Abstract
Certain HLA-B antigens have been associated with lack of progression to AIDS. HLA-B alleles can be divided into two mutually exclusive groups based on the expression of the molecular epitopes HLA-Bw4 and HLA-Bw6. Notably, in addition to its role in presenting viral peptides for immune recognition, the HLA-Bw4, but not HLA-Bw6, motif functions as a ligand for a natural killer cell inhibitory receptor (KIR). Here, we show that profound suppression of HIV-1 viremia is significantly associated with homozygosity for HLA-B alleles that share the HLA-Bw4 epitope. Furthermore, homozygosity for HLA-Bw4 alleles was also significantly associated with the ability to remain AIDS free and to maintain a normal CD4 T cell count in a second cohort of HIV-1-infected individuals with well defined dates of seroconversion. This association was independent of the presence of a mutation in CC chemokine receptor 5 (CCR5) associated with resistance to HIV-1 infection, and it was independent of the presence of HLA alleles that could potentially confound the results. We conclude that homozygosity for HLA-Bw4-bearing B alleles is associated with a significant advantage and that the HLA-Bw4 motif is important in AIDS pathogenesis.
Viruses such as HIV-1 that cause lifelong infection in humans have evolved mechanisms to elude the host immune response. Thus, the HIV-1 burden in an infected individual is a product of both HIV-1-specific immune responses of the host that eliminate virus and infected cells, and the ability of the virus to evade those mechanisms. Over the past decade, individuals have been identified who demonstrate disparate abilities to control HIV-1 infection. Although the average time to AIDS in untreated infection is ten years, a subset of infected persons has been identified who maintain low to undetectable HIV-1 viral loads, normal CD4+ T cell counts, and have no manifestations of HIV-1 clinical immunocompromise or disease despite documented infection with HIV-1, in some cases for up to 20 years (1–7). This group of individuals thus manifests a unique ability to successfully control HIV-1 viremia in the absence of antiretroviral therapy.
Immunological characterization of these individuals who persistently control viremia, representing the minority of so-called long term non-progressors (LTNP), has demonstrated vigorous HIV-1-specific CD4+ T cell proliferative responses and cytotoxic T cell responses to HIV-1 antigens (5, 6, 8). Several cohort studies examining HLA associations in HIV-1-infected persons have suggested that heterozygosity of HLA class I loci, including HLA-A, -B, and -C, is associated with the delayed onset of AIDS (9–11). It has been speculated that the broader range of antigen presentation in individuals heterozygous at class I loci confers immunological advantage (9), which would be consistent with recent studies showing the importance of host cytotoxic T lymphocytes (CTL) in disease outcome (12–14). In particular, specific HLA-B antigens, including HLA-B27, -B51, -B57, -B8, and -B35, have been associated with different patterns of HIV-1-related disease progression (10, 15–18).
All HLA-B molecules can be categorized by the presence of a Bw4 or Bw6 molecular epitope. The Bw4 and Bw6 epitopes are defined by two different amino acid sequences at residues 79–83 in the carboxyl-terminal end of the α1 helix (19) of the HLA class I binding groove. Thus, individuals who are heterozygous for two different HLA-B alleles may in fact be homozygous with respect to the HLA-Bw4 or -Bw6 epitope of their HLA-B alleles (20, 21). The amino acids that determine the Bw4 or Bw6 epitope form the wall of the peptide binding groove adjacent to the F pocket, which serves as the major anchor residue for most class I-presented antigenic peptides (19).
Notably, HLA-B molecules encoded by HLA-B alleles with the Bw4 epitope, but not the Bw6 epitope, serve as ligands for natural killer cell inhibitory receptors (KIR). HLA C alleles can also be divided into two mutually exclusive ligands for KIR. Another potential influence of HLA on disease outcome might therefore stem from the ability of certain HLA molecules to function as killer inhibitory receptor ligands, or KIR ligands. The loading of HIV-1 peptides onto HLA KIR ligands could potentially interfere with the inhibitory effect of the KIRs and thus favor the elimination of HIV-1-infected cells. Such a mechanism has been shown to be involved in the killing of cells infected with an experimentally introduced retroviral vector (22).
Here, we examine the relationship between individual HLA-B and HLA-Cw alleles and HIV-1 disease progression and identify a significant association between homozygosity for alleles that share the HLA-Bw4 molecular epitope with profound control of HIV-1 viremia. Moreover, we demonstrate an innate and independent advantage of individuals homozygous for HLA-Bw4 alleles in the ability to remain AIDS-free and to maintain normal CD4 counts 10 years after infection in a patient cohort with well-defined dates of seroconversion. This association was independent of the presence of a mutation in CC chemokine receptor 5 (CCR5) associated with resistance to HIV-1 infection and independent of the presence of HLA alleles (HLA-B27, -B51, -B57, -B8, and -B35) that could potentially confound the results. Also, our data suggest an independent association between HLA-B*44 and control of HIV-1 viremia. These results thus provide an example of the advantage of homozygosity for particular epitopes of specific HLA alleles and natural killer (NK) ligands over heterozygosity in protection from AIDS.
