Abstract
BRCA1 is a breast and ovarian cancer-specific tumor suppressor that seems to be involved in transcription and DNA repair. Here we report that BRCA1 exhibits a bona fide ubiquitin (Ub) protein ligase (E3) activity, and that cancer-predisposing mutations within the BRCA1 RING domain abolish its Ub ligase activity. Furthermore, these mutants are unable to reverse γ-radiation hypersensitivity of BRCA1-null human breast cancer cells, HCC1937. Additionally, these mutations within the BRCA1 RING domain are not capable of restoring a G2 + M checkpoint in HCC1937 cells. These results establish a link between Ub protein ligase activity and γ-radiation protection function of BRCA1, and provide an explanation for why mutations within the BRCA1 RING domain predispose to cancer. Furthermore, we propose that the analysis of the Ub ligase activity of RING-domain mutations identified in patients may constitute an assay to predict predisposition to cancer.
Germ-line mutations in breast cancer susceptibility genes BRCA1 and BRCA2 predispose carriers mostly to breast cancer but also to other cancers (1). Although breast cancer occurs mainly as a sporadic disease, genetic predisposition accounts for 5–10% of all breast cancer cases (2, 3). BRCA1 is one of two familial breast cancer genes identified to date (4, 5). Human BRCA1 encodes a 1863-aa-long nuclear phosphoprotein, which, at its extreme C terminus, contains two BRCT repeats that are present in a large number of DNA damage-responsive cell-cycle checkpoint proteins (6). The C terminus of BRCA1 also harbors a transcription activation domain (7, 8). Thus, BRCA1 has been postulated to function in transcription and DNA repair (9, 10).
Interestingly, the extreme N terminus contains a RING domain, and such domains have been documented recently to exhibit ubiquitin (Ub) protein ligase (E3) activity (10). About 36% of all BRCA1 mutations constitute missense mutations (of those, 5.2% are polymorphisms, 7.8% are deleterious, and 87% are unclassified variants), which occur throughout the whole protein sequence, including the N-terminal RING domain (A. Deffenbaugh, personal communication). In this work, we have investigated the role of the RING domain with regard to the biochemical and biological functions of BRCA1. Analyzing cancer-predisposing mutations within the RING domain by in vitro Ub ligase and in vivo γ-radiation (IR) protection assays, we have detected a good correlation and propose that mutations within the BRCA1 RING domain predispose to cancer because they inactivate BRCA1 Ub protein ligase activity.
Materials and Methods
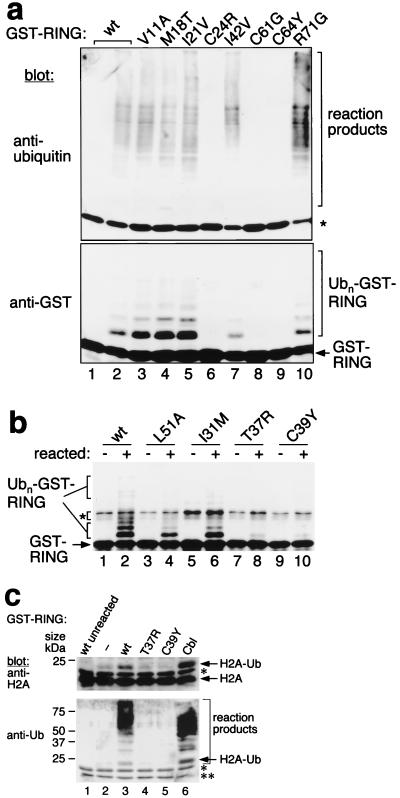
In Vitro Ubiquitination Reactions.
Expression and purification of glutathione S-transferase (GST) fusion proteins, E1 and Ubc4, as well as ubiquitination reactions, were performed as described (11), except that GST fusion proteins were not eluted from glutathione-Sepharose beads for the experiment in Fig. 2a and that 1 μg of histone H2A from calf thymus was included in the experiment presented in Fig. 2c. The monoclonal Ab to Ub was from Zymed (13–1600). The difference in distribution of Ub adducts in Fig. 2a (Top and Bottom) is likely because of the fact that any given polyubiquitinated GST-RING has only one site for anti-GST but multiple epitopes for anti-Ub Ab binding. The monoclonal Ab to histone H2A was from Santa Cruz Biotechnology (8674) and the immunoblot in Fig. 2c was developed with protein G coupled to horseradish peroxidase.
