Abstract
Ethanolamine is important for synthesis of choline, phosphatidylethanolamine (PE) and phosphatidylcholine (PC) in plants. The latter two phospholipids are the major phospholipids in eukaryotic membranes. In plants, ethanolamine is mainly synthesized directly from serine by serine decarboxylase. Serine decarboxylase is unique to plants and was previously shown to have highly specific activity to l-serine. While serine decarboxylase was biochemically characterized, its functions and importance in plants were not biologically elucidated due to the lack of serine decarboxylase mutants. Here we characterized an Arabidopsis mutant defective in serine decarboxylase, named atsdc-1 (Arabidopsis thaliana serine decarboxylase-1). The atsdc-1 mutants showed necrotic lesions in leaves, multiple inflorescences, sterility in flower, and early flowering in short day conditions. These defects were rescued by ethanolamine application to atsdc-1, suggesting the roles of ethanolamine as well as serine decarboxylase in plant development. In addition, molecular analysis of serine decarboxylase suggests that Arabidopsis serine decarboxylase is cytosol-localized and expressed in all tissue.
Keywords: serine decarboxylase, ethanolamine, choline, phosphatidylethanolamine, phosphatidylcholine, Arabidopsis thaliana
1. Introduction
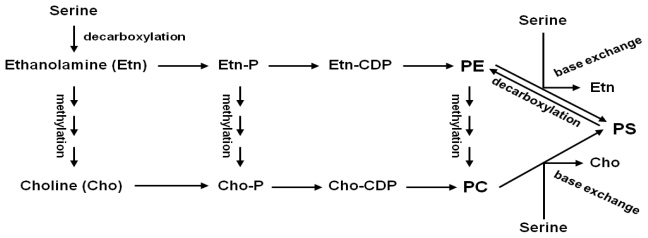
Ethanolamine is an important metabolite for synthesis of phosphatidylethanolamine (PE) and phosphatidylcholine (PC) (Figure 1), which are the two major phospholipids in eukaryotic membranes [1–5]. Unlike most animal cells, ethanolamine can be converted to choline, another precursor for PC synthesis in plant cells [2,4]. Ethanolamine is formed directly or indirectly from serine through direct decarboxylation of serine or base exchange reaction between serine and existing PE (Figure 1) [6–9]. The indirect ethanolamine production by base exchange reaction is found in most eukaryotes including animals and plants, but not fungi [6,9,10]. However, the direct ethanolamine generation by serine decarboxylation is reported only in plants and malarial parasites, Plasmodium spp. [7,8,11]. While Plasmodium serine decarboxylase gene was not definitively identified [12], plant serine decarboxylase genes were isolated and biochemically shown to be a pyridoxal phosphate-dependent soluble protein with high specific activity to L-serine [7].
Figure 1.
Synthesis of Phosphatidylethanolamine (PE) and Phosphatidylcholine (PC) through the Kennedy Pathway in Plants. Etn-P, phosphoethanolamine; Etn-CDP, cytidine-diphosphoethanolamine; Cho-P, phosphocholine; Cho-CDP, cytidine-diphosphocholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine.
Starting from ethanolamine and choline, de novo biosynthesis of PE and PC in most eukaryotic cells occurs through the “Kennedy” pathway, consisting of two parallel CDP-ethanolamine and CDP-choline pathways (Figure 1) [1,13]. PE can also be synthesized via the decarboxylation of phosphatidylserine (PS) by PS decarboxylases that is pyruvoyl cofactor-dependent and membrane-localized [9]. PC can be formed from PE through three successive methylations by PE N-methyltransferases (PEMTs). In plants, the methylation of ethanolamine moiety to form choline moiety by N-methyltransferases can occur at the levels of free ethanolamine bases, phospho-ethanolamine, and PE [4], linking two CDP-ethanolamine and CDP-choline pathways. PE and PC not only function in maintaining eukaryotic membrane structures, but also contribute to the pools of many biologically active molecules such as phosphatidic acid and diacylglycerol that act as signal molecules [1].
Through analyses of Arabidopsis mutants’ phenotypes and identification of the gene responsible for the phenotypes, the functions of many genes in Arabidopsis have been elucidated. Indeed, many functions of membrane lipids have been uncovered through many Arabidopsis lipid defective mutants [14]. For example, when phosphoethanoleamine N-methyltransferase, one enzyme that convert ethanolamine moiety to choline moiety, was silenced, the Arabidopsis mutant displayed pale-green leaves, early senescence, temperature-sensitive male sterility and salt hypersensitivity, revealing the importance of choline biosynthesis in plant growth and stress responses [15].