Methods
Study Subjects.
Thirty-nine HIV-1-seropositive subjects were recruited, of whom 20 form a group of “controllers of viremia.” This group was established at the Massachusetts General Hospital in Boston, MA, and included patients identified locally in Boston or nationally by other health care providers. Controllers of viremia were classified on the basis of documented long-term infection with HIV-1, a viral load of less than 1,000 HIV-1 RNA copies/ml plasma after 10 yr of infection in the absence of antiviral therapy (23), and an AIDS-free status on no antiretroviral therapy. Two individuals had been infected for an unknown duration on no antiretroviral therapy, but for at least 4 (C-10) and 2 (C-20) yr with repeated viral loads that were undetectable by the most sensitive HIV-1 assay available at the time of testing. These two individuals were categorized as controllers in this analysis because they had a low to undetectable viral load in the absence of therapy. These individuals could not be categorized as long-term non-progressors because the duration of their infection is unknown. That is, on presentation, they were HIV-1 seropositive and could not date their infection and had no previous HIV-1-negative tests. Data from the Multicenter AIDS Cohort Study (23) indicate that individuals with such low levels of HIV-1 RNA in the absence of therapy form a unique subset of patients with an ability to control viral load and have a strong likelihood of becoming long-term non-progressors. Viral loads were determined by the Amplicor RNA assay or by the Amplicor ultradirect HIV-1 RNA assay (Roche).
Non-controllers of viremia were HIV-1-seropositive individuals who had viral loads greater than 1,000. Although none of the non-controllers of viremia had received protease inhibitors or dual nucleoside therapy, a subset had received antiretroviral monotherapy at some point in their disease course. All available samples from controllers of viremia were analyzed, and an equivalent number of non-controllers were randomly selected on the basis of the outlined criteria and analyzed. HIV-1-seronegative control individuals were anonymous unrelated Caucasian blood donors to the American Red Cross (New England Region, Dedham, MA).
There was unambiguous DNA-based or serologic HLA-B typing available for 103 individuals from the San Francisco City Clinic Cohort (SFCCC) who had not reached the study endpoint (CD4 < 500 or diagnosis of AIDS) when they were first evaluated. The SFCCC was a study of the natural history of HIV infection and AIDS conducted among high risk men who have sex with men (MSM) in San Francisco (24, 25). The association of HLA-Bw4 and Bw6 genotype with progression to the study endpoint was first assessed by using Kaplan-Meier curves and the log-rank test. To take account of possible confounding by the HLA-B8, -B27, -B35, -B51, and -B57 alleles, the independent association of HLA-Bw4/Bw6 genotype with this endpoint was assessed by using three multivariable Cox proportional hazards models (26). In the first, Bw4 homozygotes were contrasted with the combined group of Bw4/Bw6 heterozygotes and Bw6 homozygotes; in the second, Bw4 homozygotes were compared with Bw6 homozygotes, and in the third, the linear trend in risk across the Bw6/Bw6, Bw4/Bw6, and Bw4/Bw4 groups was examined (26). Relative hazards were also adjusted for CCR5 heterozygosity.
HLA Class I Typing and Analysis and CCR5 Determination.
Genomic DNA was prepared from B cell lines or peripheral blood from HIV-1-infected subjects and controls according to standard techniques (27, 28). The HLA-B and HLA-Cw (27) loci were typed by PCR-SSOP (PCR-single-stranded oligonucleotide probes) as previously described (29). To solve ambiguous PCR-SSOP typing results, we performed PCR-SSP (28) (sequence-specific primers) by using PCR-SSP kits (Pel-Freez Clinical Systems) according to the manufacturer's instructions. The analysis of statistical significance was done by using the Fisher's exact test and 2 × 2 contingency tables, using the instat 2.01 computer program (GraphPAD, San Diego).
Characterization of the CCR5 status (wild-type or heterozygous for the variant CCR5) of these individuals is from Smith et al. (30).
Results
To determine whether specific HLA-B and HLA-Cw alleles were associated with an enhanced ability to control HIV-1 viremia, we performed PCR-based DNA typing of HLA-B and HLA-C alleles in a unique cohort of 20 individuals who were spontaneously controlling viremia. All of these chronically HIV-1-infected individuals, whom we term controllers, had achieved documented viral loads of less than 1,000 RNA copies/ml plasma in the absence of antiviral therapy. We also determined the HLA-B and HLA-C alleles in 19 randomly selected individuals who did not control HIV-1 viral load and experienced progressive HIV-1 infection, whom we term non-controllers.