Figure 2.
Ub-ligase activity of the BRCA1 RING finger. (a) Ubiquitination reactions were performed with GST fusion proteins containing BRCA1 wt or mutant RING fingers, E1, Ubc4(E2), Ub, and ATP for 0 min (lane 1) or 90 min (lanes 2–10). Ub-protein conjugates were resolved by reducing SDS/PAGE and successively analyzed by immunoblot analysis by using an Ab to Ub (Upper) or to GST (Lower). The positions of GST-RING and of reaction products (including Ubn-GST-RING) are indicated. The asterisk (Upper) indicates crossreaction between the Ab to Ub and the GST fusion proteins. (b) As in a, reactions were performed for 0 min (−) or 90 min (+) and developed with an Ab to GST. The asterisk indicates crossreacting bands. Unmodified and ubiquitinated GST-RINGs are indicated. (c) As a, reactions also contained histone H2A and were developed with Ab to H2A (Upper) or to Ub (Lower). The single asterisk in both panels indicates Ab crossreactivity to Ubc4. The double asterisk in the lower panel indicates Ab crossreactivity to H2A. Protein sizes according to prestained markers (Bio-Rad) are indicated on the left.
Plasmid Constructions.
The sequence encoding wild-type (wt) BRCA1 amino acids 1–78 was cloned into the bacterial expression vector pT7-GT by PCR. All N-terminal BRCA1 missense mutations were derived from this construct by using the CLONTECH transformer site-directed mutagenesis kit. We used the following primers (5′ to 3′): for V11A, CGCGTTGAAGAAGCACAAAATGTCAT; for M18T, GTCATTAATGCTACGCAGAAAATCTT; for I21V, GCTATGCAGAAAGTCTTAGAGTGTC; for C24R, GAAAATCTTAGAGCGTCCCATCTGTC; for I31 M, GTCTGGAGTTGATGAAGGAACCTGTCTC; for T37R, GAACCTGTCTCCAGAAAGTGTGACC; for C39Y, GTCTCCACAAAGTATGACCACATATTTT; for I42V, AGTGTGACCACGTATTTTGCAAAT; for L51A, TTGCATGCTGAAAGCTCTCAACCAGAAG; for C61G, GGGCCTTCACAGGGTCCTTTATGTA; for C64Y, GTGTCCTTTATATAAGAATGATATAAC; and for R71G, TATAACCAAAGGGAGCCTACAAG. For expression in bacteria, these BRCA1 fragments (wt and mutants) then were cloned into expression vector pET-28a (Novagen). To generate the retroviral vectors, the BRCA1 fragments were lifted from the pET-28a constructs into pCL-MFGΔ-BRCA1 (12). To generate the Δ2–76 mutant, the NcoI site at position 1 in pCL-MFGΔ-BRCA1 was blunted (with the Klenow fragment of DNA polymerase) and ligated to an RsaI site at position 227. The Δ2–473 deletion mutant was generated by ligating the blunted NcoI site at position 1 to the blunted AflII site (Klenow fragment) at position 1419 in pCL-MFGΔ-BRCA1. The Δ1528–1778 mutant was generated by ligating the Ecl136II at position 4579 to the recessed XcmI site (T4 DNA polymerase) at position 5327 in the same vector.
Abs, Immunoprecipitation, and Immunoblot Analyses.
BRCA1 Ab-D and Ab-C have been described (13). Monoclonal Ab SD 118 was a gift from R. Scully (14). Immunoprecipitation reactions (native conditions) and immunoblot analysis were performed as described (12).
Cell Culture, Retroviral Infections, and Cell-Cycle Analysis.