Although ethanolamine is thought to be synthesized mainly via serine decarboxylation [7,8], our understanding on the biological importance of ethanolamine is limited due to the lack of serine decarboxylase mutants.
Here we report the characterization of an Arabidopsis atsdc-1 (Arabidopsis thaliana serine decarboxylase-1) mutant defective in serine decarboxylase. Our analysis suggests that serine decarboxylase is important in maintenance of leaf tissue, lateral inflorescence development, floral fertility and flowering regulation in short days. Moreover, we found that serine decarboxylase is cytosol-localized and expressed in all tissue.
2. Results
2.1. Atsdc-1 Developmental Defects and Their Rescue under Short Day Conditions
One Arabidopsis mutant with growth defects was isolated from an ethyl methanesulfonate (EMS)-mutagenized Arabidopsis pool and later named atsdc-1 (Arabidopsis thaliana serine decarboxylase-1) (see below for the phenotypes and the reason for naming).
Under long day conditions, the size of atsdc-1 seedlings was much smaller than that of the wild type and the mutant never reached the height of the mature wild type (Figure 2A–G). Necrosis was observed along the edges of the mutant leaves (Figure 2C). atsdc-1 was sterile and had multiple inflorescences (Figure 2G). Interestingly, the smaller size and multiple inflorescence defects in the mutant could be rescued under short day conditions (Figure 2H–K,M). Nevertheless, the mutant leaves under short day conditions still showed necrosis along the edges, although its appearance was delayed (Figure 2J,K). Under short day conditions, atsdc-1 flowered earlier than the wild type; the mutant bolted at an average leaf number of 19, whereas the wild type bolted at an average leaf number of 28 (Figure 2L,M). Under long day conditions, there was no difference in flowering time between the wild type and atsdc-1; both bolted at an average leaf number of 8 (Figure 2L).
Figure 2.
Growth and Development of atsdc-1. (A–C): 3-week-old wild type (A) and atsdc-1 (B and C) grown under long day conditions. Bar = 1 cm. C is a magnified image of B. (D and E): 5-week-old wild type (left in D) and atsdc-1 (right in D) grown under long day conditions. Bar = 5 cm. E is a magnified image of one of the atsdc-1 seedlings. (F and G): 7-week-old wild type (left in F) and atsdc-1 (right in F) grown under long day conditions. G is a magnified image of one of the atsdc-1 seedlings. (H and I): 38-day-old wild type (H) and atsdc-1 (I) grown under short day conditions. (J and K): 43-day-old wild type (J) and atsdc-1 (K) grown under short day conditions. (L): Comparisons of leaf number upon bolting between the wild type and atsdc-1. (error bars = standard error; n = 20) (M): Difference in bolting between the wild type and atsdc-1 under short day conditions (63-day-old).
2.2. Molecular Cloning of AtSDC
None of the 25 F1 plants from a cross between the wild type and atsdc-1 showed the atsdc-1 morphological phenotypes. In the successive F2 generation, 403 seedlings tested were found to segregate into 292 wild-type phenotypes and 111 atsdc-1 phenotypes (3:1, χ2= 0.0042, p = 0.837). Taken together, these results suggest that atsdc-1 is a single recessive nuclear mutation.
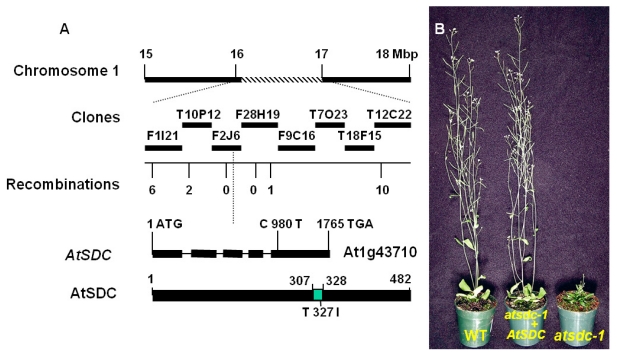
To identify the AtSDC gene responsible for the atsdc-1 phenotypes, we performed map-based cloning of the atsdc-1 mutant gene. We selected the atsdc-1 mutants in the F2 mapping population from a cross between Ler and atsdc-1. Initial mapping delimited the mutation to the middle of chromosome 1, between the nga248 and nga280 simple sequence length polymorphism (SSLP) markers. For fine mapping, new SSLP markers were developed, and the mutation was narrowed down to the F2J6 and F28H19 BAC clones (Figure 3A). Through sequencing genes in this region and comparing the wild type and atsdc-1 sequences, a mutation was found in the At1g43710 gene. The mutation was a change of C to T that took place at 980th nucleotide from the putative initiation codon of At1g43710 (Figure 3A). The mutation resulted in an alteration of the polar amino acid threonine to the nonpolar amino acid isoleucine at the putative 327th amino acid (Figure 3A).
