Abstract
Reverse phase-solid phase extraction from Cassia alata leaves (CaRP) was used to obtain a refined extract. Higher than wild-type sensitivity to CaRP was exhibited by 16 haploid Saccharomyces cerevisiae mutants with defects in DNA repair and membrane transport. CaRP had a strong DPPH free radical scavenging activity with an IC50 value of 2.27 μg mL−1 and showed no pro-oxidant activity in yeast. CaRP compounds were separated by HPLC and the three major components were shown to bind to DNA in vitro. The major HPLC peak was identified as kampferol-3-O-β-d-glucoside (astragalin), which showed high affinity to DNA as seen by HPLC-UV measurement after using centrifugal ultrafiltration of astragalin-DNA mixtures. Astragalin-DNA interaction was further studied by spectroscopic methods and its interaction with DNA was evaluated using solid-state FTIR. These and computational (in silico) docking studies revealed that astragalin-DNA binding occurs through interaction with G-C base pairs, possibly by intercalation stabilized by H-bond formation.
Keywords: Cassia alata, astragalin, DNA binding, FTIR, Saccharomyces cerevisiae, antioxidant
1. Introduction
Cassia alata L. (Fabaceae) is used in traditional medicine mainly in the tropical areas of the world, such as Malaysia, Brazil, and Indonesia. The leaves of C. alata are used as an effective treatment against ringworm and also against other skin diseases such as eczema and chronic skin impurities [1]. The main pharmacological activities cited in the literature are: anti-inflammatory, analgesic, laxative, antifungal and antimicrobial [1,2]. C. alata leaves contain emodin, kaempferol, aloe-emodin, chrysophanol and isochrysophanol, rhein, ellagitannin, phenolic acid and cassiaxanthone, amongst other substances [1].
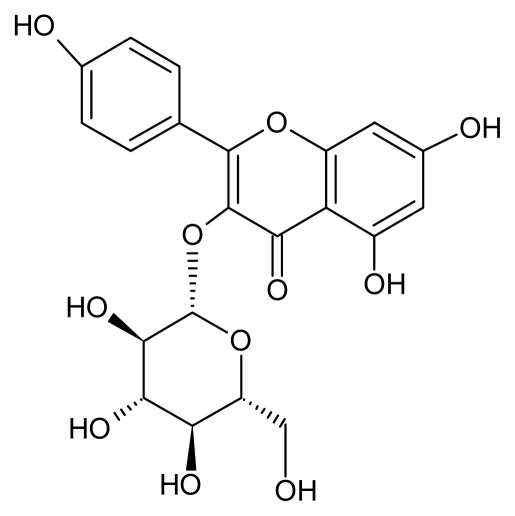
One of the important flavonoids of C. alata leaves is astragalin (AST) (Figure 1). AST is the main constituent of persimmon leaves but has also been found in different plants such as Eucommia ulmoides, Solenostemma argel and garlic, amongst others [3–6]. AST has lately aroused increased pharmaceutical interest because of its potential as anti-inflammatory agent, in addition to having antimicrobial activity with a MIC50 of 83 μg/mL against S. aureus (MRSA) [2,3] and also as an alternative medicine for allergic diseases [7].
Figure 1.
Kaempferol-3-O-β-d-glucopyranoside (astragalin).
Studies on molecular interaction of AST with some cell constituents are few and recent. An increase in activity of enzymes of red blood cell membranes (Na+K+ATPase, Ca2+ ATPase, Mg2+ ATPase and total ATPases) was observed on lead acetate-induced intoxication co-treated with AST (20 mg/kg per wt/day, 90 days) as compared to those of AST-untreated intoxicated rats [8]. AST induces proliferation of osteoblastic cells but does not have an antiestrogenic effect [6].
Deng and co-workers [9] isolated AST from Lotus leaf and showed its interaction with DNA in vitro, using fluorescence and ultraviolet absorption spectroscopy. However, detailed information on the mode of AST-DNA interaction and the type of adducts formed with nucleic acids is still not well understood and hence merits further in-depth analysis. The rareness of AST in nature has prevented its widespread study and biotechnological application and only very recently the successful synthesis of AST by enzymatic hydrolysis of tea seed extract has opened a new source for its production [3].
Since AST is known to be present in C. alata, the aim of this work was to extract a relative high concentration of the flavonoid from this plant for a possible use in alternative medicine. We first evaluated the antioxidant capacity of the RP-SPE extract from C. alata and determined the probable mechanism of action using in vitro interaction of CaRP with the eukaryotic model cell Saccharomyces cerevisiae. Identification of AST as one of the major C. alata metabolites was achieved by applying Fourier transform infra-red spectroscopy (FTIR), high-resolution mass spectrometry (ESI-qTOF-MS/MS) and H1 and C13 NMR analysis. An in- depth study of AST interaction with DNA was performed using FTIR spectroscopy and in silico docking studies. To our knowledge this is the first report that shows C. alata extract/compound in interaction with nucleic acids using spectroscopic methods and Saccharomyces cerevisiae null mutants as a model of study.
2. Results and Discussion
2.1. Antioxidant Activity of CaRP
Antioxidant and anti-infectious properties of C. alata extracts are well documented in the literature [1,2,10,11]. In order to increase the relative amount of probable bioactive compounds of the aqueous extract of C. alata leaves, we decided to refine the aqueous extract using Reverse Phase (C18)-Solid Phase Extraction obtaining a 17-fold concentrated extract named CaRP that showed much more intensive bands in TLC than the original aqueous extract (data not shown). CaRP also presented antimicrobial activity against Staphylococcus aureus, S. epidermidis, Bacillus subtilis and Pseudomonas aeruginosa (data not shown). We used the DPPH assay to also search for antioxidant properties of CaRP.
In order to calculate the IC50, calibration curves of each standard (ascorbic acid and Trolox) and of CaRP were determined by 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay as described in Material and Methods. The curves proved to be linear in the concentration range shown in Table 1 using linear regression and resulted in a correlation coefficient (r) >0.99 to standards and CaRP.