HCC1937 cells were grown in RPMI medium 1640 (GIBCO/BRL)/10% (vol/vol) FBS in a 5% CO2 atmosphere. HCC1937 cells stably expressing wt BRCA1 as well as various mutant forms were generated by use of retrovirus-mediated gene transfer. Briefly, 150 μg of the retroviral vectors harboring BRCA1 inserts were cotransfected transiently with 100 μg of a plasmid encoding gag pol (a gift from Nik Somia, University of Minnesota, Minneapolis) and 50 μg of a plasmid encoding VSV-G (15) proteins into four 15-cm plates of 293T cells by using the calcium phosphate method. The supernatants were harvested at about 24 and 48 h after medium change after transfection and were subjected to a 0.45-μm filtration. Viruses were concentrated by ultracentrifugation at 19,400 rpm in an SW-28 rotor (Beckman) for 2 h at 21°C. For V11A, I21V, I31M, I42V, R71G, Δ2–76, Δ2–473, Δ1528–1778, and wt BRCA1, four 15-cm plates of 293T were transfected (1 virus preparation). The concentrated virus from each preparation was divided into 4 aliquots, and HCC1937 cells were infected in succession 4 times with 1 aliquot each. For M18T, C24R, T37R, C39Y and C61G, two sets of four 15-cm plates of 293T were transfected. Cells were infected 8 times with 1 aliquot each to obtain similar steady-state levels of BRCA1 protein compared with the other cultures. Cell-cycle analyses were performed as described (13).
IR Protection Assay.
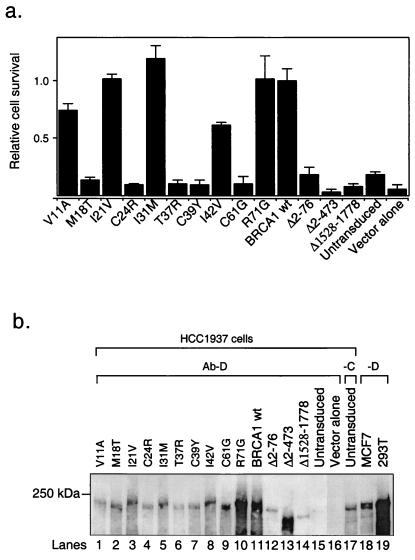
Cultures were passaged at the same passage number and the same confluency before each experiment. For the assay, cells were seeded at 20,000 cells per T25 tissue-culture flask, irradiated with 4 Gy (J/kg) at least 24 h after seeding, and kept subsequently in culture for 21 days before colonies were stained and counted (irradiated cultures). In parallel, 1,000 cells from each culture were seeded into T25 flasks and kept in culture for the same length of time without IR (nonirradiated cultures). Typically, about 30–50 colonies survived per flask for BRCA1 wt-reconstituted cells in both cases. For each experiment, cells were plated in triplicate or quadruplicate flasks. To determine the relative IR resistance (relative cell survival), the ratios of colony numbers per flask of irradiated cells to colony numbers of nonirradiated cells were calculated (pairwise) for individual cultures in each experiment. The mean and SEM values of all ratios (n = number of ratios per culture) were calculated for each reconstituted culture (V11A, M18T, I21V, C24R, T37R, C39Y, I42V, C61G, R71G and empty virus-transduced HCC1937 cells, n = 3; I31 M, n = 10; BRCA1 wt, n = 16; Δ2–76, Δ2–473, and Δ1528–1778, n = 7; untransduced HCC1937 cells, n = 13).
Results
Ub Protein Ligase Activity.
