reconstituted. Their translocation activities were assayed by incubating them with radiolabeled proOmpA, SecA, and ATP. Under the conditions used, 25 and 2% of proOmpA was translocated into the SecY+- and SecY(R357E)-containing proteoliposomes, respectively (Fig. 5C, lanes 1 and 4). Thus, the SecYEG translocase activity is compromised markedly by the R357E substitution of SecY. When the purified SecA329 suppressor form of SecA was used to drive translocation, the translocation yield increased 2.5- and 7.5-fold, respectively, for the wild-type and mutant proteoliposomes (Fig. 5C, lanes 2 and 5). No significant translocation occurred into liposomes without added protein components (Fig. 5C, lane 8), showing that the superactive SecA only drives translocation in conjunction with SecYEG but not by itself or via some other pathway of translocation. The fact that the mutated SecY(R357E)EG responded in vitro to the superactive form of SecA to exhibit significant translocation activity indicates that it was not inactivated nonspecifically. Thus, the in vivo assessments used in our analyses faithfully represented the molecular functionality of SecY in facilitating protein translocation, in which Arg-357 plays a crucial role.
Discussion
It is unlikely that a SecYEG channel is just a static structure in the membrane. It should be under precise gate control such that it is open only to allow transport of selected polypeptides without significant leakage of nonspecific solutes. Ribosomes, in the eukaryotic system, and SecA, in the prokaryotic system, may induce major subunit arrangements of the channel complex (20, 21). It is conceivable that there are specific regions in SecY that are crucial for the dynamic functionality of the channel. To identify such a region at the residue level, we worked out efficient strategies of mutagenesis and functional assignments of mutants. We focused the present analysis on a cytosolic domain of SecY, because cytoplasmic events such as interaction with a translocation substrate and with SecA for initiation of translocation will be fundamental for all the subsequent translocation reactions. We choose the Ser-349 to Tyr-365 interval as the target of mutagenesis on the basis of the locations of the previously isolated cold-sensitive secY mutations: secY39 (R357H), secY110 (R357C), secY122 (G359R), and secY40 (or secY115, A363T). All these mutations impair the abilities of inverted membrane vesicles to activate the translocation ATPase of SecA (10).
In our screenings, simple knocking-out and destabilizing mutations must have been excluded, because such mutations will never complement a loss of function of the chromosomal secY gene or interfere with its normal functioning. In addition, mutations that cause extreme toxicity, if any, must have escaped the isolation. Partly because of the screening design and partly because of the nonexhaustive nature of our isolation, we did not obtain all of the possible changes of each residue. Nevertheless, the results were sufficiently informative to tell which residues are important for SecY functionality.
Newly synthesized SecY molecules are under mutual competition with respect to their binding with SecE, which determines stability of SecY (8, 22). Thus, those molecules that have failed in the binding are eliminated rapidly by the FtsH protease (23). We examined cellular accumulation of three weakly complementing and four dominant-negative SecY variants (including R357E) when they were overproduced together with SecE. They were all as stable as the wild-type SecY that was expressed similarly (data not shown). Thus, the different phenotypes of SecY variants cannot be explained primarily by stability difference.
Residues other than Arg-357, Pro-358, Gly-359, and Thr-362 can be mutated to a large variety of different amino acids without causing major defects (Fig. 3A), although basic amino acids are less favored at residues 351 and 352, and acidic amino acids are less favored at residue 353. Even the latter substitutions only mildly compromised the SecY function (Fig. 4B). In contrast, almost any changes at Arg-357 inactivated SecY and conferred dominant-negative properties (Figs. 3B and 4B), although dominant-negative substitutions were isolated also at Pro-358, Gly-359, Thr-362, and Ala-363. From these results, we surmise that Arg-357 is absolutely required, whereas the two residues C-terminally adjacent to it as well as Thr-362 and Ala-363 are important also.
We identified a total of 10 different substitutions of Arg-357 as dominant-negative mutations. Taken together with the two cold-sensitive mutations (secY39 and secY110) as well as with the two site-directed mutations (R357F and R357K), a total of 14 substitutions have been characterized. Only the Lys substitution retained some weak complementation activity. In contrast, Asp and Glu substitutions caused extremely severe dysfunction of SecY. Thus this position should at least be basic, but Arg is required specifically for the full functionality. Our reconstitution experiments showed compellingly that the R357E mutated translocase was virtually inactive in translocation activity. However, this mutational defect, but not the defect caused by a deletion of an Ala-343/Arg-357 interval (data not shown), was still suppressible by the superactive forms of SecA. The suppression seems to require a preserved overall architecture of the SecYE complex and possibly represents a recovery of a productive SecY–SecA interaction (7, 12). The limited suppression specificity observed in vivo with respect to the secY alleles supports this notion. The R357E SecY alteration may impair an aspect of SecA–SecY interaction, in which SecA is activated to energize translocation.