The viral loads of the controllers of viremia ranged from <40 to 999 RNA copies/ml, and CD4 counts ranged from 551 to 1234. The viral loads of the non-controllers ranged from 6,073 to 641,000, and CD4 counts ranged from 16 to 750 (Table 1). The duration of documented infection at the time of evaluation ranged from 2 to 20 yr for the controllers versus 1 to 15 yr in the non-controllers, a subset of whom received single agent antiretroviral therapy (Table 1). There were no statistically significant differences between age at diagnosis, gender, and race of the controllers and non-controllers of viremia (Table 1).
Table 1.
Characterization of HLA-B and HLA-Cw alleles in HIV-1-infected individuals with variable abilities to control viremia
| Patient ID | Race | Gender | Age at diagnosis | Years of documented infection | Viral load (RNA copies/ml) | CD4 count | HLA-B alleles | HLA-B epitopes | HLA-Cw alleles | HLA-C KIR ligands |
|---|---|---|---|---|---|---|---|---|---|---|
| C-1 | C | M | 26 | 16 | 260 | 839 | 27/5701-4 | Bw4/Bw4 | 0202/0602 | 1/1 |
| C-2 | C | M | 33 | 16 | 360 | 608 | 27/4402 | Bw4/Bw4 | 0102-3/0501-2 | 2/1 |
| C-3 | C | M | 25 | 20 | 999 | 421 | 4402/51 | Bw4/Bw4 | 0501-2/14 | 1/2 |
| C-4 | C | M | 22 | 16 | <500 | 684 | 27/5701-4 | Bw4/Bw4 | 0602/12 | 1/2 |
| C-5 | AA | M | 25 | 16 | <50 | 898 | 4403/5201 | Bw4/Bw4 | 14/15 | 2/1 |
| C-6 | C | M | 35 | 12 | <40 | 1,009 | 5701-4/5701-4 | Bw4/Bw4 | 0602/0602 | 1/1 |
| C-7 | H | F | 29 | 8 | <400 | 1,016 | 4402/5701-4 | Bw4/Bw4 | 0501-2/07 | 1/2 |
| C-8 | C | F | 24 | 12 | <50 | 1,573 | 2701/5701-4 | Bw4/Bw4 | 0102-3/0602 | 2/1 |
| C-9 | C | M | 38 | 8 | <50 | 911 | 4402/5701-4 | Bw4/Bw4 | 0501-2/1801-2 | 1/1 |
| C-10 | C | M | 41 | 4 | <400 | 1,234 | 1524/27 | Bw4/Bw4 | 0202/0303 | 1/2 |
| C-11 | AA | M | 45 | 18 | 380 | 551 | 4403/18 | Bw4/Bw6 | 04/12 | 1/2 |
| C-12 | C | M | 27 | 20 | 300 | 778 | 4402/14 | Bw4/Bw6 | 0501-2/0801-4 | 1/2 |
| C-13 | C | M | 39 | 10 | <50 | 604 | 4402/18 | Bw4/Bw6 | 0501-2/07 | 1/2 |
| C-14 | C | M | 29 | 15 | <100 | 873 | 1301-3/4101-3 | Bw4/Bw6 | 0602/07 | 1/2 |
| C-15 | C | M | NA | 10 | 100 | 1,196 | 1301-3/0702 | Bw4/Bw6 | 0602/15 | 1/1 |
| C-16 | AA | F | 35 | 12 | <400 | 620 | 07/1503 | Bw6/Bw6 | 0202/15 | 1/1 |
| C-17 | C | M | 19 | 21 | <50 | 668 | 07/4001 | Bw6/Bw6 | 03/07 | 2/2 |
| C-18 | C | M | 24 | 13 | 720 | 790 | 39/4001 | Bw6/Bw6 | 03/07 | 2/2 |
| C-19 | C | M | 27 | 10 | 150 | 679 | 08/14 | Bw6/Bw6 | 07/0801-4 | 2/2 |
| C-20 | H | M | 42 | >2 | <40 | 642 | 35/4004 | Bw6/Bw6 | 03/04 | 2/1 |
| NC-1 | C | M | 47 | 12 | 57,000 | 430 | 27/07 | Bw4/Bw6 | 0202/07 | 1/2 |
| NC-2 | C | M | 28 | 8 | 53,000 | 612 | 1301-3/07 | Bw4/Bw6 | 0602/15 | 1/1 |
| NC-3 | C | M | 53 | 11 | 33,000 | 741 | 27/39 | Bw4/Bw6 | 0102-3/1701-2 | 2/1 |
| NC-4 | C | M | 25 | 4 | 247,000 | 449 | 3801-3/35 | Bw4/Bw6 | 04/12 | 1/2 |
| NC-5 | C | M | 26 | 9 | 248,000 | 217 | 1301-3/35 | Bw4/Bw6 | 04/0602 | 1/1 |
| NC-6 | C | M | 27 | 6 | 70,000 | 473 | 3801-3/35 | Bw4/Bw6 | 04/1202 | 1/2 |
| NC-7 | C | M | 27 | 7 | 6,073 | 302 | 4403/14 | Bw4/Bw6 | 0801-4/0102-3 | 2/2 |
| NC-8 | C | M | 49 | 3 | 45,000 | <50 | 3701-2/18 | Bw4/Bw6 | 14/0602 | 2/1 |
| NC-9 | C | M | 32 | 6 | 284,000 | 454 | 27/14 | Bw4/Bw6 | 0801-4/16 | 2/1 |
| NC-10 | H | M | 24 | 5 | 700,000 | <50 | 5201/15 | Bw4/Bw6 | 0102-3/0303 | 2/2 |
| NC-11 | C | M | 35 | 9 | 35,500 | 269 | 5201/14 | Bw4/Bw6 | 0801-4/1202 | 2/2 |
| NC-12 | C | M | 31 | 1 | 55,000 | 300 | 1503/35 | Bw6/Bw6 | 0303/04 | 2/1 |
| NC-13 | C | M | 49 | 11 | 641,000 | 613 | 07/08 | Bw6/Bw6 | 07/07 | 2/2 |