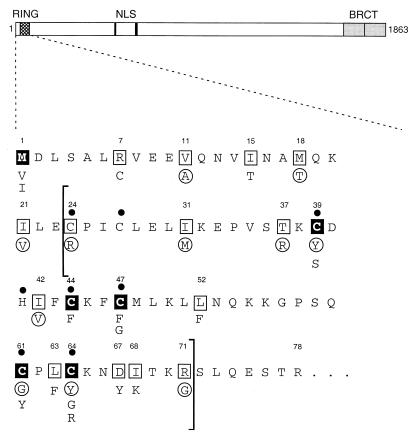
Fig. 1 displays the N-terminal 78 aa of BRCA1 encompassing the RING domain (amino acids 24–71). The functional consequences of missense mutations within the BRCA1 RING domain are unclear but some of them are known to predispose to cancer. Interestingly, all five cancer-predisposing mutations identified so far within the RING domain affect the RING-finger consensus motif (www.nhgri.nih.gov/Intramural_research/Lab_transfer/bic/; last date accessed August 15, 2000). We therefore sought to determine whether the BRCA1 RING domain harbors intrinsic E3 Ub protein ligase activity and, if so, whether cancer-predisposing mutations within that domain abrogate the ligase activity. Consequently, these mutations were analyzed (i) in vitro for their effects on Ub protein ligase activity, and (ii) in vivo with regard to their effects on the capability of BRCA1 to confer resistance to radiation hypersensitivity.
Figure 1.
BRCA1 N-terminal protein sequence (amino acids 1–78). A line diagram showing the domains of the BRCA1 protein is depicted at the top. The N-terminal sequence (amino acids 1–78) is represented below, as indicated by the dashed lines. The RING domain [residues 24–71 defined by comparison to the c-Cbl RING finger structure (11)] is bracketed. Numbers above individual amino acids denote the position of each residue. Filled circles indicate putative zinc-binding residues, filled squares represent cancer-predisposing mutations, and open squares represent unclassified variants. Amino acid substitutions found in patients are indicated below each residue. Circled residues denote the mutation analyzed (see also Table 1).
To determine the E3 Ub-ligase activity of the RING finger of BRCA1, GST fusion proteins encompassing residues 2–78 of wt human BRCA1 were generated. Purified, bacterially expressed GST-RING fusion protein was assayed for its ability to stimulate the synthesis of stable Ub conjugates (11). Reaction products were analyzed by immunoblot analysis using Abs to Ub (Fig. 2a Upper) or to GST (Fig. 2a Lower). High molecular weight Ub protein conjugates, including conjugates with the GST fusion protein itself (Fig. 2a Lower), were formed in the presence of the wt BRCA1 RING finger (Fig. 2a lanes 1 and 2), in an E1- and E2-dependent manner (data not shown), consistent with recently published results (16). This assay then was used to examine the effect of BRCA1 RING finger mutations found in cancer patients.
All three cancer-predisposing mutations analyzed abolished the E3 activity of the BRCA1 RING finger (Fig. 2 a, C61G and C64Y, and b, C39Y). Mutation of a fourth putative zinc-binding residue (Fig. 2a, C24R) was also detrimental to ubiquitination. Interestingly, the T37R mutant was similarly inactive, even though there is no amino acid conservation at the corresponding position in other RING fingers. Two additional cancer-predisposing mutations have been identified recently (C44F and C47F/G; Fig. 1), and because they both comprise putative zinc-binding residues, they also are expected to abolish E3 activity. Conversely, mutations at three positions that are not conserved among RING fingers (Fig. 2 a, I42V and R71G, and b, I31M) as well as at positions N-terminal to the RING finger (Fig. 2a, V11A, M18T, and I21V) did not impair E3 activity. These results therefore represent a strong correlation between loss of E3 Ub ligase activity of BRCA1 RING finger mutants and cancer predisposition.
A tryptophan (Trp) residue that is conserved in a subset of RING fingers harboring E3 activity is critical for E2 binding and ubiquitination (11). However, this Trp residue is not present in the BRCA1 RING domain. Instead, another bulky hydrophobic residue occupies the corresponding position leucine (Leu)-51 in BRCA1, and its mutation to alanine had deleterious effects that ranged from severe to moderate in different experiments (Fig. 2b, lane 4, and data not shown), suggesting that this Leu residue is also important for optimal E3 Ub ligase activity. We conclude that all four amino acid substitutions analyzed that affect the RING finger consensus motif of BRCA1 (including three cancer-predisposing mutations) as well as T37R, abolish ligase activity, whereas the other mutations analyzed within and N-terminal of the RING domain constitute unclassified variants and behave like wt BRCA1.
