Abstract
Although the pathogenesis of BCR–ABL1-positive acute lymphoblastic leukemia (ALL) is mainly related to the expression of the BCR–ABL1 fusion transcript, additional cooperating genetic lesions are supposed to be involved in its development and progression. Therefore, in an attempt to investigate the complex landscape of mutations, changes in expression profiles and alternative splicing (AS) events that can be observed in such disease, the leukemia transcriptome of a BCR–ABL1-positive ALL patient at diagnosis and at relapse was sequenced using a whole-transcriptome shotgun sequencing (RNA-Seq) approach. A total of 13.9 and 15.8 million sequence reads was generated from de novo and relapsed samples, respectively, and aligned to the human genome reference sequence. This led to the identification of five validated missense mutations in genes involved in metabolic processes (DPEP1, TMEM46), transport (MVP), cell cycle regulation (ABL1) and catalytic activity (CTSZ), two of which resulted in acquired relapse variants. In all, 6390 and 4671 putative AS events were also detected, as well as expression levels for 18 315 and 18 795 genes, 28% of which were differentially expressed in the two disease phases. These data demonstrate that RNA-Seq is a suitable approach for identifying a wide spectrum of genetic alterations potentially involved in ALL.
Keywords: sequencing, ALL, BCR-ABL1
Introduction
The Philadelphia (Ph) chromosome1 arises from a reciprocal translocation between chromosome 9 and 222 and it was the first cytogenetic abnormality linked to both chronic myeloid leukemia and Ph-positive acute lymphoblastic leukemia. This translocation fuses the ABL1 oncogene on chromosome 9 to a breakpoint cluster region (BCR) from chromosome 22, generating the constitutively activated Bcr–Abl tyrosine kinase that is responsible for both acute and chronic diseases.3, 4, 5 BCR–ABL1-positive ALL represents the most frequent and prognostically unfavorable subtype of ALL in adults.6 Driven by technological advances, copy-number alterations have been identified in such disease using single-nucleotide polymorphism (SNP) array platforms, suggesting that additional cooperating genetic lesions are involved in its pathogenesis.7, 8 Nowadays, high-throughput ‘next-generation sequencing technologies', overcoming the limited scalability of traditional Sanger sequencing, are revolutionizing genomics and transcriptomics by providing a cost-efficient and single-base resolution tool for a unified deep analysis of the cancer complexity.9, 10, 11, 12 There is a vast diversity of next-generation technologies, but these sequencing approaches generally use massively parallel amplification and detection strategies. The first whole-cancer genome sequence was reported in 2008 with the description of the nucleotide sequence of DNA from a patient with acute myeloid leukemia (AML) compared with DNA from normal skin from the same patient.13 Since then, the number of complete sequences of cancer genomes and/or transcriptomes identified has been rapidly growing. In the present study, Illumina technology was used to perform a whole-transcriptome shotgun sequencing (RNA-Seq)9 on leukemia cells from a Ph+ ALL patient at diagnosis and at the time of hematologic, cytogenetic and molecular relapse. A transcriptional picture of the examined genome was drawn by mapping complementary DNA sequence reads to the reference sequence of the human genome, identifying expressed annotated and novel transcripts, single-nucleotide variants (SNVs), alternative splicing (AS) events and related absolute expression levels. A unified picture of a Ph+ ALL transcriptome was thus provided for the first time, supporting the belief that RNA-Seq may represent one of the most suitable approaches to identify the genetic alterations harbored by leukemia clones.
Materials and methods
The case of a 56-year-old man affected by Ph+ ALL diagnosed in April 2007 is herein reported.
Double-stranded complementary DNA libraries were prepared from his primary and relapsed RNA samples and sequenced using the Genome Analyzer II platform, generating 36-base-pair (bp) sequence reads. These reads were mapped to the reference sequence of the human genome (NCBI Build 36.1) using the ELAND software to assign them to exons, splice junctions, introns/untranslated regions, external exons or intergenic regions. Mapping reads were subsequently used for discovery of expressed candidate SNVs by means of the BOWTIE and ERANGE software. Reads that failed to directly align to the human genome reference sequence were instead used to assess the splicing extent by mapping them to an in silico-generated data set of all possible exon splice junctions. The number of reads corresponding to RNA from known exons, canonical splice events and new candidate genes was also estimated and a normalized measure of gene expression level (RPKM)11 was computed to define gene expression profiles.