Table 1.
IC50 (mean ± standard deviation), ascorbate and Trolox equivalent antioxidant capacity of Cassia alata leaves (CaRP) using 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay.
| Compound | IC50 (μg.mL−1) | AEAC (g) | TEAC (g) | linear range (μg.mL−1) | slope | intercept | Correlation coefficient (r) |
|---|---|---|---|---|---|---|---|
| ascorbic acid | 3.99 ± 0.09 | 1.0000 | 0.8867 | 2.0–4.7 | −12.131 | 98.414 | 0.9975 |
| Trolox | 4.50 ± 0.08 | 1.1278 | 1.0000 | 2.7–5.3 | −12.275 | 105.230 | 0.9965 |
| CaRP | 2.25 ± 0.28 | 0.5639 | 0.5000 | 2.0–4.7 | −14.733 | 83.457 | 0.9989 |
AEAC, ascorbate equivalent antioxidant capacity; TEAC, Trolox equivalent antioxidant capacity.
At the conditions used, antioxidant activity of ascorbic acid was higher than that of Trolox. Interestingly, CaRP had higher antioxidant activity than ascorbic acid and Trolox, i.e., 0.5639 g of antioxidant activity of CaRP is equivalent to 1 g of ascorbic acid, or 0.5000 g of CaRP is equivalent to 1 g of Trolox. Compared to the literature, the IC50 of CaRP was about 50× less (IC50 of CaRP: 2.25 μg/mL; IC50 of ethanolic C. alata extract: 112.46 μg/mL) than ethanol-extracted without any refined method [11].
2.2. Yeast Sensitivity Assay
Since CaRP may be a valuable source of compounds, especially of AST, with its different biological and pharmacological effects, we decided to evaluate the effect of CaRP using Saccharomyces cerevisiae mutants lacking synthesis of endogenous antioxidants, DNA repair enzymes or membrane constituents as model cells to find possible targets that could hint at mechanisms of action.
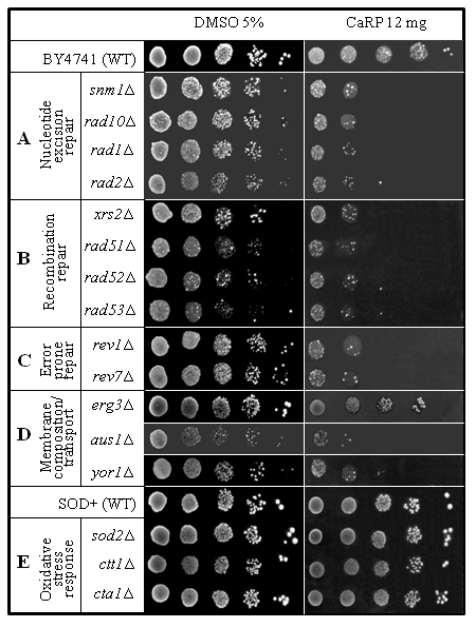
S. cerevisiae null mutants that lack only one gene in comparison to the isogenic wild type offer the possibility to study the relevance of a missing specific metabolic pathway on the response to the defined treatment with the chemical studied. The absence of the protein codified by that gene leads to specific responses described as phenotypes [12] that characterize the metabolic pathway in which it is involved, i.e., mutants in proteins involved in DNA repair are more sensitive to drugs that cause DNA damage that is repaired by that biological pathway. Therefore, this is a very powerful system to rapidly identify a putative mechanism of drug action in a complex cell [13]. Here we used a set of 10 yeast null mutants involved in DNA repair, representing the three so-called epistasis groups, which are thought to reflect three major DNA repair pathways: RAD3 (involved in nucleotide excision repair [NER], Figure 2A), RAD52 (recombinational repair, Figure 2B) and RAD6 (error-prone repair, Figure 2C) epistasis group genes [14–16]; 3 yeast mutants defective in membrane composition or transport across the membrane (Figure 2D); and 3 yeast mutants defective in oxidative stress protection mechanisms (Figure 2E).
Figure 2.
Sensitivity to CaRP of haploid yeast strains.
Although CaRP (12 mg/plate) did not show any cytotoxic effect in wild type strains (BY4741, SOD2), isogenic DNA repair mutants showed higher sensitivity when treated with CaRP (Figure 2A–C). Interestingly, all repair mutants of the three epistasis groups of DNA repair (RAD3, RAD52 and RAD6 epistasis group) showed some degree of sensitivity. These results suggest that CaRP contains one or more substances that interact with DNA individually or simultaneously in distinct ways, considering that the repair pathway seemed not specific. Additionally, all mutant strains in the RAD3 (nucleotide excision repair—NER, Figure 2A), RAD52 (homologous recombination, Figure 2B) and RAD6 (translesion repair, Figure 2C) epistasis group had similar sensitivity. The higher than wild-type sensitivity to pso2Δ (snm1Δ) (Figure 2B) suggested that some substance of CaRP might intercalate with DNA and induce lesions that link the two DNA strands. Evidence of this comes from some studies that showed pso2Δ mutant specifically blocked in repair of interstrand cross-links [13]. These results could be promising because many antitumoral drugs act by intercalating into DNA and forming interstrand cross-links, e.g., photoactivated psoralens, mitomycin C and cis-platinum [17].
Mutants defective in membrane transport such as aus1Δ (lacking the function of uptake of exogenous sterols and its subsequent incorporation into the plasma membrane) and yor1Δ (lacking multidrug transporter which mediates export of many different organic anions) had the same sensitivity as the DNA repair-mutant strains except for erg3Δ (lacking C-5 sterol desaturase) that was almost unaffected, i.e., showed a wild type response (Figure 2D). Erg3p is known to protect cells specifically from drugs inducing lipid peroxidation [18]. Lack of sensitivity in erg3Δ mutant, therefore, indicates that CaRP did not induce lipid peroxidation.