It has been well established that RING-type E3s can ubiquitinate substrates and themselves in vitro and in vivo. However, it can be expected that interaction between a RING finger and an E2 occasionally may serve solely for modification of the RING- finger protein itself. To determine whether BRCA1 is a bona fide E3, we tested its ability to stimulate ubiquitination of a heterologous protein, histone H2A. The results in Fig. 2c show that E1 and E2 alone can ubiquitinate H2A weakly, and that GST fusions with either the wt BRCA1 RING or the c-Cbl RING (11) significantly stimulate that reaction, whereas the BRCA1 mutants T37R and C39Y fail to do so.
Protection from IR.
We next wanted to correlate the effects of the mutations on the intrinsic E3 Ub ligase activity of the BRCA1 RING domain to their effects on BRCA1 function in vivo. To this end, we stably reconstituted HCC1937 cells, which have truncated BRCA1 alleles and are hypersensitive to IR, with wt or mutant BRCA1 harboring individual N-terminal mutations (17–19). As controls, we analyzed deletion mutants of BRCA1 lacking either the entire RING domain (Δ2–76), a larger region extending to amino acid 473 (Δ2–473), or a region close to the C terminus, including the N-terminal and part of the C-terminal BRCT repeat (Δ1528–1778; ref. 20). The mutations specified in Fig. 1 were engineered into the viral expression vector pCL-MFGΔ-BRCA1 containing full-length BRCA1 (12). Viral stocks were prepared by transient transfection of human embryonic kidney (293T) cells. HCC1937 cells were infected with the various recombinant viruses and assayed for increased IR resistance. Fig. 3a shows that infection with wt BRCA1 as well as the mutants V11A, I21V, I31 M, I42V, and R71G increased IR resistance of HCC1937 cells. Importantly, M18T, T37R, the two cancer-predisposing mutations (C39Y and C61G), and C24R (which also affects the RING finger motif), as well as all deletion mutants (Δ2–76, Δ2–473, and Δ1528–1778) failed to reverse IR hypersensitivity. Similarly, a mutation at cysteine residue 64 (C64G) also has been reported to be deleterious in this assay (18). The inability of the deletion mutants to enhance IR resistance further underscores the importance of the RING and BRCT domains for BRCA1 function. Taken together, the RING-domain mutants reveal a strong correlation between in vitro Ub protein ligase activity of BRCA1 and in vivo function for protection from IR hypersensitivity. All mutants that abolish Ub ligase activity are nonfunctional in reversing IR hypersensitivity, whereas wt and all other RING-domain mutants are active in ubiquitination and confer IR resistance (Table 1).
Figure 3.
Effect of BRCA1 RING mutations on protection from radiation hypersensitivity. HCC1937 cells were stably reconstituted with either wt BRCA1 or different mutants by using retroviral vectors. (a) Relative cell survival after IR. The mean values (and SEM) of the ratio of the number of colonies with and without IR are shown (1.0 is defined arbitrarily for the mean value of wt BRCA1). (b) Steady-state levels of the BRCA1 proteins in HCC1937 cells. BRCA1 was immunoprecipitated from lysates of cultures that were harvested at comparable confluence (between 70% and 100% confluent). MCF7 cells were only about 50% and 293T cells about 70% confluent when harvested; therefore, BRCA1 from MCF7 cells is relatively underrepresented (lane 18). BRCA1 was immunoprecipitated by using Ab-D (lanes 1–16) or Ab-C (lane 17) from lysates of HCC1937 cells or Ab-D from lysates of MCF7 (lane 18) and 293T cells (lane 19). All immunoprecipitates were separated by SDS/PAGE and analyzed by Western blot analysis using Ab SD 118. Lanes 1–10, individual missense mutations within the N terminus of BRCA1, as indicated; lane 11, wt BRCA1; lanes 12–14, truncated BRCA1 species used as controls; lanes 15 and 17, untransduced HCC1937 cells; lane 16, HCC1937 cells infected with the empty vector; and lanes 18 and 19, endogenous BRCA1 from MCF7 and 293T cells, respectively. The 250-kDa protein marker is indicated on the left.
Table 1.