| NC-14 | C | M | 35 | 13 | 80,508 | 16 | 08/15 | Bw6/Bw6 | 04/07 | 1/2 |
| NC-15 | C | M | 33 | 4 | 290,000 | <50 | 07/39 | Bw6/Bw6 | 07/07 | 2/2 |
| NC-16 | C | M | 33 | 6 | 20,000 | 438 | 07/07 | Bw6/Bw6 | 07/07 | 2/2 |
| NC-17 | NA | M | NA | 5 | 225,000 | 700 | 35/35 | Bw6/Bw6 | 04/04 | 1/1 |
| NC-18 | NA | M | NA | 9 | 80,000 | 750 | 07/50 | Bw6/Bw6 | 0602/07 | 1/2 |
| NC-19 | C | F | 23 | 15 | 315,000 | 150 | 08/4001 | Bw6/Bw6 | 03/07 | 2/2 |
HIV-1 seropositive individuals who control HIV-1 viremia (controllers) or who do not control viremia (non-controllers) were typed for HLA-B and HLA-Cw alleles. Race (Caucasian, C; African American, AA; hispanic, H), gender, and age at diagnosis is noted. The designation NA indicates that this data was not available. The number of years of documented infection at the time that the viral load and CD4 count were evaluated is also noted. The HLA-B alleles were grouped into HLA-Bw4 (Bw4) and HLA-Bw6 (Bw6) epitopes based on residues 79–83 at the carboxyl-terminal end of the α1 helix (19). The HLA-Bw4 alleles are shown in bold. HLA-C allotypes (group 1 or group 2) are based on residues 77 and 80 of HLA-C previously described (31). All genotyping was performed without knowledge of the disease status of the individuals analyzed.
The HLA-B antigens HLA-B27, -B51, and -B57 have been associated with delayed progression to AIDS (1, 10, 11, 18). Of these alleles, only HLA-B*57 was found significantly increased in our controller of viremia group (Table 1). In this cohort, we detected 7 HLA-B*57 alleles of a total of 40 alleles in the controller group and found none among the 38 alleles in the non-controller group (P = 0.012, Table 1). Strikingly, we also found a significant association between HLA-B*44 and control of viremia; 8 HLA-B*44 alleles were detected of a total of 40 alleles in the controller group, as compared with 1 of 38 in the non-controller group (P = 0.03, Table 1). By contrast, HLA-B8 and -B35 have been associated with AIDS progression (1, 11, 15, 17). Among the non-controller group, we found 8 of 40 alleles that are within HLA-B8 and -B35 antigen groups, whereas we found only 1 among those controlling viremia (P = 0.012, Table 1). Thus, our data support the previously reported association of HLA-B*57 with delayed progression and the association of HLA-B8 and -B35 with progression to AIDS in our cohort. Moreover, we have detected a significant association between HLA-B*44 and control of HIV-1 viremia. No clear associations between HLA-Cw alleles with control or non-control of viremia were observed (Table 1).
We next classified the HLA-B alleles of the controllers and non-controllers according to their Bw4 or Bw6 epitope; HLA-Bw4 but not -Bw6 serves as a KIR ligand that inhibits NK lysis of autologous cells (31, 32). Remarkably, 10 of the 20 individuals who control viremia were homozygous for alleles that share the HLA-Bw4 epitope, and we found no homozygous Bw4 individuals among the 19 individuals who do not control viremia (Table 1). Furthermore, the allele frequency of Bw4 is significantly increased in those controlling viremia, as compared with the non-controllers (P < 0.0001) or to its frequency in 108 HIV-1 negative blood donors (P = 0.0001; Table 2). Moreover, the proportion of individuals with the HLA-Bw4 homozygous genotype is also significantly increased in the controllers, as compared with non-controllers of viremia (P = 0.0004) or to the HIV-1-negative control group (P = 0.001, Table 2).