Conversely, mutants lacking endogenous antioxidants such as superoxide dismutase (sod2Δ) or catalase (ctt1Δ and cta1Δ) had identical response to CaRP as the isogenic wild type that is proficient in all these enzymes (Figure 2E). This result corroborates with the DPPH assay (Table 1) and suggests that CaRP in the applied concentrations is not pro-oxidant in these eukaryotic cells.
2.3. Bioactive DNA-Binding Compounds from CaRP
Since CaRP showed cytotoxicity in DNA repair-deficient yeast mutants and in strains with defects in membrane transport, we decided to test if CaRP compounds could bind to DNA in vitro. For this, centrifugal ultrafiltration sampling followed by HPLC analysis was used, a fast screening for bioactive compounds binding to DNA [19].
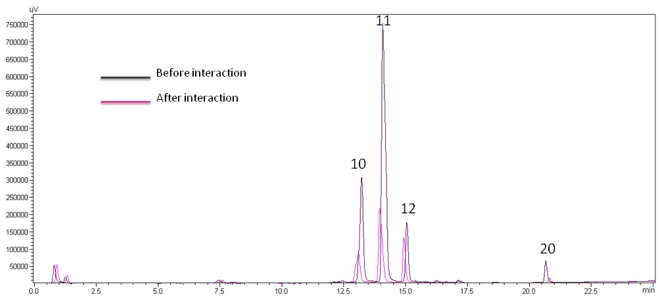
The Amicom filtrates from CaRP extract and from the mixed solution of CaRP + DNA were collected and analyzed by HPLC (Figure 3). Because the free concentration of ligands decreases in the solution after the interaction (DNA remains in the Amicon membrane and only unbound metabolites with medium polarity can pass through) the peak areas of these compounds decrease in the chromatogram; for those not DNA binding, there should be no or little change in their peak areas before and after the interaction. Therefore, comparing the chromatograms of the Amicon filtrates after the reaction of the control (without DNA) and with DNA, the DNA-binding compounds in the CaRP can be easily distinguished from those that do not have affinity to DNA.
Figure 3.
Bio fingerprinting chromatograms for CaRP at 254 nm using 50 μL CaRP (400 μg mL−1) (black line) and the mixed CaRP-DNA solution (red line) at a final concentration of 510 μg mL−1 in BPES buffer. Peaks are numbered according to the LC-microfractionation.
The binding degree of any component to DNA was calculated according to Zhou et al. [19]:
where the peak areas of a compound before (Ab) and after (Ac) the interaction with DNA in the HPLC chromatograms, were measured.
Figure 3 shows the bio fingerprinting chromatograms at 254 nm. The integration of the peak areas showed that 3 peaks in CaRP decreased significantly after interaction with DNA: the respective decreases were 64% for peak 10, 72% for peak 11, and 78% for peak 20.
2.4. Structural Identification
We chose to study by FTIR spectroscopy the CaRP fraction equivalent to peak 11 (Figure 3) in its interaction with calf thymus DNA since it is the major fraction (yield of 52%) and also showed cytotoxicity against prokaryotic cells, such as S. epidermidis and s. aureus [20].
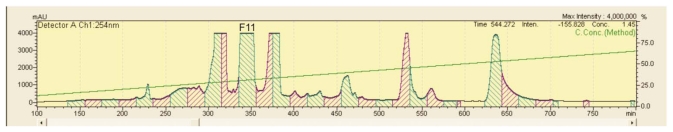
The equivalent fraction of HPLC peak 11 was obtained by LC-microfractionation (Figure 4). Identification was carried out using FTIR, high-resolution mass spectrometry (ESI-qTOF-MS/MS) and H1 and C13 NMR.
Figure 4.
Semi-preparative LC chromatogram of CaRP. F11 is equivalent to peak 11 in the analytical method (HPLC DNA binding).
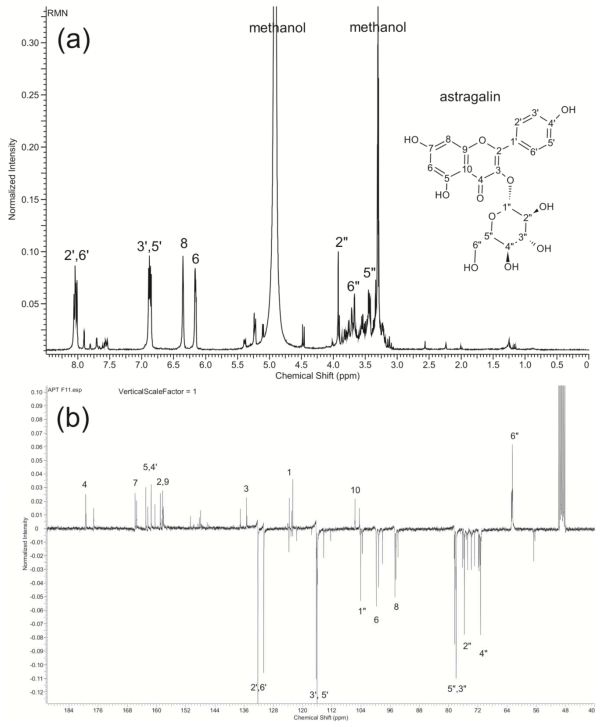
Fraction 11 (F11): yield of 0.523 mg/mg. Green powder, IR (KBr) cm−1: 3364 (OH), 1782 (C=O), 1656, 1607, 1505, 1452 (C=C, Ar), 1286 (=C–O–C), 1118 (C–OH); Negative high resolution ESI-qTOF-MS m/z 447.1187 [M-H]− (32%), MS-MS fragmentation of m/z 447.1187: 284.0699 [M-H-Glu]−, 255.0648, 227.0681; H1 NMR (MeOD) and C13 NMR (MeOD) were carried out and after comparison to data in the literature [3,4], F11 was identified as kaempferol-3-O-β-d-glucoside (astragalin) (Figure 5).
Figure 5.