A full description of the examined Ph+ ALL patient, as well as of bioinformatic analyses performed to produce the described results, is provided in the Supplementary Materials and Methods.
Results
Whole-transcriptome sequencing
The RNA-Seq technique generated 13.9 and 15.8 million 36-bp sequence reads from de novo and relapsed Ph+ ALL samples, respectively. The total number of processed reads, as well as of those successfully mapped to the human genome reference sequence, is shown in Table 1.
Table 1. Summary of RNA-Seq genomic mapping results from the primary and relapse BCR–ABL1-positive ALL samples.
| Primary ALL | Relapse | |
|---|---|---|
| Reads processed | 13 913 719 | 15 782 973 |
| Aligned genomic readsa | 11 999 193 | 14 467 276 |
| Unique readsb | 5 265 914 | 7 470 979 |
| Multiple readsc | 6 733 279 | 6 996 297 |
| Unaligned reads | 1 914 526 | 1 315 697 |
| AS junctions reads | 25 119 | 22 859 |
| Expressed RefSeq transcripts | 18 315 | 19 796 |
| Putative novel exons in annotated genes | 6637 | 2541 |
| Putative novel genes | 18 | 23 |
| Coverage (%) | 86.24 | 91.66 |
Abbreviations: ALL, acute lymphoblastic leukemia; AS, alternative splicing; BCR, breakpoint cluster region.
Reads mapped to the human genome reference sequence (NCBI Build 36.1).
Reads matching with a unique genomic location.
Reads matching with a multiple genomic location.
Identification of SNVs
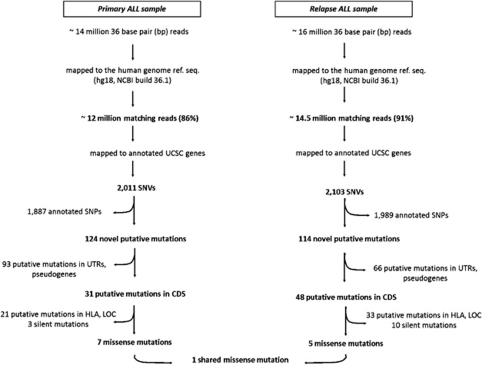
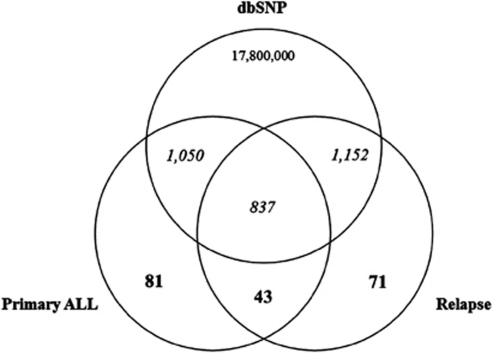
SNV discovery was performed by mapping the 12 million primary ALL and 14.5 million relapse sequence reads that matched the reference sequence of the human genome (Table 1) to all annotated human genes and applying stringent criteria for reducing the relative false-positive rate. The adopted filter led to the identification of 2011 and 2103 SNVs in the primary ALL and relapse samples, respectively (Figure 1). Approximately 94% of these variants have been already reported in the dbSNP build 130 (Supplementary Table S1), whereas 124 and 114 were putative novel SNVs in the primary ALL and relapse samples, respectively. Of these, 43 affected both samples, 81 were found only in the primary ALL sample and 71 were relapse private substitutions (Figure 2). These putative novel mutations were further subdivided into four groups according to their genomic location: group 1 contained 60 changes located in the amino-acid-coding regions of annotated exons, group 2 contained 38 changes located in untranslated regions, group 3 contained 26 changes found on annotated pseudogenes and group 4 contained 71 variants for which no information about their functional annotation was available (Table 2).
Figure 1.
Flow chart for identification of somatic point mutations in the examined BCR-ABL1-positive ALL transcriptome at diagnosis and at relapse.
Figure 2.
Venn diagram of primary ALL and relapse-detected SNVs. Numbers in bold are putative novel SNVs, while numbers in italics are known SNPs annotated on dbSNP Build 130.