Table 2.
Genotype and allele frequencies of HLA-Bw4 and HLA-Bw6 in HIV-1-infected individuals with variable abilities to control viremia and in HIV-1-negative control individuals
| HLA | Total | Genotypes
|
Alleles
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bw4/Bw4 | f | P | Bw4/Bw6 | f | P | Bw6/Bw6 | f | P | Bw4 | Bw6 | P | ||
| Controllers | 20 | 10 | 0.5 | 5 | 0.25 | 5 | 0.25 | 0.62 | 0.38 | ||||
| Non-controllers | 19 | 0 | 0 | 0.0004 | 11 | 0.57 | 0.05 | 8 | 0.33 | 0.3 | 0.29 | 0.71 | <0.0001 |
| HIV-1-negative | 108 | 16 | 0.15 | 0.001 | 57 | 0.53 | 0.02 | 35 | 0.32 | 0.6 | 0.41 | 0.59 | 0.0001 |
The frequencies (f) of HLA-Bw4 and HLA-Bw6 in 20 individuals who control viremia (controllers), 19 individuals who do not control viremia (non-controllers), and 108 uninfected control (HIV-1-negative) individuals are shown. P values compare genotype and allele frequencies of controllers versus non-controllers and controllers versus HIV-1-negative individuals. The 108 uninfected controls are from a Caucasian database from New England. Two of the controller individuals were infected for an unknown duration on no antiretroviral therapy, but for at least 4 (C-10) or 2 (C-20) years with repeated viral loads that were undetectable by the most sensitive HIV-1 assay available at the time of testing. We note that, even if these two individuals are excluded from the analysis, the association between Bw4 homozygosity and control of viremia remains statistically significant. The frequency of HLA-Bw4 and HLA-Bw6 alleles that we found is reflective of their frequencies in other ethnic groups (41).
Two mutually exclusive groups of HLA-C alleles also serve as KIR ligands (31, 32). We did not, however, find an association between a particular HLA-C KIR ligand group (group 1 or group 2) and the control of viremia (Table 1). Thus, homozygous expression of the HLA-Bw4 NKB1 KIR ligand was significantly associated with profound suppression of HIV-1 viremia and protection from AIDS, whereas HLA-C KIR ligands were not.
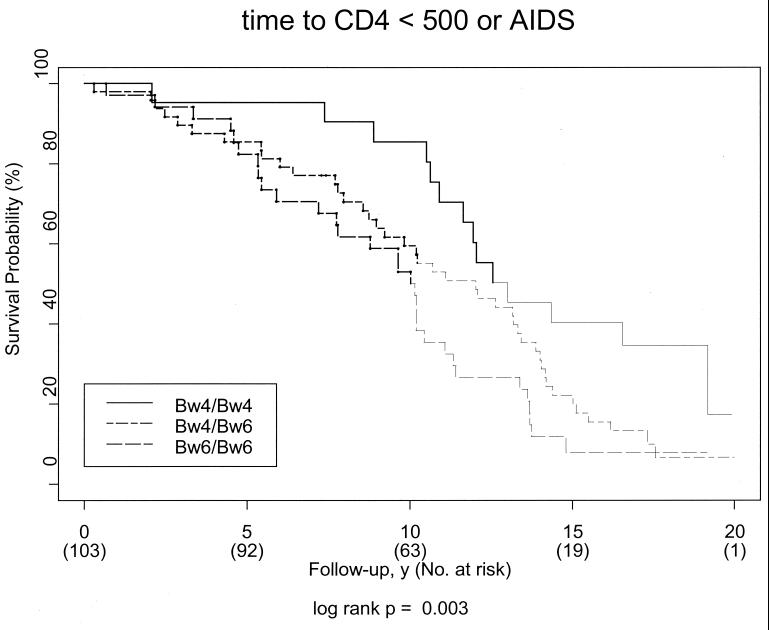
Our data led us to hypothesize that HLA-Bw4 homozygous individuals would have an intrinsic advantage in delaying HIV-1 disease progression. We therefore next investigated whether HLA-Bw4 homozygosity was predictive of the ability to maintain a normal CD4 count and to remain AIDS free in the SFCCC, a cohort of high-risk men who had had sex with men (MSM) with well defined dates of seroconversion. We determined the presence of HLA-Bw4 or -Bw6 epitopes in 103 individuals from the SFCCC, and assessed their association with time from seroconversion to disease progression. We chose first CD4 count less than 500 or a diagnosis of AIDS as the endpoints because these criteria have been used to assess long-term non-progressor status at 10 yr after infection in this cohort (25). In the course of follow-up, 87 (84%) study participants reached these endpoints. As shown in Fig. 1, Kaplan-Meier survival curves demonstrate that HLA-Bw4 homozygosity confers a significant advantage in avoiding HIV-1 disease progression (P = 0.003, log-rank test), as compared with Bw4/Bw6 heterozygosity and especially as compared with HLA-Bw6 homozygosity.