(a) 1H-NMR and (b) 13C-NMR (APT) of astragalin in MeOD.
Only a little quantity of AST was isolated from various plants such as persimmon leaves, Eucommia ulmoides leaves, Nelumbo nucifera (Lotus leaves), Solenostemma argel, garlic leaf and shoot, Flaveria bidentis, Cuscuta chinensis, and the seeds of Centaurea schischkinii [3–7].
Although this compound has been isolated from C. alata leaves before [1,2,21], our results showed that solid phase extraction is an improved technique to obtain the enriched extract CaRP, having also the advantage of an easy and rapid purification method.
2.5. FTIR Spectra of AST-DNA Complex
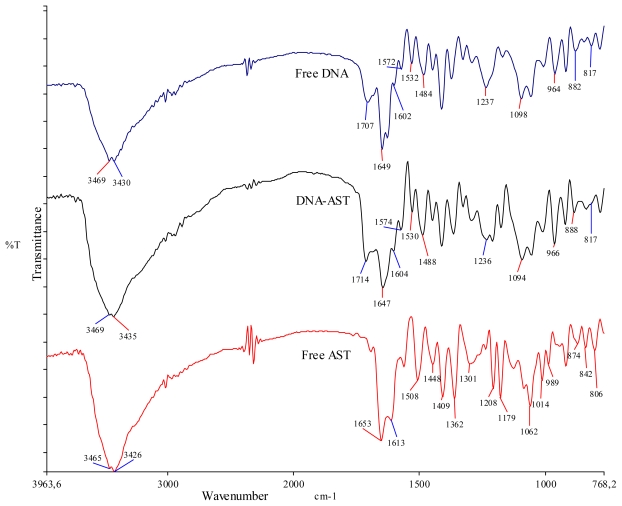
The infrared spectrum of AST-DNA mixture offered evidence for the direct binding of this compound to DNA (Figure 6 and Table 2). The vibrational frequencies of NH and OH groups of free DNA appeared around 3469–3430 cm−1, which is in accordance to Usha and co-workers [22], and in the AST-DNA complex had a little shift to 3469–3435 cm−1. Therefore, the vibrational frequencies of NH band in DNA and OH in the fraction has changed in the complex, evidencing the effective interaction of AST OH with NH of DNA bases, possibly through H-bond interaction.
Figure 6.
Fourier transform intra-red spectroscopy (FTIR) spectra of free DNA (top), DNA-AST (middle) and free astragalin (AST) (bottom).
Table 2.
The vibrational frequencies of major functional groups in free AST, free DNA and DNA-AST complexes.
| Free AST, Free DNA and complexes | Observed changes in the vibrational frequency of the functional groups and bases (cm−1) | |||||||
|---|---|---|---|---|---|---|---|---|
| OH | C=O | NH | PO−2 | Gua | Thy | Ade | Cyt | |
| Free AST | 3465–3426 | 1653 | - | - | - | - | - | - |
| Free DNA | 3469–3430 | 1707, 1649 | 3469–3430 | 1237, 1098 | 1707 | 1649 | 1602 | 1484 |
| DNA-AST | 3469–3435 | 1714, 1647 | 3469–3435 | 1236, 1094 | 1714 | 1647 | 1604 | 1488 |
The vibrational frequency of C=O of the free DNA at 1707 cm−1 shifted to 1714 cm−1 in the complex. According to Alex and Dupuis [23] further indication for H-bonding interaction of AST with DNA base pairs such G-C and A-T and with phosphate groups comes from major spectral changes of DNA in-plane-vibrations in the region of 1707–1080 cm−1 (Figure 6, Table 2). The band at 1707 cm−1 (Gua, Thy) related to mainly Gua, 1602 cm−1 (Ade, Cyt) related to mainly Ade, and 1484 cm−1 (Cyt, Gua) related to mainly Cyt, shifted towards higher frequency with the change in intensity in complex at 1714, 1604 and 1488 cm−1, respectively. The band at 1649 cm−1 (Thy, Gua, Cyt), mainly for Thy, shifted to lower frequency at 1647 cm−1. The observed changes in frequency can be related to AST interaction with Gua, Cyt, Ade and Thy bases. But there is some evidence that AST binds preferentially to Gua beyond the major shift at 1707 cm−1 (Gua C6=O stretching) to 1714 cm−1, where we can see also the shift of the band at 1572 cm−1 (purine N7 stretching) to 1574 cm−1 and shift of the band at 1532 cm−1 (in-plane vibration of C≡G) to 1530 cm−1.
Additional evidence for DNA-AST interaction is obtained from the spectral shifting of AST vibrational frequencies upon DNA binding. Besides the functional OH group of AST, the symmetrical and asymmetrical stretching is also essential in the binding of the aromatic ring to DNA. The frequency vibration of C=C stretching of free AST at 1653 (asymmetrical C=C stretching) and 1613 cm−1 (symmetrical C=C stretching) shifted to 1686 and 1628 cm−1, respectively. The out-of-plane C–H bending at 806 cm−1 in AST shifted to 795 cm−1 in the complex. Other minor changes (shifting/intensity variation) were also observed in AST at 1362 cm−1 (in plane OH bend), 1208 cm−1 (C–O stretching), 1508 and 1448 cm−1 (C–C stretching (in ring)) [24].
Minor variation in PO2 − asymmetric and symmetric vibration at 1237 to 1236 cm−1 and 1098 to 1094 cm−1, infrared B-marker bands at 882 cm−1 (sugar-phosphate stretch) and 817 cm−1 (phospho-diester mode) present in free DNA did not appreciably shift in the DNA-AST complex. This result shows that the DNA remains in the B-DNA conformation.
Deng and co-workers [9], using fluorescence and UV-vis absorption spectroscopy analyzed the interaction between Lotus leaf derived AST and DNA and computed the binding constant to K = 3.41 × 104 M−1 and concluded that AST could interact with DNA by intercalative binding. Their data corroborate with ours. By applying FTIR spectroscopy we could obtain the additional information that AST binding to DNA occurs preferentially with Gua-Cyt rather than Ade-Thy base pairs.