Table 2. Putative novel SNVs detected in the primary and relapsed BCR–ABL1-positive ALL samples.
| SNV location | Diagnosisa | Relapseb | Both phasesc |
|---|---|---|---|
| Coding sequences | 12 | 29 | 19 |
| Untranslated regions (UTRs) | 11 | 17 | 10 |
| Pseudogenes | 4 | 13 | 9 |
| Unknown | 54 | 12 | 5 |
| Total | 81 | 71 | 43 |
Abbreviations: ALL, acute lymphoblastic leukemia; BCR, breakpoint cluster region; SNVs, single-nucleotide variants; UTRs, untranslated regions.
Private primary ALL SNVs.
Private relapse SNVs.
Common SNVs identified at both diagnosis and relapse.
As mutations affecting amino-acid-coding regions may impair gene function, downstream analyses were focused on SNVs belonging to group 1. From this group, mutations in human leukocyte antigen genes, immunoglobulin heavy variable chain genes and those in genes encoding for hypothetical proteins (LOC728238, LOC654340, LOC285299, LOC441581, LOC644937) were removed. This approach identified 11 non-synonymous changes: one affecting the PLXNB2 gene on both primary ALL and relapse samples, six affecting genes involved in metabolic processes (PDE4DIP, EIF2S3, DPEP1, ZC3H12D, TMEM46) or transport (MVP) at diagnosis and two affecting genes involved in cell cycle regulation (CDC2L1) and catalytic activity (CTSZ), as well as one affecting a gene (CXorf21) encoding for an uncharacterized protein, at relapse (Table 3). Furthermore, the T315I mutation in the Bcr–Abl kinase domain, which is known to be responsible for insensitivity to current tyrosine kinase inhibitors,14, 15, 16 was also identified.
Table 3. Putative novel non-synonymous SNVs detected in the primary and relapsed BCR–ABL1-positive ALL samples.
| Chr | Gene | Mutation type | AA change | Primary ALL wt:ma | Relapse wt:ma | Validationb | UPD/CNAc | Mutations in other ALL patients |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC2L1 | Missense | V97A | 16:0 | 7:9(21) | n.a | No | n.a |
| 1 | PDE4DIP | Missense | R921Q | 0:5(7) | 15:0 | No | No | n.a |
| 6 | ZC3H12D | Missense | P406S | 0:8(15) | 8:0 | n.a | No | n.a |
| 9 | ABL1 | Missense | T315I | 18:0 | 1:6(8) | Yes | No | n.a |
| 13 | TMEM46 | Missense | G59D | 1:8(8) | 5:0 | Yes | No | 0/24 |
| 16 | DPEP1 | Missense | R20Q | 6:11(19) | 18:0 | Yes | No | 1/24 |
| 16 | MVP | Missense | P620S | 1:5(5) | 13:0 | Yes | No | 0/24 |
| 20 | CTSZ | Missense | R183Q | 16:0 | 2:7(11) | Yes | No | 0/24 |
| 22 | PLXNB2 | Missense | N759Dd | 0:9(13) | 0:11(31) | Yes | No | 0/24 |
| X | EIF2S3 | Missense | Q39K | 4:5(14) | 27:0 | No | No | No |
| X | CXorf21 | Missense | V230Md | 4:0 | 0:6(8) | Yese | No | 0/24 |
Abbreviations: ALL, acute lymphoblastic leukemia; AA, amino-acid; BCR, breakpoint cluster region; Chr, chromosome; CNA, copy number alteration; n.a, not applicable; UPD, uniparental disomy; wt, number of reads showing the wild-type allele; m, number of unique reads showing the mutated allele. SNVs in italics are RNA-Seq false-positive calls.
In brackets is the number of multiple reads (i.e., reads matching the human genome reference sequence with a multiple genomic location) showing the mutated allele.
Sanger sequencing of PCR-generated amplicons.
Detected by means of Affymetrix SNP chip 6.0.
Inherited variant observed in the primary, remission and relapsed genomic DNA samples.
In contrast with RNA-seq, this mutation was also found at diagnosis.