Summary of Ub-ligase activity and in vivo IR protection function of wt and mutant BRCA1
| Mutant | Intrinsic Ub protein ligase activity | Increase in IR resistance |
|---|---|---|
| V11A | + | + |
| M18T | + | − |
| I21V | + | + |
| RING domain | ||
| C24R | − | − |
| I31M | + | + |
| T37R | − | − |
| C39Y | − | − |
| I42V | + | + |
| C61G | − | − |
| C64Y | − | ND (−*) |
| R71G | + | + |
| wt | + | + |
ND (−*), not done in the current analysis, but a mutation at this amino acid position (C64G) has been reported previously to impair the IR protection function of BRCA1 (18).
The inability of some mutants to confer IR resistance was not caused by low or absent protein levels (Fig. 3b). BRCA1 was immunoprecipitated from lysates of the various HCC1937 cultures by using Ab-D, which recognizes an epitope at the very C terminus of BRCA1 (13), and subsequently was subjected to immunoblot analysis using Ab SD 118, which recognizes an epitope within the BRCA1 region 758-1313 (14). Fig. 3b shows that BRCA1 protein was detectable in all reconstituted HCC1937 cells (lanes 1–14). The levels of various mutant proteins are different but there is no apparent correlation between the amount of the protein and its biological effect. No BRCA1 protein was detected by Ab-D in lysates of untransduced (lane 15) or vector alone-infected cells (lane 16). Ab-C, which is directed against epitope 768–793 (13), detected a slightly faster migrating form of BRCA1 in untransduced cells, probably representing the endogenous, C-terminal-truncated form present in HCC1937 cells (lane 17; refs. 18 and 19). For comparison, BRCA1 was immunoprecipitated from lysates of MCF7 and 293T cells (lanes 18 and 19, respectively). BRCA1 reconstitution in HCC1937 cells also was confirmed in each case by Northern blot analysis (data not shown).
Cell-Cycle Checkpoint Reconstitution.
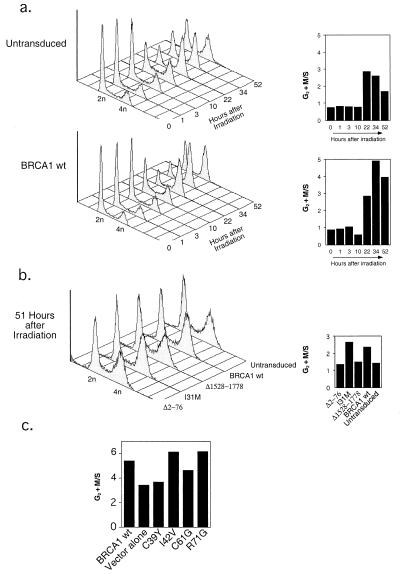
DNA damage-responsive cell-cycle checkpoints are important for maintaining genomic stability (21). It has been reported previously that primary mouse cells carrying two mutant Brca1 alleles lacking exon 11 (encoding nearly 60% of the BRCA1 protein) are defective in a G2 + M cell-cycle checkpoint (22). Furthermore, ectopic expression of BRCA1 in certain cell lines leads to accumulation in G2 + M (23). To test whether expression of BRCA1 in HCC1937 cells affects cell-cycle progression, we compared the cell-cycle profiles of untransduced or BRCA1-reconstituted HCC1937 cells at various time points after IR with 4 Gy. As shown in Fig. 4a, cells expressing wt BRCA1 progress slower through G2 + M compared with untransduced cells (or cells that were transduced with the green fluorescent protein; data not shown). The ratio of numbers of cells in G2 + M to S is shown in each case. Similar results were obtained after IR with higher doses (data not shown). In addition, the I31M mutant, which is positive for Ub protein ligase activity and increase of IR resistance, also delayed progression through mitosis, whereas BRCA1 forms carrying the Δ2–76 or Δ1528–1778 deletions failed to do so (Fig. 4b). We also tested the cell-cycle progression of cancer-predisposing mutants C39Y and C61G. The results shown in Fig. 4c reveal that HCC1937 cells reconstituted with BRCA1 species containing these mutations progress more rapidly through cell cycle as compared with mutants (I42V and R71G) that are not conserved in RING finger motifs. These results demonstrate that stable expression of BRCA1 by retrovirus-mediated gene transfer in HCC1937 cells is capable of restoring a G2 + M checkpoint, which may contribute to IR resistance.