It is generally accepted that small molecules might bind to a DNA double helix by three modes: electrostatic binding, groove binding and intercalative binding [25]. Normally, small molecules have to some extent selectivity except for their binding to DNA by electrostatic interaction (along the external DNA double helix) [19]. Intercalative binding is usually affected by planarity of the bound candidates and groove binding is related to the two types of grooves in DNA, i.e., major and minor groove. In fact, each small molecule has a particular structure, which possibly has different binding modes with DNA and each mode resulting in a characteristic distortion of DNA, and this in theory giving rise to a specific pharmacological effect [26]. Indeed, in vitro studies of compound-DNA interaction allow valid predictions to the possible in vivo interactions with DNA and their cytotoxic/genotoxic consequences (Figure 2).
However, it should be stressed that we have no direct prove that AST reaches nuclear DNA. TOXNET database [27] has so far no entry related either to mutagenicity, genotoxicity nor to carcinogenicity of this compound. Our S. cerevisiae data (Figure 2) give some hint that nuclear uptake of CaRP compounds may occur. Whether it is AST or one of the metabolites cannot be answered by our experiments, although the significantly higher sensitivity of yeast DNA repair mutants and the abundance of AST (> 50%) in CaRP suggests this.
2.6. Docking Study
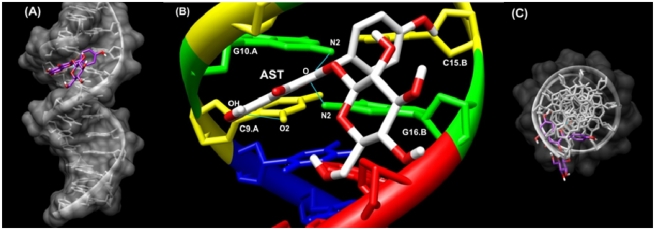
To determine its preferred binding sites on sequence of DNA [CGCGAATTCGCG (PDB ID:1D30)], AST was docked to DNA using HexServer [28]. The docking result is shown in Figure 7. The model shows that AST is surrounded by C9.A, G10.A, C11.A, C15.B, G16.B and A17.B of the oligonucleotide 1D30 (Figure 7). For the AST-1D30 complex, docking results indicated the presence of a hydrogen bond between H hydroxyl (C7) of AST and O2 of C9.A and O (C ring), which can form a hydrogen bond to both N2 of G10.A and N2 of G16.B (Figure 7). The binding energy of AST (Etotal −289.30) was found to be higher than that of kaempferol (Etotal −238.29) when docking with the same method/form and this result could be due to the fact that kaempferol only can form a hydrogen bond between O (C ring) and N2 of G10.A and N2 of G16.B (figure not shown).
Figure 7.
Docking structure between d(CGCGAATTCGCG)2 (PDB ID:1D30) and AST. (A) Surface representation of d(CGCGAATTCGCG)2 complexes with AST (Display sideways); (B) Close up view of d(CGCGAATTCGCG)2 complexes with AST represented in the stick style. Hydrogen bonds between d(CGCGAATTCGCG)2 and AST were calculated by Chimera software and are drawn in blue lines; (C) Surface representation of d(CGCGAATTCGCG)2 complexes with AST (Display top).
These docking data are compatible with our FTIR results that showed intercalation of AST into DNA (mainly with GC base pairs) stabilized by hydrogen bonds.
3. Experimental Section
3.1. Extraction Protocol
Fresh leaves (950 g) were freeze-dried and ground to obtain 320 g of dry material, which was extracted by decoction (1:20 p/v, 80 °C per 20 min) followed by filtration, rotary evaporation and freeze-drying to obtain 80 g of crude extract. Reverse phase-solid phase extraction was performed using the column Strata C18E 5 g/20 mL Giga Tubes (Phenomenex, USA). For each batch, 1 g of crude extract was diluted in 100 mL of distilled water. The cartridge was conditioned with acetone (25 mL) and washed with 5 mL of water before loading the sample (100 mL). It was then washed with 20 mL of water and eluted with 25 mL of ethyl acetate. After evaporation this eluent yielded the CaRP refined extract. The 80 g of crude extract yielded 4 g of CaRP that was stored in desiccators at room temperature in the dark until further use.
3.2. DPPH Assay
The free radical scavenging capacity of CaRP and of reference substances was determined using 2,2-diphenyl-1-picrylhydrazyl (DPPH) method according to Brand-Willians and co-workers [29]. Five concentrations of CaRP, ascorbic acid (AA), and Trolox in methanol were used to obtain the curves (Table 1). CaRP and standards (200 μL in the range of 2.66 to 7.09 μg mL−1) in methanol were added to 100 μL of DPPH (10 mg/100 mL MeOH) and allowed to stand for 30 min before absorbance was measured at 515 nm using a Molecular Devices (Sunnyvale, USA) spectrophotometer model Versamax Tunable Plate Reader. The DPPH solution (100 μL) and methanol (200 μL) were used as a negative control. This experiment was conducted in triplicate. Antioxidant activity was expressed as IC50 (inhibitory concentration in μg/mL necessary to reduce the absorbance of DPPH by 50% compared to the negative control). The lower the IC50, the higher was the antioxidant activity. Results were also expressed as AEAC (AA equivalent antioxidant capacity) or TEAC (Trolox equivalent antioxidant capacity) in grams and calculated as follows:
3.3. Strains and Media
The relevant genotypes of the yeast strain used in this work are given in Table 3. Media and solutions were prepared according to [30]. Complete medium (YPD) was used for routine growth of yeast cells and minimal medium (MM) was supplemented with the appropriate amino acids and uracil to yield SC (synthetic complete medium). Stationary (STAT) phase cultures with 2 × 108 cells/mL were obtained after inoculation of an isolated colony into liquid YPD and after 72 h incubation at 30 °C with aeration by shaking. Cells were washed and diluted in saline (0.9% NaCl, pH 5.0) before all treatments.