Figure 3.
Molecular Cloning and Complementation of atsdc-1. (A): Map-based cloning of AtSDC. Simple sequence length polymorphism (SSLP) markers used were, from left to right above the recombination mark; F1I21-6K, T10P12-20K, F2J6-92K, F28H19-10K, F28H19-83K, and T12C22-11K. The number of recombination events was out of a total of 948 chromosomes. The mutation in AtSDC gene is a C to T change at 980 nt, which resulted in threonine to isoleucine change at the 327th amino acid. Green box indicates the PLP binding domain; (B): Molecular complementation of atsdc-1 developmental defects in 5-week-old atsdc-1 transformed with the wild type AtSDC gene (T1).
To confirm the authenticity of the gene, a genomic DNA fragment that spans from 1276 base pair 5′ upstream of the initiation codon to 410 base pair 3′ downstream of the stop codon of the At1g43710 open reading frame was subcloned into a binary vector and transferred into the atsdc-1 mutant through Agrobacterium-mediated transformation. The resulting T1 transformants did not exhibit the atsdc-1 developmental defects (Figure 3B). These results confirmed that At1g43710 is indeed AtSDC, and the atsdc-1 mutation is responsible for the atsdc-1 morphological defects.
2.3. AtSDC Encodes a Serine Decarboxylase
Results of BLAST searches of the Arabidopsis genome database suggested that AtSDC is a single-copy gene in the Arabidopsis genome. An AtSDC cDNA with a complete coding sequence was available from the Arabidopsis Biological Resource Center under the clone id U09195 [16]. Comparison of the genomic and cDNA sequences suggested that the AtSDC gene contains 5 exons and 4 introns with a 1449-bp coding sequence that would result in a 482-amino acid protein. The AtSDC protein did not have any detectable cellular targeting domains. It contains a putative pyridoxal phosphate (PLP) binding site spanning from 307 to 328 amino acids (Figure 3A). The PLP binding site is a conserved domain among many PLP-dependent decarboxylases, including glutamate-, histidine-, tryptophan-, and tyrosine-decarboxylases [17]. The atsdc-1 mutation occurred in the putative PLP binding site (Figure 3A). Therefore, the mutation might affect PLP binding to the AtSDC protein. AtSDC was annotated in the Arabidopsis database [18] as a histidine decarboxylase. However, Rotein et al. (2001) showed that AtSDC is in fact a serine decarboxylase. The authors expressed Arabidopsis AtSDC recombinant protein in E. coli using AtSDC cDNA and found specific decarboxylase activity on l-serine. They also showed that AtSDC does not have significant activity in histidine decarboxylation.
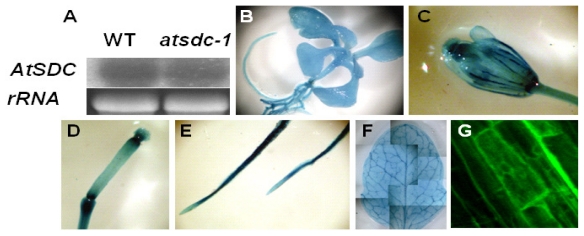
We obtained an Arabidopsis line (SALK_070968) with a T-DNA insertion [19] in the second intron of AtSDC. Through PCR genotyping out of 12 plants from this line (data not shown), we isolated two plants heterozygous for the T-DNA insertion. In the generation following self-pollination of the two heterozygous plants, we still could not identify any plants that were homozygous for the T-DNA insertion (data not shown). This suggested that the T-DNA insertion (potentially full knockout of function) in AtSDC is lethal, and our atsdc-1 EMS mutant is a weak allele. In support of the essential role of AtSDC in plant viability, another atsdc-1 T-DNA allele discovered by the Seed Gene Project [20] is a recessive-embryo-defective mutant (hence named embryo defective 1075), and the germplasm (ABRC id: CS16096) is maintained as a heterozygote [21]. RNA blot analysis showed that the AtSDC transcript level was not affected in atsdc-1 (Figure 4A), suggesting that the atsdc-1 mutation does not affect the steady state level of the transcript.