Nine exons containing putative novel non-synonymous changes were analyzed using direct Sanger sequencing on samples collected at diagnosis, hematological remission and relapse time. Seven SNVs identified by RNA-Seq were confirmed (Table 3 and Supplementary Figure S1), whereas two non-synonymous changes in PDE4DIP and EIF2S3 resulted in false-positive calls (Table 3). In accordance with RNA-seq results, conventional Sanger sequencing confirmed that the TMEM46 G59D, DPEP1 R20Q and MVP P620S mutations were specific for diagnosis, whereas the ABL1 T315I and CTSZ R183Q were limited to relapse, thus demonstrating that they are somatic mutations. On the contrary, the PLXNB2 N759D and CXorf21 V230M mutations were identified in all the examined phases of the disease (diagnosis, hematological remission, relapse), suggesting their germline origin. In the case of CXorf21 V230M, the Sanger sequencing method contrasted with RNA-seq, identifying the mutation also in the diagnosis sample.
Rare or common mutations?
To determine whether validated novel missense SNVs were ‘private' mutations of the analyzed patient or recurrent variants in Ph+ ALL, they were investigated in 24 additional Ph+ ALL samples and two different cell lines (BV-173 and SD-1) by means of Sanger sequencing of amplified genomic-DNA target regions. All mutations were not confirmed in this set of leukemia patients and cell lines, with the exception of the R20Q substitution on the DPEP1 gene. This change was identified in one of the additional Ph+ ALL patients, suggesting that it may be not a ‘private' mutation. However, constitutional/remission DNA was not available for this additional case, preventing us from assessing whether the R20Q mutation was an inherited alteration in such an ALL patient.
Moreover, confirmed mutated genes were searched out from a list of 649 genes with potential roles in cancer susceptibility,13 compiled on the basis of recently published data and the Cancer Genome Project database (http://www.sanger.ac.uk/genetics/CGP/Census/). Only one specific primary ALL variant and one specific relapse variant were identified by RNA-Seq on genes that have already been associated with cancer susceptibility (MVP, ABL1), even though none of the detected mutated genes were included in a list of 41 ALL-related genes extracted from the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/), which includes an exhaustive collection of genes presenting recurrent mutations in several cancer types.
Inherited polymorphisms
The Cancer Genome Project and COSMIC databases were also investigated in search of genes that in the present study show inherited annotated SNPs, as it has been recently suggested that such polymorphisms, if they occur in somatically mutated genes, may act as low-penetrance susceptibility alleles in some non-acute lymphoblastic leukemias.13 Of the detected annotated SNPs, 13 were represented by missense mutations; however, none of the affected genes were included in the list of ALL-related genes from the COSMIC database. By comparing the detected genes with missense SNPs with the whole COSMIC list we have instead observed that seven of them have already been found to display also somatic mutations, whereas 14 were known cancer-related genes, but without previously reported somatic mutations (Supplementary Table S2).
SNP array correlation
To investigate whether somatic copy-number alterations and uniparental-disomy events could characterize regions containing the observed mutations, Affymetrix Genome Wide Human SNP 6.0. array data were also analyzed. None of the genes carrying confirmed non-synonymous substitutions lie within abnormal genomic regions (Table 3).
Detection of AS events
Approximately 14 and 8% of the total number of processed reads from the primary and relapse Ph+ ALL samples failed to directly align to the human genome reference sequence (Table 1). These reads were mapped to an in silico data set of all possible splice junctions, created by pairwise combination of annotated human exons, in order to investigate potential AS events. According to this approach, 6390 and 4671 putative AS events were identified within 4334 and 3651 annotated transcripts (Supplementary Figure S2), concerning 3833 and 3327 genes, which represent 21% and 17% of primary ALL and relapse expressed transcripts, respectively.
A total of 1269 putative AS events were shared between the primary ALL and relapse samples, whereas 80 and 73% of them were found to be private ALL primary and relapse events. These private putative AS events showed lower expression levels compared to putative AS events shared between both samples, with 93 and 91% of private ALL primary and relapse AS, which showed a very small number of reads.
All identified alternatively spliced genes were compared with a list of 729 cancer-related genes showing AS.17 A total of 99 primary ALL and 85 relapse genes were found to belong to such list, with 40 genes showing putative AS in both samples (Supplementary Data 1). Among these latter genes, 17 (43%) had no reference to a specific functional class according to Ingenuity pathway analysis results (Ingenuity Systems, Redwood City, CA, USA, http://www.ingenuity.com), whereas 11 (28%) were kinases and only 2 (5%) were transcription regulators. As regards primary ALL and relapse, private alternatively spliced genes, 21 (36%) and 20 (44%), respectively, had no reference to a specific functional class, 7 (12 and 16%) were kinases, as well as 6 (10%) and 5 (11%) were transcription regulators (Supplementary Data 1).