The Arg-357, Pro-358, Gly-359, and Thr-362 residues, which were shown to be important in this study, are all well conserved among SecY homologues from prokaryotes as well as from chloroplasts, in which SecA acts as the primary “motor.” In addition, Pro-354 and Gly-355 are well conserved (Fig. 6, Left). This segment did not show highly significant homology with the eukaryotic Sec61α sequences (Fig. 6, Right). The Sec61α sequences themselves contain several absolutely conserved residues in this segment including Gly-404, Arg-406, Glu-413, and Leu-414 (numbering for S. cerevisiae Sec61p; Fig. 6, Right). These observations are consistent with a notion that this segment of SecY/Sec61 is involved in some functions that are specific for prokaryotes and eukaryotes, respectively. The most likely function might be the SecA binding for the former and the ribosome binding for the latter.
Figure 6.
Conservation of the Ser-349/Tyr-365 segment in different organisms. (Left) The amino acid sequence of the C5 domain was subjected to a homology search by using the BLAST program, and the Ser-349/Tyr-365 region is shown. The number in parenthesis indicates amino acid residue number of the residue that is equivalent to E. coli Ser-349 in each organism. Residues identical with those of E. coli SecY are shown in reverse font. (Right) Saccharomyces cerevisiae Sec61p sequence was aligned with E. coli SecY by using the CLUSTALW program. The region that corresponds to C5 of E. coli SecY was subjected to BLAST homology searches, and the Ser-349/Tyr-365 equivalent regions are shown. Residues identical with those of S. cerevisiae Sec61p are shown in reverse font.
Interestingly however, sequence alignment indicated that the Arg-406 residue of S. cerevisiae Sec61p may correspond to Arg-357 of E. coli SecY, making this Arg residue conserved in almost all organisms sequenced thus far (Fig. 6, *). It is conceivable that this residue is involved in some fundamental characteristic of the protein translocation channel. One essential element common between prokaryotes and eukaryotes may be the signal sequence of presecretory proteins (24). Our characterization of the secY39 (R357H) mutant suggests that the mutation impairs the initiation step of translocation that occurs after precursor protein binding and before the signal sequence cleavage (unpublished data). Thus, Arg-357 may be involved in the initiation of translocation that occurs under the mutual interactions of SecYEG, SecA, and the signal sequence region of the precursor protein. The present results will guide further characterization of this interesting molecular machinery found in the membranes of all organisms.
Acknowledgments
We thank Ei-ichi Matsuo, Yoshinori Akiyama, Hitoshi Nakatogawa, Tohru Yoshihisa, and Gen Matsumoto for plasmids, E. coli strains, and stimulating discussion, and Yusuke Shimizu and Kiyoko Mochizuki for technical supports. This work was supported by grants from Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, and from the Ministry of Education, Science and Culture, Japan.
Abbreviation
- MBP
maltose-binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A E, van Waes M A. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 3.Ito K. Genes Cells. 1996;1:337–346. doi: 10.1046/j.1365-2443.1996.34034.x. [DOI] [PubMed] [Google Scholar]
- 4.Joly J C, Wickner W. EMBO J. 1993;12:255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lill R, Dowhan W, Wickner W. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 6.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto G, Yoshihisa T, Ito K. EMBO J. 1997;16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba T, Taura T, Shimoike T, Akiyama Y, Yoshihisa T, Ito K. Proc Natl Acad Sci USA. 1994;91:4539–4543. doi: 10.1073/pnas.91.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimoike T, Akiyama Y, Baba T, Taura T, Ito K. Mol Microbiol. 1992;6:1205–1210. doi: 10.1111/j.1365-2958.1992.tb01559.x. [DOI] [PubMed] [Google Scholar]
- 10.Taura T, Yoshihisa T, Ito K. Biochimie. 1997;79:512–517. doi: 10.1016/s0300-9084(97)82744-7. [DOI] [PubMed] [Google Scholar]
- 11.Taura T, Akiyama Y, Ito K. Mol Gen Genet. 1994;243:261–269. doi: 10.1007/BF00301061. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto G, Nakatogawa H, Mori H, Ito K. Genes Cells. 2000;5:991–1000. doi: 10.1046/j.1365-2443.2000.00388.x. [DOI] [PubMed] [Google Scholar]
- 13.Akiyama Y, Ito K. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo E, Ito K. Mol Gen Genet. 1998;258:240–249. doi: 10.1007/s004380050728. [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H, Mori H, Ito K. J Biol Chem. 2000;275:33209–33212. doi: 10.1074/jbc.C000550200. [DOI] [PubMed] [Google Scholar]
- 16.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 17.Driessen A J, Wickner W. Proc Natl Acad Sci USA. 1991;88:201–210. doi: 10.1073/pnas.88.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baba T, Jacq A, Brickman E, Beckwith J, Taura T, Ueguchi C, Akiyama Y, Ito K. J Bacteriol. 1990;172:7005–7010. doi: 10.1128/jb.172.12.7005-7010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs P D, Derman A I, Beckwith J. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 21.Manting E H, van Der Does C, Remigy H, Engel A, Driessen A J. EMBO J. 2000;19:851–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taura T, Baba T, Akiyama Y, Ito K. J Bacteriol. 1993;175:7771–7775. doi: 10.1128/jb.175.24.7771-7775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kihara A, Akiyama Y, Ito K. Proc Natl Acad Sci USA. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]