Figure 4.
Expression Patterns of AtSDC and AtSDCp-GUS, and Localization of GFP-AtSDC Fusion Protein. (A): AtSDC expression in the wild type and atsdc-1. rRNA gel picture was shown as loading control (B–F): AtSDC promoter-driven GUS activity in the whole seedling (B), flower (C), silique (D), roots (E) and leaf (F). (G): Subcellular localization of the GFP-AtSDC fusion protein in root cells.
To study the expression pattern of AtSDC, we investigated its promoter activity by fusing an 1126-bp genomic DNA fragment upstream of the AtSDC ORF to the β-glucuronidase (GUS) reporter gene (AtSDCp-GUS). Transgenic Arabidopsis plants expressing the AtSDCp-GUS reporter were then subjected to histochemical GUS analysis. GUS activity was detected in whole seedlings and in all organs tested from adult plants, including flowers, sliques, roots, and leaves (Figure 4B–F). This result suggests a ubiquitous expression of AtSDC.
Transgenic Arabidopsis plants expressing the GFP-AtSDC fusion protein were generated to study AtSDC subcellular localization. Confocal microscopy analysis with the roots of the GFP-AtSDC transgenic Arabidopsis detected green fluorescence in the cytosol and plasma membrane, suggesting a cytosolic and plasma membrane localization of the AtSDC protein (Figures 4G). However, database searches showed no detabtable transmembrane domains in AtSDC. Also, some known cytosolic proteins displayed peripheral fluorescence when fused with GFP [22]. Although further analysis will clarify the exact AtSDC subcellular localization, it is likely that AtSDC is cytosolic localized.
2.4. Ethanolamine Rescues the atsdc-1 Developmental Defects
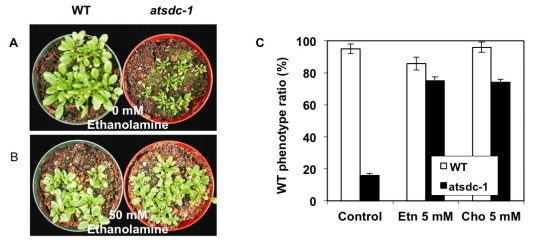
Because AtSDC is involved in the conversion of serine to ethanolamine and the eventual biosynthesis of choline [7], the atsdc-1 mutant seedlings should be deficient in these metabolites. We therefore applied ethanolamine or choline to atsdc-1 mutant plants growing in soil or MS agar plates. Although the application of 50 mM ethanolamine on soil grown plants had only a slight effect on the growth of the wild type, it remarkably rescued the atsdc-1 developmental defects (Figure 5A,B).
Figure 5.
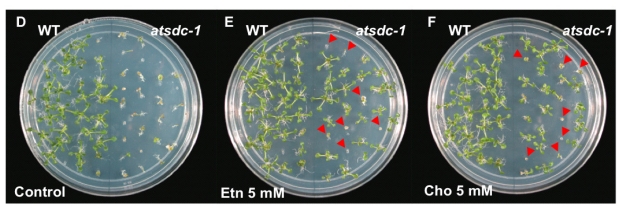
atsdc-1 Morphology After Ethanolamine or Choline Treatment. (A-B): Comparison of the wild type and atsdc-1 with or without 50 mM ethanolamine treatment. (C): atsdc-1 morphology recovery with ethanolamine (Etn) or choline (Cho) treatment in MS agar plates. WT phenotype ratios were obtained by dividing numbers of normally grown WT seedlings divided with total germinated seeds. Results are from three replicates (error bar = standard error). (D-F): Comparison of the wild type and atsdc-1 on MS agar plates supplemented with or without 5 mM ethanolamine or 5 mM choline. Red arrow heads indicate unrecovered seedlings. Small recovered atsdc-1 seedlings on ethanolamine and choline plates were eventually grown to the wild type size.
It also rescued atsdc-1 sterility (data not shown). Similarly, atsdc-1 morphology recovered almost to that of the wild-type in MS agar plates with ethanolamine or choline (Figure 5C–F). Although choline application also rescued the defects in the atsdc-1 mutant, the effect was not as efficient on mutant plants grown in soil as compared with those on MS plates (data not shown). The rescue by ethanolamine or choline suggests that the atsdc-1 mutant is indeed defective in the conversion of serine to ethanolamine and might accumulate lower amounts of these metabolites than the wild type.