Exon-skipping events, in which exons are alternatively included or spliced out of the mature mRNA, affected mostly one or few exons, especially those involving putative AS shared between both samples (Supplementary Figure S3).
The known AS pattern in the IKZF1 gene7, 18, 19, 20 was detected both at diagnosis and at relapse by this approach, supporting its validity.
Quantitative measurement of transcripts expression
In all 12 and 14.5 million reads matched with the reference sequence of the human genome (Table 1), ensuring a read density sufficient for quantitative gene-expression analysis.12 These reads were subsequently mapped to exon sequences from annotated human genes and counted to estimate the number of reads corresponding to RNA from each known exon or putative novel gene. A detailed gene expression profile was thus obtained, with a normalized measure of gene expression for each transcript (RPKM; reads per kb of gene model per million of reads). According to this procedure, the expression of 18 315 and 18 795 known transcripts was detected in the primary ALL and relapse samples, respectively (Supplementary Data 2), showing that 62 and 64% of annotated human genes were transcribed in the examined stages of the disease. However, very low RPKM estimates (0.01 <RPKM< 10) were computed for the majority of active genes (78% at diagnosis and 73% at relapse), whereas moderate expression (10 <RPKM< 100) was observed for 20–24% of active genes and only 2–3% of detected transcripts had high RPKM values (100<RPKM<8000) (Figure 3).
Figure 3.
Distribution of annotated human transcripts in classes of expression level based on the RPKM estimates. n.e., not expressed; 0.01<RPKM<10, scarcely expressed; 10<RPKM<100, moderately expressed; 100<RPKM<8000 highly expressed.
Differential gene-expression analysis
Fisher's exact test was used to compare read-count log ratios derived from RPKM values to statistically validate differences observed in gene expression levels between the primary ALL and relapse samples.21 Among genes for which expression was detected in both the examined samples, 31% were differentially expressed, 73% of which were upregulated (fold change >2, Fisher's exact test P<0.01 after Bonferroni correction) and 27% were downregulated (fold change <−2, Fisher's exact test P<0.01 after Bonferroni correction) at relapse compared with diagnosis (Supplementary Data 3).
A functional analysis was also carried out on differentially regulated genes using the GeneGo software (http://www.genego.com). In the list of the most overexpressed genes at relapse, the most significant GeneGo Pathway Map was the ‘Cell cycle: the metaphase checkpoint' pathway (P value=3.94E−10), including genes such as AURORA Kinase A (AURKA), AURORA Kinase B (AURKB), SURVIVIN (BIRC5), BUB1, RAD51, CENPA, INCENP and PLK1 (Supplementary Figure S4). The most significant GeneGo Pathway Map representing underexpressed genes at relapse with respect to diagnosis was instead that of the ‘Signal transduction PKA signaling' (P-value=2.60E−07), including PDE3B, PDE4A and PDE4D phosphodiesterases (Supplementary Figure S5).
In order to determine whether the observed differential expression was due to a random variation in our data or due to biologically relevant factors, some key genes (AURORA Kinase B and SURVIVIN) were investigated in an additional set of eight matched diagnosis-relapse BCR–ABL1-positive ALL samples by quantitative RT-PCR analysis. A significant difference in gene-expression levels between diagnosis and relapse was found for AURORA Kinase B (P=0.04), confirming RNA-seq results, whereas a positive, but not significant (P=0.08), trend of overexpression at relapse with respect to diagnosis was observed for SURVIVIN (Supplementary Figure S6).
Moreover, in order to further validate RNA-seq results, we performed a gene expression analysis by Affymetrix Human Exon 1.0 ST arrays on 22 BCR–ABL1-positive ALL patients at diagnosis and 6 patients at the relapse. A list of differentially expressed genes (Supplementary Data 4) was obtained between the diagnosis and relapse phases performing the analysis of variance (ANOVA) on data from the 22 diagnosis samples versus the six relapse samples and including genes with a P-value below 0.05. A concordance of 97% was found considering over- and underexpressed genes at relapse with respect to diagnosis in both the RNA-Seq and human exon 1.0 ST results. In other words, the great majority of transcripts (140 genes) differentially expressed in both the experiments were differentially expressed in the same direction in the sequenced and in the additional set of BCR–ABL1-positive ALL samples.