Figure 1.
HLA-Bw4 homozygosity confers a significant advantage in avoiding HIV-1 disease progression at all time points after seroconversion. Kaplan-Meier survival curves for time to CD4 count less than 500 or diagnosis of AIDS according to HLA-Bw4 homozygosity, HLA-Bw4/Bw6 heterozygosity, or HLA-Bw6 homozygosity among 103 SFCCC study participants. Each curve becomes fainter at the point where less than half the subgroup remains at risk.
The HLA-B antigens previously associated with slower disease progression (HLA-B27, -B51, and -B57), all share the HLA-Bw4 epitope whereas antigens previously associated with rapid progression (HLA-B8 and -B35) share the HLA-Bw6 epitope. To investigate any potential confounding effects created by the presence of HLA-B27, -B51, -B57, -B8, and -B35 on the observed association of Bw4 homozygosity and protection from HIV-1 disease progression, we performed multivariable Cox proportional hazards models to adjust for their presence in the SFCCC cohort. The number of individuals in this group was large enough to perform this analysis whereas the numbers of controllers and non-controllers analyzed in Table 1 did not allow us to investigate confounding effects.
After correction for the potential confounding effects of HLA-B27, -B57, -B51, -B35, and -B8 in the SFCCC, we found that Bw4 homozygotes were at lower risk for HIV-1 disease progression than the other two genotypes as a group (Table 3, Model 1), and in particular at lower risk than Bw6 homozygotes (Models 2 and 3). Both the contrast with Bw6 homozygotes and the trend in risk across the three genotypes are highly significant (Table 3). Because a polymorphism in the chemokine receptor gene CCR-5 is associated with resistance to HIV-1 infection and disease progression (30, 33, 34), we next investigated whether this polymorphism was potentially confounding our observation that Bw4 homozygotes were at lower risk for HIV-1 disease. As shown in Table 3, when our data were adjusted for the presence of the variant CCR5 gene, the HLA-Bw4 advantage remained significant (Table 3). Moreover, the effect of HLA-Bw4 homozygosity that we detected in the Kaplan-Meier analysis was slightly larger than the protective effect of CCR5 heterozygosity in this cohort (data not shown). We conclude that homozygosity for HLA-Bw4 is strongly and independently associated with slower HIV-1 disease progression.
Table 3.
HLA-Bw4 is independently associated with the ability to maintain a normal CD4 count and to remain AIDS free
| Model | Not adjusted for
CCR5
|
Adjusted for CCR5
|
||||
|---|---|---|---|---|---|---|
| RH | 95% CI | P | RH | 95% CI | P | |
| Bw4/Bw4 vs. Bw4/Bw6 + Bw6/Bw6 | 0.43 | 0.21–0.87 | 0.02 | 0.39 | 0.18–0.85 | 0.02 |
| Bw4/Bw4 vs. Bw6/Bw6 | 0.31 | 0.14–0.70 | 0.005 | 0.29 | 0.12–0.71 | 0.007 |
| Trend across genotypes | 0.34 | 0.16–0.72 | 0.004 | 0.35 | 0.16–0.77 | 0.009 |
Adjusted relative hazards (RH) for progression to CD4 count <500 or diagnosis of AIDS. Results are shown for 103 participants in the SFCCC who had well-defined dates of seroconversion and unambiguous HLA-B typing, but had not reached the endpoint when first evaluated. In model 1, Bw4 homozygotes are compared with the other two groups combined. In model 2, the Bw4/Bw4 and Bw6/Bw6 homozygote groups are compared. In model 3, the trend in risk from Bw6/Bw6 to Bw4/Bw6 to Bw4/Bw4 is examined. All estimated RH are adjusted for the presence of HLA-B8, -B27, -B35, -B51, and -B57 alleles. Characterization of the CCR5 status (wild-type or heterozygous for the variant CCR5) of these individuals is from Smith et al. (30), and relative hazards on the right are also adjusted for CCR5 heterozygosity. CI, confidence interval.
Discussion
Previous studies have identified genetic polymorphisms in chemokine receptor genes that encode molecules critical for HIV-1 entry into host cells (30, 33–35). Other studies have identified independent HLA associations with susceptibility or resistance to HIV-1 disease progression after infection with the virus has occurred. Here, we have shown a significant association between HLA-Bw4 homozygosity and the ability to spontaneously control HIV-1 replication in vivo and the maintenance of a normal CD4 count and an AIDS-free clinical status. Moreover, this effect was independent of the presence of a mutation in CCR5 associated with resistance to HIV-1 infection and independent of the presence of HLA alleles that could potentially confound the results.