Figure 4.
Restoration of a G2 + M checkpoint in HCC1937 cells by BRCA1. (a) Cell-cycle profiles of untransduced and BRCA1 wt-reconstituted cells at various times after IR. Cells were irradiated with 4 Gy 2 days after plating and analyzed for cell-cycle distribution at the times indicated (hours after irradiation). The 2n and 4n DNA contents are shown. To better illustrate the progression of the BRCA1-reconstituted cells through G2 + M, we expressed the ratio of cells in G2 + M to cells in S phase (G2 + M/S) for each time point (Right). The difference in G2 + M progression between untransduced (or GFP- or vector alone-transduced) and BRCA1-reconstituted HCC1937 cells was obtained reproducibly in independent experiments. (b) To test the specificity of a G2 + M checkpoint restoration by BRCA1, we analyzed the cell-cycle profiles of HCC1937 cells reconstituted with wt BRCA1 or the I31M, Δ2–76, and Δ1528–1778 mutants at similar time points after IR, as in a. Shown are the cell-cycle profiles (Left) and the G2 + M/S ratios (Right) at 51 h after IR. (c) Ratios of G2 + M/S of HCC1937 cells reconstituted with wt BRCA1 or C39Y, C61G, I42V, and R71G mutants at 46 h after IR (as in a and b, cell-cycle progression of the cell cultures was analyzed at various time points after IR). Similar results were obtained in an independent experiment.
Discussion
The results presented here establish a direct link between the Ub ligase activity of the BRCA1 RING domain and IR hypersensitivity protection by BRCA1. This correlation prompted us to speculate that mutations such as C39Y, C61G, and C64Y (Fig. 1) predispose to cancer because they inactivate BRCA1 Ub protein ligase activity. We therefore predict that Ub ligase activity is needed for BRCA1 to execute its function as a caretaker in the DNA damage-response pathway. Mice homozygous for Brca1 exon 11 deletion exhibit genetic instability caused by defects in DNA damage repair, loss of a G2 + M checkpoint, and centrosome amplification (22). Accordingly, we find decelerated progression through mitosis in irradiated HCC1937 cells caused by BRCA1 reconstitution. It is conceivable that transcription activation function of BRCA1 is required in the DNA repair pathway. Indeed, it has been shown that BRCA1 is involved in the transcription-coupled repair of oxidative DNA damage (24). In response to various types of DNA damage such as IR, BRCA1 is hyperphosphorylated, possibly leading to interaction with other proteins (refs. 25–29; H.R., unpublished data). The Ub protein ligase activity may be required for executing a function in the repair process, for example, by targeting protein(s) for degradation. Alternatively, phosphorylated BRCA1 protein itself could become the target of ubiquitination and subsequent degradation. Furthermore, mutations within the RING domain of BRCA1 could affect the interaction with other proteins such as BAP1, which is a Ub hydrolase, and BARD1, which also contains a RING finger (30, 31). It has been suggested that at least one of the cancer-predisposing mutations abrogates homodimer formation of the BRCA1 RING domain (32). RING finger proteins (Rad18 and Rad5) and interacting E2s (Rad6/Ubc2 and Ubc13, respectively) have been implicated in DNA repair in budding yeast as well. It may be interesting to determine whether these proteins and BRCA1 operate via similar pathways—targeting themselves or specific substrates for modification with Ub or Ub-like proteins as a means to regulate protein levels, activity, or subcellular localization.
Finally, we present a simple in vitro assay in which a negative result (lack of Ub protein ligase activity) may be predictive for cancer predisposition. This predictive value can be tested by establishing the relationship between the unclassified variant mutations and cancer predisposition. The Ub ligase activity associated with the RING domain of BRCA1 further adds to the biochemical diversity of the BRCA1 protein, ranging from transcription to DNA repair.