Discussion
Ph+ ALL is the most frequent and prognostically unfavorable subtype of ALL in adults,6, 22, 23, 24 with pathogenesis long since shown to be closely related to the BCR–ABL1 fusion-transcript expression. Nevertheless, high-resolution SNP array-based studies have recently suggested that additional genetic lesions may be involved in its development.7 The present study marks for the first time the whole transcriptome of Ph+ ALL cells, which was sequenced using the RNA-Seq technique9 in an effort to identify as many alterations as possible. RNA-Seq overcomes the limitations of array-based experiments25, 26 by providing a more exhaustive approach that is able to draw a reliable qualitative and quantitative picture of the transcriptome complexity. Thus, two samples from a Ph+ ALL patient at diagnosis and at the time of hematological, cytogenetic and molecular relapse were sequenced in search of genetic alterations that potentially cooperate with the BCR–ABL1 fusion transcript.
RNA-Seq generated approximately 15 million of 36-bp sequence reads from each sample, most of which successfully mapped the reference sequence of the human genome. With the exclusion of T315I mutation in the Bcr–Abl kinase domain, five missense mutations were detected in the Ph+ ALL cells after applying stringent criteria to reduce the SNVs discovery false-positive rate and validating novel substitutions by Sanger sequencing. Three of these non-synonymous changes were found in the primary ALL sample and affected genes involved in metabolic processes (DPEP1, TMEM46) or transport (MVP). The role of these alterations is not clear and no clues can be derived from the literature as evidence of tumor association has not been reported for most of them. The sole gene that has already been associated with malignant disorders is MVP, encoding the major vault protein (lung-resistance related protein) and involved in nucleocytoplasmic transport. Overexpression of MVP is a potential useful marker of clinical drug resistance in lung cancer. Moreover, MVP has been described to have a role in cervical carcinoma, affecting the non homologous end-joining repair system and apoptosis through Ku70/80 and Bax downregulation.27, 28, 29, 30 However, a point mutation in this gene has never been described and the occurrence in a single case suggests that it could be a ‘passenger' rare mutation in Ph+ ALL. As regards the other primary ALL specific mutations, substitution in the DPEP1 gene,31 which encodes for a kidney membrane enzyme, was the sole gene to be found in an additional Ph+ ALL patient, although constitutional/remission DNA was not available for this additional case. The two validated missense mutations specific to the relapse sample instead affected genes involved in catalytic activity (CTSZ) and impaired drug responsiveness, such as the case of the T315I mutation in the kinase domain of BCR–ABL1. Differences in mutational patterns of primary ALL and relapse samples may suggest that the leukemia clone from which relapsed cells have been developed was not the predominant one at diagnosis or, more plausibly, that most of the relapse-specific changes are ‘passenger' mutations acquired by chance during Ph+ ALL progression by the clone harboring the BCR–ABL1 T315I mutation responsible for resistance to tyrosine kinase inhibitor treatments.14, 32
Although a greater number of samples would be necessary for a more stringent quantitative analysis, a detailed gene-expression profile was nonetheless obtained by taking advantage of a normalized measure of gene expression for each transcript (RPKM). This quantification of transcript abundance indicated that slightly more than 60% of annotated human genes were transcribed in leukemia cells in both diagnosis and relapse phases. Approximately 23% of genes for which expression was detected by RNA-Seq in both samples were upregulated at relapse with respect to diagnosis. Many of these genes affect cell-cycle progression, suggesting that the loss of cell-cycle control and the subsequent increased proliferation have a role in the disease progression. Conversely, only 9% of active genes in both samples were downregulated at relapse with respect to diagnosis. In particular, transcripts belonging to the PKA signaling pathway, such as PDE3B, PDE4A and PDE4D phosphodiesterases, turned out to be the most overrepresented genes in such list, with differential expression patterns, which were confirmed also in the additional set of paired diagnosis-relapse BCR–ABL1-positive ALL samples analyzed with Human Exon 1.0 ST array. Proteins encoded by these genes have 3′,5′-cyclic-AMP phosphodiesterase activity, being directly involved in the process of cAMP degradation and thus having the potential to modulate signal transduction in multiple cell types.33 Although further candidate-gene studies will be required for an exhaustive characterization of the roles of these differentially expressed genes, our results prove that the RNA-Seq estimate of transcript abundance is sensitive enough to draw an accurate differential gene-expression profile and to deepen the description of transcriptional changes potentially involved in ALL progression.