Heterozygosity at HLA loci is thought to confer on an individual an advantage in immune containment of infectious diseases, including HIV-1 (9), because of the larger array of peptides that can be potentially presented by diverse HLA molecules. Our data indicate that not all homozygotes are at a disadvantage in fighting an infectious disease challenge. In the case of protection from AIDS, homozygosity for HLA-Bw4-bearing B alleles is associated with a significant advantage.
We note that, among the controllers of viremia group, there are five individuals who are homozygous for HLA-Bw6 alleles. Only one of these Bw6 homozygous individuals (C-17), who has been infected for 21 years, was typed for the CCR5 variant that confers advantage in HIV-1 infection and disease progression and was found to be negative for this CCR5 variant (data not shown). Taken together, these observations emphasize that the ability to control viremia is multifactorial and that additional genetic advantages remain to be discovered.
Intriguingly, whereas the HIV-1 viral product Nef selectively down-regulates the expression of HLA-A and HLA-B molecules in vitro (36, 37), this phenomenon has not been demonstrated in vivo in HIV-1-infected patients. Furthermore, HIV-1-specific CTL clones isolated from HIV-1-seropositive patients carrying different class I phenotypes have significant activity against several HIV proteins, including Nef (38). Thus, the role of Nef in CTL and in HLA class I regulation in vivo remains to be determined. Moreover, there is no evidence to date that Nef differentially modulates the activity of HLA-Bw4 and -Bw6 alleles. Taken together, we conclude that the HLA-Bw4 advantage does not involve a Nef-dependent mechanism.
We have also found in the context of this small case-control study a novel association between HLA-B44 and control of HIV-1 viremia. This association was not observed in the SFCCC, and thus it needs to be confirmed in other cohorts. Notably, HLA-B44 and the other HLA-B alleles associated with better outcome in HIV infection (HLA-B27, -B51, and -B57) are all of the Bw4 specificity, whereas the HLA-B alleles (HLA-B8 and -B35) associated with a worse outcome are of the Bw6 specificity. There are a number of different potential mechanisms for the apparent protective effect. It is possible that viral peptides that bind with greater affinity to B alleles that share the Bw4 specificity elicit more effective HIV-1-specific CTL responses than alleles that share the Bw6 specificity. Support for this hypothesis is provided by studies of HLA B*0801 and B*0802, which differ only in their Bw4 or Bw6 specificity. HLA-B*0802, which has the Bw4 epitope, is associated with a broader repertoire of endogenously bound peptides interacting with the F pocket (39) than is B*0801, which has the Bw6 epitope. Thus, peptides that bind to HLA-Bw4 alleles may prove useful in peptide-based vaccine development.
In addition to their role in peptide presentation, another potential advantage of homozygosity of HLA-Bw4 is related to the role of this motif as a specific ligand for NK cell inhibitory receptors, or KIRs, which inhibit NK lysis of autologous targets (40). Interestingly, a recent report demonstrated that binding of a virally encoded peptide presented in the context of HLA-Bw4 alleles, but not HLA-Bw6 alleles, blocks the NK inhibitory receptor, which resulted in the NK-mediated autologous lysis of the infected cell (22). It is thus intriguing to speculate that the presentation of HIV-1 peptides by HLA-Bw4 alleles may block the inhibition of NK lysis, resulting in the enhanced NK lysis of infected cells, which could thereby influence HIV-1 viremia and subsequent progression to AIDS.
Acknowledgments
We thank Alvaro Muñoz and Marvin Zellen for helpful discussions, anonymous reviewers for helpful suggestions, and Ann Corbett for assistance with manuscript preparation. This work was supported by grants from the National Institutes of Health to A.E.G (HL-59838), E.J.Y (HL-29582), and B.D.W (AI-28568); an Established Investigator Award from the American Heart Association to A.E.G; a Cooperative Agreement (U64/CCU900523-09) from the Centers for Disease Control to E.V.; and a Bridge Award from Harvard Medical School (to P.O.F.-V.).