Table 3.
Yeast strains used.
| strain | genotype | Protein lacking | source |
|---|---|---|---|
| BY4741 | Matα his3Δ1 leu2Δ0 lys2Δ0 ura 3Δ0 | none | EUROSCARF |
| snm1Δ | Like BY4741 except snm1Δ::KanMx4 | 5′-3′ exonuclease activity | EUROSCARF |
| rad10Δ | Like BY4741 except rad10Δ::KanMx4 | Single-stranded DNA endonuclease (with Rad1p) | EUROSCARF |
| rad1Δ | Like BY4741 except rad1Δ::KanMx4 | Single-stranded DNA endonuclease (with Rad10p) | EUROSCARF |
| rad2Δ | Like BY4741 except rad2Δ::KanMx4 | Single-stranded DNA endonuclease | EUROSCARF |
| xrs2Δ | Like BY4741 except xrs2Δ::KanMx4 | Protein required for DNA repair; Mre11 complex component | EUROSCARF |
| rad5Δ | Like BY4741 except rad5Δ::KanMx4 | DNA helicase proposed to promote replication fork regression during postreplication repair | EUROSCARF |
| rad52Δ | Like BY4741 except rad52Δ::KanMx4 | Protein that stimulates strand exchange by facilitating Rad51p binding to single-stranded DNA | EUROSCARF |
| rad53Δ | Like BY4741 except rad53Δ::KanMx4 | Protein kinase, required for cell-cycle arrest in response to DNA damage | EUROSCARF |
| rev1Δ | Like BY4741 except rev1Δ::KanMx4 | Deoxycytidyl transferase; involved in repair of abasic sites and adducted guanines in damaged DNA by translesion synthesis (TLS) | EUROSCARF |
| rev7Δ | Like BY4741 except rev7Δ::KanMx4 | Accessory subunit of DNA polymerase zeta, involved in translesion synthesis during post-replication repair | EUROSCARF |
| erg3Δ | Like BY4741 except erg3Δ::KanMx4 | C-5 sterol desaturase, catalyzes the introduction of a C-5(6) double bond into episterol, a precursor in ergosterol biosynthesis | EUROSCARF |
| aus1Δ | Like BY4741 except aus1Δ::KanMx4 | Plasma membrane sterol transporter of the ATP-binding cassette family | EUROSCARF |
| yor1Δ | Like BY4741 except yor1Δ::KanMx4 | Plasma membrane ATP-binding cassette (ABC) transporter, multidrug transporter mediates export of many different organic anions | EUROSCARF |
| SOD+ | Matα his3Δ1 leu2Δ0 trp1-289 ura3-52 | None | E. B. Gralla, Los Angeles |
| sod2Δ | Like SOD + except sod2::TRP1 | Mn superoxide dismutase | E. B. Gralla, Los Angeles |
| ctt1Δ | Like SOD + except ctt1Δ::TRP1 | cytosolic catalase T | E. B. Gralla, Los Angeles |
| cta1Δ | Like SOD + except cta1Δ::TRP1 | catalase A present in peroxissomal matrix | E. B. Gralla, Los Angeles |
3.4. Sensitivity Assay in Haploid Yeast to CaRP
The sensitivity of different yeast strains (STAT cells of wild type [WT] and of isogenic mutants) was evaluated on freshly prepared solid SC medium containing either no drug or 12 mg of CaRP. Cells were diluted in 1:10 steps in sterile saline and 5 μL of each dilution (suspensions containing from 107 to 103 cell/mL) was placed on agar surface of SC solid medium containing the appropriate compound. Cellular growth on SC was determined after 5 days incubation at 30 °C. Photos were taken using a Canon PowerShot G10 camera. Photos represent one of at least three independent experiments with similar results.
3.5. HPLC Analyses
3.5.1. LC-UV DNA Binding of CaRP
This method was performed according to Zhou and co-workers [19] with slight modification. The centrifugal ultrafiltration was performed on an Eppendorf Refrigerated Centrifuge 5415R (Eppendorf, Germany). A 200 μL of CaRP (4 mg mL−1) and 300 μL of DNA solution (850 μg mL−1) were mixed in an Amicon centrifugal filter from Millipore (Bedford, MA, USA) with molecular weight cut-off of 10,000 Da and incubated at 37 °C in a thermo-mixer for 15 min. A blank sample was prepared with the addition of 300 μL of BPES buffer pH 7.0 (6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM EDTA, 185 mM NaCl) instead of DNA. Samples were centrifuged at 4000 rpm for 30 min at 10° C, and the filtrates were analyzed by HPLC Shimadzu LC-20 Prominence series (Japan). Separations were achieved on a Kinetex C-18 column Phenomenex (100 × 3 mm I.D.; 2.6 μm 100 Å) with MeCN-water (10:90 to 90:10; 30 min). The flow-rate was 1.0 mL.min−1; UV absorbance was measured at 254 and 347 nm. The sample injection volume was set at 5 μL.
3.5.2. LC-UV Microfractionation and Identification of the Fractions
The microfractionation of CaRP was performed according to Queiroz and co-workers [31]. CaRP was submitted to fractionation by LC performed on a Shimadzu LC-20AT Prominence with detector UV-Vis SPD-20A and collector FRC-10A (Japan). The separations were achieved on a Gemini C-18 column Phenomenex (150 × 10 mm I.D.; 5 μm 110Å) with MeCN-water (5:95 to 95:5; 1200 min). The sample injection volume was set at 250 μL (10 mg). The flow-rate was 0.2 mL min−1; UV absorbance was measured at 254 and 347 nm. Twenty fractions were collected for every peak with level >80,000 μV and width 10 s in plastic tubes (4 mL). After collection, all fractions were distributed in Eppendorf tubes (1.5 mL) and evaporated to dryness on a speedvac system (Eppendorf concentrator 5301, Germany). The fraction corresponding to peak 11 was identified using the LC analytical method described above. The semi-preparative chromatogram is shown in Figure 4.