The approximately three million sequence reads that failed to directly align to the human genome reference sequence have allowed us to identify thousands of putative AS events, which have contributed to the high transcriptional complexity of Ph+ ALL primary and relapse samples. Interestingly, 144 genes showing putative AS events were known to be cancer-related alternatively spliced genes, of which kinases and transcription regulators were the most represented functional classes. As tyrosine kinases are good drug targets, the possibility that some of the alternatively spliced kinases may be subjected to therapeutic inhibition is clearly attractive. Unfortunately, the lack of a comparison with RNA-Seq data from non-leukemic B cells did not allow us to identify whether some of the observed AS events are enriched in the malignant cells relative to normal cells. Nevertheless, the obtained wide ALL transcriptional picture suggests that a whole-transcriptome approach might have the potential to lay the foundation for future improvement of diagnostic and prognostic tools based on AS recognition, as well as for the discovery of additional therapeutic targets for the leukemia under consideration.
In the meantime that the present work was under editorial revision, a number of papers focused on new bioinformatic pipelines for RNA-Seq data analyses flourished in literature. In particular, several innovative tools have been developed for the detection of AS events both at the gene34, 35, 36 and at the inter-chromosomal level,37, 38, 39, 40, 41 thus enabling the reliable discovery of gene fusions derived from genomic rearrangements that are quite frequent in many cancer types. Although the exploitation of such new approaches might have the potential to improve our analyses, it was beyond the actual scope of this work and we believe that it could be much more effective for processing RNA-Seq data sets made up of hundreds of millions of paired-end reads that turned out to be more suitable starting points for the identification of chimeric transcripts with respect to single-end read data sets.
In conclusion, the adopted RNA-Seq approach provided, for the first time, an overview of a Ph+ ALL transcriptome, identifying novel mutations, changes in gene-expression levels and putative AS events potentially involved in ALL manifestation and progression. This descriptive study demonstrates that the RNA-Seq technique, if supported by adequate bioinformatic resources, provides promising new opportunities for a cost-efficient, single-base resolution analysis of the transcriptome complexity of leukemia cells, from both mutational and gene-expression perspectives. This could lead to the identification of novel target candidate genes. Therefore, such an approach may represent one of the most effective tools for discovering genetic rules of Ph+ ALL and of many other cancer types.
The authors declare no conflict of interest.
Author contributions
GI MAR and II: project conception; II and MS: equally contributed to manuscript writing; GI MAR and MD: project leaders and analysis coordination; AF and LX: alternative splicing algorithm development; EG: library optimization and construction; MS, AF, AB, PG, II and LX: single-nucleotide variant analysis, alternative splicing analysis; MS and GI MAL: gene expression analysis; AL, AF and II: sequencing validation analysis; CP, AV: clinical data collection; DL, SS, FP, MB: data interpretation; II and AL: SNP array analysis; GI MAR and MD: final approval.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- Rowley JD. Acquired trisomy 9. Lancet. 1973;2:390. doi: 10.1016/s0140-6736(73)93247-9. [DOI] [PubMed] [Google Scholar]
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Chan LC, Karhi KK, Rayter SI, Heisterkamp N, Eridani S, Powles R, et al. A novel abl protein expressed in Philadelphia chromosome positive acute lymphoblastic leukaemia. Nature. 1987;325:635–637. doi: 10.1038/325635a0. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Ray M, Jiang C, Clark AB, Rogozin IB, Diaz M. Known components of the immunoglobulin A:T mutational machinery are intact in Burkitt lymphoma cell lines with G:C bias. Mol Immunol. 2007;44:2659–2666. doi: 10.1016/j.molimm.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Li H, Lovci MT, Kwon YS, Rosenfeld MG, Fu XD, Yeo GW. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc Natl Acad Sci U S A. 2008;105:20179–20184. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soverini S, Iacobucci I, Baccarani M, Martinelli G. Targeted therapy and the T315I mutation in Philadelphia-positive leukemias. Haematologica. 2007;92:437–439. doi: 10.3324/haematol.11248. [DOI] [PubMed] [Google Scholar]