Acknowledgments
We thank Tal Kafri (University of North Carolina, Chapel Hill, NC) and Nik Somia for scientific advice and reagents; Ralph Scully (Dana–Farber Cancer Institute, Boston) for BRCA1 Ab SD 118; Gail Tomlinson (University of Texas Southwestern Medical Center, Dallas) for providing HCC1937 cells; David Chambers, Barbara Kahn, and Ann Atzberger for technical help; Amie Deffenbaugh of Myriad Genetics (Salt Lake City) for discussion; and Beth Coyne for help with the manuscript. We are also grateful to CellZome GmbH (Heidelberg, Germany) for their support. This work was supported by fellowships from the California Breast Cancer Research Program (4FB-0102 to H.R.); the American Cancer Society, California Division (2-41-98) and Leukemia and Lymphoma Society (formerly Leukemia Society of America) (3104-01 to C.A.P.J.); the Doris A. Howell Foundation (to D.H.); and by support from the U.S. Public Health Service Grant CA82683 (to T.H.). T.H. is a Frank and Else Schilling American Cancer Society Research Professor. I.M.V. is an American Cancer Society Professor of Molecular Biology and is also supported by grants from the National Institutes of Health, the March of Dimes, the Wayne and Gladys Valley Foundation, and the H. N. and Frances C. Berger Foundation.
Abbreviations
- Ub
ubiquitin
- IR
irradiation
- wt
wild-type
- GST
glutathione S-transferase
References
- 1.Rahman N, Stratton M R. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 2.Claus E, Risch N, Thompson W. Am J Hum Genet. 1991;48:232–242. [PMC free article] [PubMed] [Google Scholar]
- 3.Easton D F, Bishop D T, Ford D, Crockford G P, Consortium T B C L. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 4.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 5.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 7.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 8.Monteiro A N, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scully R, Livingston D M. Nature (London) 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freemont P S. Curr Biol. 2000;10:84–87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 11.Joazeiro C A P, Wing S S, Huang H-K, Leverson J D, Hunter T, Liu Y-C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 12.Ruffner H, Jiang W, Craig A G, Hunter T, Verma I M. Mol Cell Biol. 1999;19:4843–4854. doi: 10.1128/mcb.19.7.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruffner H, Verma I M. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 15.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 16.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott D W, Thompson M E, Robinson-Benion C, Tomlinson G, Jensen R A, Holt J T. J Biol Chem. 1999;274:18808–18812. doi: 10.1074/jbc.274.26.18808. [DOI] [PubMed] [Google Scholar]
- 18.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston D M. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 19.Tomlinson G E, Chen T T, Stastny V A, Virmani A K, Spillman M A, Tonk V, Blum J L, Schneider N R, Wistuba I I, Shay J W, et al. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 20.Koonin E V, Altschul S F, Bork P. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 21.Weinert T. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Wagner K-U, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng C-X. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 23.MacLachlan T K, Somasundaram K, Sgagias M, Shifman Y, Muschel R J, Cowan K H, El-Deiry W S. J Biol Chem. 2000;275:2777–2785. doi: 10.1074/jbc.275.4.2777. [DOI] [PubMed] [Google Scholar]
- 24.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 25.Cortez D, Wang Y, Qin J, Elledge S J. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 26.Lee J S, Collins K M, Brown A L, Lee C H, Chung J H. Nature (London) 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 27.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 28.Thomas J E, Smith M, Tonkinson J L, Rubinfeld B, Polakis P. Cell Growth Differ. 1997;8:801–809. [PubMed] [Google Scholar]
- 29.Tibbetts R S, Cortez D, Brumbaugh K M, Scully R, Livingston D M, Elledge S J, Abraham R T. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen D E, Proctor M, Marquis S T, Gardner H P, Ha S I, Chodosh L A, Ishov A M, Tommerup N, Vissing H, Sekido Y, et al. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 31.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 32.Brzovic P S, Meza J, King M C, Klevit R E. J Biol Chem. 1998;273:7795–7799. doi: 10.1074/jbc.273.14.7795. [DOI] [PubMed] [Google Scholar]