Chromatographic preparation was repeated (about 50 injections) until obtaining 5 mg of dried mass of Fraction 11. The structural elucidation of this fraction was carried out by FTIR (KBr), HR MS/MS, 1H NMR and 13C NMR. A high resolution mass spectrum was recorded on a Waters ESI-Q-TOF mass spectrometer using the MassLynx V4.1 software package. 1H NMR and 13C NMR (APT) were recorded on a Bruker (300 MHz) NMR.
3.6. FTIR Analysis
Infrared spectra of dried fraction 11, obtained from the LC-microfractionation, were recorded on a Perkin Elmer Spectrum 400 FT-IR/FT-NIR spectrometer (Perkin Elmer, UK). Absorbance spectra were acquired at 4 cm−1 resolution and signal-averaged over 8 scans. For DNA interaction analysis, the fraction 11 (F11) was mixed with DNA separately in 1:2 ratio in BPES buffer pH 7.0 in sterile Eppendorf tubes; after a thorough mixing and overnight incubation at 37 °C, the samples were concentrated by speed-vac centrifugation (Eppendorf concentrator mod. 5301). DNA-F11 complexes were prepared and studied in solid-state using KBr pellet according to Usha and co-workers [22]. Spectra were processed using the SPECTRUM software from Perkin-Elmer. The de-convoluted parameters were set with a gamma value of 8.0 and a smoothing length of 90%.
3.7. Docking Study
The crystal structure of DNA-ligand complex was selected from Protein Data Bank PDB ID: 1D30 [32]. We docked AST onto the oligonucleotide extracted from the crystal structure. To determine the preferred binding sites on DNA and the binding energy of AST, docking studies were performed using HexServer [28]. HexServer is a fast Fourier transform (FFT)-based protein docking server to be powered by graphics processors. FFT-based approaches assume that the proteins (receptor) to be docked are rigid, but they sample densely all possible rigid-body orientations in the 6D search space [26]. The ligand structure was extracted from Pubchem (SDF: CID 5282102) file. To display docking results, we used UCSF Chimera 1.5.3 [33].
4. Conclusions
This study demonstrates that the C. alata leaf extract CaRP, obtained by reverse phase (C18)-solid phase extraction, exhibited potent DPPH free radical scavenging activity, was cytotoxic to 12 different S. cerevisiae DNA repair mutants and two mutants deficient in membrane transport. Using HPLC, three fractions of CaRP showed great affinity to bind DNA as revealed by a bio-fingerprinting chromatogram. Also, CaRP showed to be a good source of astragalin, which was shown, by FTIR and docking studies to interact with Gua-Cyt bases probably by intercalation stabilized mainly through H-bond formation.
Acknowledgments
Research supported by CNPq grant #480162/2007-7, #474460/2009-6, #555106/2010-1. S.T.S. and R.X. hold doctoral fellowship by CNPq, G.S. a fellowship of Iniciação Cientifica by ICB/UESC. G.G. is grateful for financial support by CNPq.
Abbreviations
- CaRP
Reverse phase-solid phase extract from Cassia alata leaves
- AST
astragalin
- MRSA
Methicillin-resistant Staphylococcus aureus
References
- 1.Hennebelle T., Weniger B., Joseph H., Sahpaz S., Bailleul F. Senna alata. Fitoterapia. 2009;80:385–393. doi: 10.1016/j.fitote.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Hazni H., Ahmad N., Hitotsuyanagi Y., Takeya K., Choo C.Y. Phytochemical constituents from Cassia alata with inhibition against methicillin-resistant Staphylococcus aureus (MRSA) Planta Med. 2008;74:1802–1805. doi: 10.1055/s-0028-1088340. [DOI] [PubMed] [Google Scholar]
- 3.Lee H.-B., Kim E.-K., Park S.J., Bang S.-G., Kim T.G., Chung D.-W. Isolation and anti-inflammatory effect of astragalin synthesized by enzymatic hydrolysis of tea seed extract. J. Sci. Food Agric. 2011;91:2315–2321. doi: 10.1002/jsfa.4457. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y., Xie Q., Fisher D., Sutherland I.A. Separation of patuletin-3-O-glucoside, astrgalin, quercetin, kaempferol and isorhamnetin from Flaveria bidentis (L.) Kuntze by elution-pump-out high-performance counter-current chromatography. J. Chromatogr. A. 2011;1218:6206–6211. doi: 10.1016/j.chroma.2011.01.058. [DOI] [PubMed] [Google Scholar]
- 5.Huang B., Ban X., Jingsheng H., Tong J., Tian J., Wang Y. Hepatoprotective and antioxidant activity of ethanolic extract of edible lotus (Nelumbo nucifera Gaertn.) leaves. Food Chem. 2010;120:873–878. [Google Scholar]
- 6.Yang L., Chen Q., Wang F., Zhang G. Antiosteoporotic compounds from seeds of Cuscuta chinensis. J. Ethnolpharmacol. 2011;135:553–560. doi: 10.1016/j.jep.2011.03.056. [DOI] [PubMed] [Google Scholar]
- 7.Kotani M., Matsumoto M., Fujita A., Higa S., Wang W., Suemura M., Kishimoto T., Tanaka T. Persimmon leaf extract and astragalin inhibit development of dermatitis and IgE elevation in NC/Nga mice. J. Allergy Clin. Immunol. 2000;106:159–166. doi: 10.1067/mai.2000.107194. [DOI] [PubMed] [Google Scholar]
- 8.Anuradha R., Krishnamoorthy P. Effect of astragalin on the activities of membrane bound enzymes on leadacetate toxicity in albino rats. J. Chem. Pharm. Res. 2011;3:616–620. [Google Scholar]
- 9.Deng S.-G., Deng Z.-Y., Fan Y.-W., Shan B., Xiong D.M. Spectroscopic investigation on the interaction between astragalin from Lotus leaf and DNA. Spectrosc. Spect. Anal. 2010;30:476–480. [PubMed] [Google Scholar]
- 10.Akinmoladun A.C., Obuotor E.M., Farombi E.O. Evaluation of antioxidant and free radical scavenging capacities of some Nigerian indigenous medicinal plants. J. Med. Food. 2010;13:444–451. doi: 10.1089/jmf.2008.0292. [DOI] [PubMed] [Google Scholar]
- 11.Chomnawang M.T., Surassmo S., Nukoolkarn V.S., Gritsanapan W. Effect of Garcinia mangostana on inflammation caused by Propionibacterium acnes. Fitoterapia. 2007;78:401–408. doi: 10.1016/j.fitote.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 12.SGD—Saccharomyces Genome Database. [accessed on 20 February 2012]. Available online: www.yeastgenome.org.