3. Discussion
In this study, we used a forward genetics screen to identify a locus, AtSDC, that encodes a serine decarboxylase and is crucial for plant growth. Serine decarboxylase is involved in the direct conversion of serine to ethanolamine [7]. Plants form ethanolamine through either serine decarboxylation or base exchange reaction of PS, but serine decarboxylation is thought to be the major source of ethanolamine in plants [7,8]. Ethanolamine is an important metabolite in plants for the synthesis of choline and membrane lipids, such as PE and PC, as well as choline betaine in some species. Thus, a deficiency in these ethanolamine-derived metabolites might undermine the membrane system. Many lipid-related Arabidopsis mutants display varying degrees of abnormal morphological phenotypes, which indicates a crucial role of lipid biosynthesis in plant growth and development [14]. Indeed, many Arabidopsis mutants impaired in functions of genes involved in the Kennedy pathway displayed abnormal growth and development. PE N-methyltransferase (PEMT)-silenced Arabidopsis mutant (t365) plants were pale green, early aging, and exhibited temperature-sensitive male sterility [15]. Mutations in CTP:PE cytidylyltransferase caused embryo abortion, dwarfism, and reduced fertility in Arabidopsis [23]. Defects in a base exchange type PS synthase also resulted in embryo lethality and sterile dwarfism [24]. These abnormalities were also observed in atsdc-1 confirming the important functions of ethanolamine-derived metabolites in these developmental processes.
The cytosolic localization of GFP-AtSDC is consistent with the soluble property of AtSDC protein [7]. The ubiquitous AtSDCp-GUS activity is also in agreement with the universal requirement of serine decarboxylase in membrane lipid biosynthesis. The defect in serine decarboxylase in atsdc-1 was confirmed by the rescue of atsdc-1 morphological defects by ethanolamine (Figure 5) or choline application. Thus, it appears that the atsdc-1 defects are due to the deficiency of ethanolamine and/or ethanolamine-derived metabolites in atsdc-1.
The nearly normal growth and development in atsdc-1 under short day conditions is intriguing. Apparently, the ethanolamine requirement is either low, or other compensation pathways might become activated under short day conditions. An example of a conditional phenotype is that found in the fab2 Arabidopsis mutant, which is defective in the desaturation of stearic acid (18:0) to oleic acid (18:1) [25]. The fab2 mutation results in an elevated level of stearic acid in membrane lipids. At 22 °C, fab2 shows a dramatic dwarf phenotype, which can be restored to a normal phenotype with high temperatures (i.e., 36 °C) [25]. This phenotype recovery did not include changes in fatty acid composition. Thus, this recovery was proposed to be due to the increased fluidity of lipids with stearic acid at high temperatures. pect1-4, defective in CTP:PE cytidylyltransferase and PEMT-silenced Arabidopsis were also shown to display temperature-sensitive developmental defects [15,20].
The atsdc-1 growth temperature was set at 22 °C under both long and short day conditions. Therefore, it appears that the different photoperiods somehow brought about metabolic differences in plants. Since plants are photosynthetic organisms, it is not difficult to hypothesize that their metabolism depends on the duration of light illumination. The induction of different genes and physiological changes under different photoperiods are well documented [26–28]. Interestingly, the early bolting phenotype of atsdc-1 was observed only under short day conditions. This observation suggests that short day inhibition of flowering may require metabolism that involves proper AtSDC activity and probably ethanolamine-derived metabolites in plants.
4. Experimental Section
4.1. Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia gl1 plants were mutated by use of ethyl methanesulfonate to generate the mutant seed pool (M2 generation). For Arabidopsis cultivation, Arabidopsis seeds were put on soil or MS (Murashige and Skoog salt base, JRH Biosciences, Lenexa, KS) agar (0.6%) plates supplemented with 3% sucrose and placed at room temperature (22 ± 1 °C) under continuous light after 2–3 day cold stratification. When necessary, seedlings on MS were transferred to soil pots in a growth chamber under 22 °C cycles of 16-h light/8-h dark. For short day conditions, 22 °C cycles of 8-h light/16-h dark were used. Stock solutions of 1 M ethanolamine and choline chloride (Sigma, St. Louis, MO, USA) were prepared, and the pH was adjusted to 5.8 with HCl when necessary. Appropriate working concentrations were then prepared. Ethanolamine or choline was added into the MS agar media or sprayed every 4 to 5 days on soil-grown Arabidopsis plants.