Abbreviations
- KIR
natural killer cell inhibitory receptor
- CTL
cytotoxic T lymphocytes
- NK
natural killer
- CCR5
CC chemokine receptor 5
- SFCCC
San Francisco City Clinic Cohort
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 3.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L K, et al. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 4.Rinaldo C, Huang X L, Fan Z F, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, et al. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalams S A, Buchbinder S P, Rosenberg E S, Billingsley J M, Colbert D S, Jones N G, Shea A K, Trocha A K, Walker B D. J Virol. 1999;73:6715–6720. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 7.Harrer T, Harrer E, Kalams S A, Barbosa P, Trocha A, Johnson R P, Elbeik T, Feinberg M B, Buchbinder S P, Walker B D. J Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- 8.Lisziewicz J, Rosenberg E, Lieberman J, Jessen H, Lopalco L, Siliciano R, Walker B, Lori F. N Engl J Med. 1999;340:1683–1684. doi: 10.1056/NEJM199905273402114. [DOI] [PubMed] [Google Scholar]
- 9.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 10.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O'Brien S J, Rinaldo C, Detels R, Blattner Z W, Phair J, Erlich H, Mann D L. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 11.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O'Brien S, Andrieu J M, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson G W, Zagury J F. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 12.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 13.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 14.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brettle R P, McNeil A J, Burns S, Gore S M, Bird A G, Yap P L, MacCallum L, Leen C S, Richardson A M. AIDS. 1996;10:419–430. [PubMed] [Google Scholar]
- 16.Klein M R, Keet I P, D'Amaro J, Bende R J, Hekman A, Mesman B, Koot M, de Waal L P, Coutinho R A, Miedema F. J Infect Dis. 1994;169:1244–1249. doi: 10.1093/infdis/169.6.1244. [DOI] [PubMed] [Google Scholar]
- 17.Tomiyama H, Miwa K, Shiga H, Moore Y I, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. J Immunol. 1997;158:5026–5034. [PubMed] [Google Scholar]
- 18.Migueles S A, Sabbaghian M S, Shupert W L, Bettinotti M P, Marincola F M, Martino L, Hallahan C W, Selig S M, Schwartz D, Sullivan J, Connors M. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. . (First Published February 29, 2000; 10.1073/pnas.050567397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salter R D, Parham P. Hum Immunol. 1989;26:85–89. doi: 10.1016/0198-8859(89)90093-1. [DOI] [PubMed] [Google Scholar]
- 20.Van Rood J J. Dissertation. Leiden, The Netherlands: University of Leiden; 1962. [Google Scholar]
- 21.Muller C A, Engler-Blum G, Gekeler V, Steiert I, Weiss E, Schmidt H. Immunogenetics. 1989;30:200–207. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 22.Liberatore C, Capanni M, Albi N, Volpi I, Urbani E, Ruggeri L, Mencarelli A, Grignani F, Velardi A. J Exp Med. 1999;189:1855–1862. doi: 10.1084/jem.189.12.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 24.Katz M H, Chang S W, Buchbinder S P, Hessol N A, O'Malley P, Doll L S. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:58–63. [PubMed] [Google Scholar]
- 25.Koblin B A, van Benthem B H, Buchbinder S P, Ren L, Vittinghoff E, Stevens C E, Coutinho R A, van Griensven G J. Am J Epidemiol. 1999;150:1026–1030. doi: 10.1093/oxfordjournals.aje.a009926. [DOI] [PubMed] [Google Scholar]
- 26.Kalbfleisch J D, Prentice R L. The Statistical Analysis of Failure Time Data. New York: Wiley; 1980. [Google Scholar]
- 27.Clavijo O P, Delgado J C, Awdeh Z L, Fici D, Turbay D, Alper C A, Truedsson L, Yunis E J. Tissue Antigens. 1998;52:282–285. doi: 10.1111/j.1399-0039.1998.tb03045.x. [DOI] [PubMed] [Google Scholar]
- 28.Yu N, Ohashi M, Alosco S, Salazar M, Cao K, Fernandez V M, Yunis E J. Tissue Antigens. 1998;52:260–269. doi: 10.1111/j.1399-0039.1998.tb03041.x. [DOI] [PubMed] [Google Scholar]
- 29.Cereb N, Maye P, Lee S, Kong Y, Yang S Y. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 30.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, et al. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 31.Lanier L L. Immunity. 1997;6:371–378. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- 32.Moretta A. Cell. 1997;90:13–18. doi: 10.1016/s0092-8674(00)80309-8. [DOI] [PubMed] [Google Scholar]
- 33.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, et al. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 34.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 35.Faure S, Meyer L, Costagliola D, Vaneensberghe C, Genin E, Autran B, Delfraissy J F, McDermott D H, Murphy P M, Debre P, et al. Science. 2000;287:2274–2277. doi: 10.1126/science.287.5461.2274. [DOI] [PubMed] [Google Scholar]
- 36.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. Nature (London) 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 37.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 38.Shankar P, Xu Z, Lieberman J. Blood. 1999;94:3084–3093. [PubMed] [Google Scholar]
- 39.Arnett K L, Huang W, Valiante N M, Barber L D, Parham P. Immunogenetics. 1998;48:56–61. doi: 10.1007/s002510050400. [DOI] [PubMed] [Google Scholar]
- 40.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips J H, Lanier L L. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 41.Schreuder I, D'Amaro J, Hackbarth S, Duquesnoy R. In: Report of the Eighth International Histocompatibility Workshop. Terasaki P, editor. Los Angeles, CA: UCLA Tissue Typing Laboratory; 1980. pp. 346–352. [Google Scholar]