- 13.Pungartnik C., Picada J., Brendel M., Henriques J.A.P. Further phenotypic characterization of pso mutants of Saccharomyces cerevisiae with respect to DNA repair and response to oxidative stress. Gen. Mol. Res. 2002;1:79–89. [PubMed] [Google Scholar]
- 14.Friedberg E.C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Game J.C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin. Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 16.Kunz B.A., Haynes R.H. Phenomenology and genetic control of mitotic recombination in yeast. Annu. Rev. Genet. 1981;15:57–89. doi: 10.1146/annurev.ge.15.120181.000421. [DOI] [PubMed] [Google Scholar]
- 17.Noll D.M., Mason T.M., Miller P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt C.L., Grey M., Schmidt M., Brendel M., Henriques J.A. Allelism of Saccharomyces cerevisiae genes PSO6, involved in survival after 3-CPs+UVA induced damage, and ERG3, encoding the enzyme sterol C-5 desaturase. Yeast. 1999;15:1503–1510. doi: 10.1002/(sici)1097-0061(199910)15:14<1503::aid-yea481>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J.L., Qian Z.M., Luo Y.D., Tang D., Chen H., Yi L., Li P. Screening and mechanism study of components targeting DNA from the Chinese herb Lonicera japonica by liquid chromatography/mass spectrometry and fluorescence spectroscopy. Biomed. Chromatogr. 2008;22:1164–1172. doi: 10.1002/bmc.1040. [DOI] [PubMed] [Google Scholar]
- 20.Saito S.T., Trentin D.S., Macedo A.J., Pungartnik C., Gosmann G., Silveira J.D., Guecheva T.N., Henriques J.A.P., Brendel M. Bio-guided fractionation shows Cassia alata extract to inhibit Staphylococcus epidermidis and Pseudomonas aeruginosa growth and biofilm formation. Evid. Based Complement. Alternat. Med. 2012 doi: 10.1155/2012/867103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu A., Xu L., Zou Z., Yang S. Studies on chemical constituents from leaves of Cassia alata. Zhongguo Zhong Yao Za Zhi. 2009;34:861–863. [PubMed] [Google Scholar]
- 22.Usha S., Johnson I.M., Malathi R. Interaction of resveratrol and genistein with nucleic acids. J. Biochem. Mol. Biol. 2005;38:198–205. doi: 10.5483/bmbrep.2005.38.2.198. [DOI] [PubMed] [Google Scholar]
- 23.Alex S., Dupuis P. FT-IR and Raman investigation of cadmium binding by DNA. Inorg. Chim. Acta. 1989;157:271–281. [Google Scholar]
- 24.Pavia D.L., Lampman G.M., Kriz G.S. Introduction to Spectroscopy: A Guide for Students of Organic Chemistry. Chapter 2. Brooks/Cole; Belmont, CA, USA: 2001. Infrared Spectroscopy; pp. 14–101. [Google Scholar]
- 25.Kumar C.V., Asuncion E.H. DNA binding studies and site selective fluorescence sensitization of an anthryl probe. J. Am. Chem. Soc. 1993;115:8547–8553. [Google Scholar]
- 26.Singer B., Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenun Press; New York, NY, USA: 1983. [Google Scholar]
- 27.TOXNET database. [accessed on 20 February 2012]. Available online: http://toxnet.nlm.nih.gov/cgi-bin/sis/search.
- 28.Macindoe G., Mavridis L., Venkatraman V., Devignes M.-D., Ritchie D.W. HexServer: Na FFT-based protein docking Server powered by graphics processors. Nucleic Acids Res. 2010;38:445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand-Williams W., Cuvelier M.E., Berset C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995;28:25–30. [Google Scholar]
- 30.Ruhland A., Haase E., Siede W., Brendel M. Isolation of yeast mutants sensitive to the bifunctional alkylating agent nitrogen mustard. Mol. Gen. Genet. 1981;181:346–351. doi: 10.1007/BF00425609. [DOI] [PubMed] [Google Scholar]
- 31.Queiroz E.F., Wolfender J.L., Atindehou K.K., Traore D., Hostettmann K. On-line identification of the antifungal constituintes of Erythrina vogelii by liquid chromatography with tandem mass spectrometry, ultraviolet absorbance detection and nuclear magnetic resonance spectrometry combined with liquid chromatographic micro-fractionation. J. Chromatogr. A. 2002;974:123–134. doi: 10.1016/s0021-9673(02)01224-4. [DOI] [PubMed] [Google Scholar]
- 32.Larsen T.A., Goodsell D.S., Cascio D., Grzeskowiak K., Dickerson R.E. The structure of DAPI bound to DNA. J. Biomol. Struct. Dyn. 1989;7:477–491. doi: 10.1080/07391102.1989.10508505. [DOI] [PubMed] [Google Scholar]
- 33.Couch G.S., Hendrix D.K., Ferrin T.E. Nucleic acid visualization with UCSF Chimera. Nucleic. Acids Res. 2006;34:2–5. doi: 10.1093/nar/gnj031. [DOI] [PMC free article] [PubMed] [Google Scholar]