- Pricl S, Fermeglia M, Ferrone M, Tamborini E. T315I-mutated Bcr-Abl in chronic myeloid leukemia and imatinib: insights from a computational study. Mol Cancer Ther. 2005;4:1167–1174. doi: 10.1158/1535-7163.MCT-05-0101. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Kurosu T, Kakihana K, Mizuchi D, Miura O. The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem Biophys Res Commun. 2004;319:1272–1275. doi: 10.1016/j.bbrc.2004.05.113. [DOI] [PubMed] [Google Scholar]
- Xi L, Feber A, Gupta V, Wu M, Bergemann AD, Landreneau RJ, et al. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic Acids Res. 2008;36:6535–6547. doi: 10.1093/nar/gkn697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I, Lonetti A, Messa F, Cilloni D, Arruga F, Ottaviani E, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112:3847–3855. doi: 10.1182/blood-2007-09-112631. [DOI] [PubMed] [Google Scholar]
- Mullighan C, Downing J. Ikaros and acute leukemia. Leuk Lymphoma. 2008;49:847–849. doi: 10.1080/10428190801947500. [DOI] [PubMed] [Google Scholar]
- Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27:5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom EA, Hansen KD, Durinck S, Dudoit S. Statistical inference in mRNASeq: exploratory data analysis and differential expression. UC Berkeley Division of Biostatistics Working Paper Series. 2009;247 [Google Scholar]
- Fielding A. The treatment of adults with acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program) 2008. pp. 381–389. [DOI] [PubMed]
- Ottmann OG, Wassmann B. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program) 2005. pp. 118–122. [DOI] [PubMed]
- Telegeev GD, Dubrovska AN, Dybkov MV, Maliuta SS. Influence of BCR/ABL fusion proteins on the course of Ph leukemias. Acta Biochim Pol. 2004;51:845–849. [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret M, Lara PC, Bordon E, Fontes F, Rey A, Pinar B, et al. Major vault protein may affect nonhomologous end-joining repair and apoptosis through Ku70/80 and bax downregulation in cervical carcinoma tumors. Int J Radiat Oncol Biol Phys. 2009;73:976–979. doi: 10.1016/j.ijrobp.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Oue T, Yoneda A, Uehara S, Yamanaka H, Fukuzawa M. Increased expression of multidrug resistance-associated genes after chemotherapy in pediatric solid malignancies. J Pediatr Surg. 2009;44:377–380. doi: 10.1016/j.jpedsurg.2008.10.088. [DOI] [PubMed] [Google Scholar]
- Lee E, Lim SJ. The association of increased lung resistance protein expression with acquired etoposide resistance in human H460 lung cancer cell lines. Arch Pharm Res. 2006;29:1018–1023. doi: 10.1007/BF02969286. [DOI] [PubMed] [Google Scholar]
- Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EA. Vaults: a ribonucleoprotein particle involved in drug resistance. Oncogene. 2003;22:7458–7467. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Inazawa J, Inoue K, Misawa S, Kashima K, Adachi H, et al. Assignment of the human renal dipeptidase gene (DPEP1) to band q24 of chromosome 16. Cytogenet Cell Genet. 1992;59:258–260. doi: 10.1159/000133263. [DOI] [PubMed] [Google Scholar]
- Nicolini FE, Mauro MJ, Martinelli G, Kim DW, Soverini S, Muller MC, et al. Epidemiologic study on survival of chronic myeloid leukemia and Ph(+) acute lymphoblastic leukemia patients with BCR-ABL T315I mutation. Blood. 2009;114:5271–5278. doi: 10.1182/blood-2009-04-219410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D, Jimmo SL, Raymond DR, Wilson LS, Carter RL, Maurice DH. Protein kinase A phosphorylation of human phosphodiesterase 3B promotes 14-3-3 protein binding and inhibits phosphatase-catalyzed inactivation. J Biol Chem. 2007;282:9411–9419. doi: 10.1074/jbc.M606936200. [DOI] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A, Wetterbom A, Feuk L, Gyllensten U. Global and unbiased detection of splice junctions from RNA-seq data. Genome Biol. 2010;11:R34. doi: 10.1186/gb-2010-11-3-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, et al. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M, Harismendy O, Nakano M, Frazer KA, Bafna V. Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics. 2011;27:1068–1075. doi: 10.1093/bioinformatics/btr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chien J, Smith DI, Ma J. FusionHunter: identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011;27:1708–1710. doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.