4.2. Gene Expression Analysis
Nine-day-old seedlings grown on MS agar plates were used for RNA analysis. Total RNA was extracted and analyzed as described previously [29]. Briefly, total RNA was extracted with extraction buffer containing 50 mM Tris-HC1, pH 8.0, 300 mM NaCl, 5 mM EDTA, 2% SDS, 2 mM aurintricarboxylic acid, and 10 mM beta-mercaptoethanol. Extracted total RNA was separated on formaldehyde-agarose gel and the resulting gel was blotted onto nylon membrane. The membrane was then hybridized with 32P labeled DNA fragment probe of the specific genes. As a loading control, the β-tubulin gene was amplified by PCR with the following primer pairs: Tubulin-F (5′-CGTGGATCACAGCAATACAGAGCC-3′) and Tubulin-R (5′-CCTCCTGCACTTCCACTTCGTCTTC-3′). For the AtSDC probe, AtSDC full-length cDNA was used. After washing, the membrane was exposed to X-ray film.
4.3. Positional Cloning
For genetic mapping of the atsdc-1 mutation, atsdc-1 was crossed with the wild type in the ecotype Landsberg erecta. The resulting F1 plants were allowed to self and the F2 seeds collected. Homozygous atsdc-1 mutations in the segregated F2 population were selected on the basis of their developmental defects. The mutation was mapped with use of SSLP markers [30]. If necessary, new SSLP markers were developed by use of the Monsanto Arabidopsis Polymorphism collection [31]. Newly developed SSLP markers are as follows: F1I21-6K (F1I21-6KF, 5′-TCCAGTCCTT AAGCGGATTT-3′; F1I21-6KR, 5′-GCATAACACAATGTTCAGACAAA-3′), T10P12-20K (T10P12-20KF, 5′-TAGCAAAGCTTTCGATCCAT-3′; T10P12-20KR, 5′-ATTCTGTTGGGTTG CTATGC-3′), F2J6-92K (F2J6-92KF, 5′-GTGCGGGAGTGTGATAGAAT-3′; F2J6-92KR, 5′-TCC TCGAAAGATTCATTGATTT-3′), F28H19-10K (F28H19-10KF, 5′-GTGCGGGAGTGTGAT AGAAT-3′; F28H19-10KR, 5′-TCCTCGAAAGATTCATTGATTT-3′), F28H19-83K (F28H19-83KF, 5′-GGAGCCAAAACCAGCAACTA-3′; F28H19-83KR, 5′-GTTCTTGGTTGAGTGGAACG-3′), T12C22-11K (T12C22-11KF, 5′-TCCACCGGAGTAAACTCCATA-3′; T12C22-11KR, 5′-TCCG GAGATAGCTCAACAGC-3′).
4.4. Plasmid Construction and Plant Transformation
The F2J6 BAC clone and U09195 cDNA clone were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA) and used as a PCR template for AtSDC genomic DNA or ORF amplification. For atsdc-1 complementation, the 3451-bp genomic DNA fragment of AtSDC covering 1276 bp upstream of the translation start codon to 410 bp downstream of the stop codon was amplified by LA taq polymerase (Takara Shuzo CO., Shiga, Japan) with use of F2J6 BAC DNA as a template and the following primers: L126GKpnI-F (5′-CGGGGTACCT CATGTTCTCCGAGAGTTAGGGATA-3′) and L126GXbaI-R2 (5′-GCTCTAGACTTTTGTGT TGTTAGCGTCTATGCAG-3′). The resulting fragment was cloned into pCAMBIA1200 between the KpnI and XbaI sites, resulting in pCAM1200-AtSDCg. For the AtSDC promoter-driven GUS (β-glucuronidase) construct, a 1126-bp fragment spanning −1134~−9 upstream of the AtSDC ORF was amplified by PCR with use of F2J6 BAC DNA as a template and the primer pair L126pEcoRI-F (5′-GAAAGAATTCCTTTGGGGTCTTTGGTTG-3′) and L126pBamHI-R (5′-TAAAGGATCCTGT AGATCTGAGTGATTGGA-3′). The AtSDC promoter fragment was then subcloned into pCAMBIA1381 between the EcoRI and BamHI sites, which resulted in pCAM1381-AtSDCp-GUS. For the construct for the GFP-AtSDC fusion protein, AtSDC ORF was amplified by PCR with U09195 cDNA used as a template and the primer pair L126GFP-XhoI-F (5′-TTAAACTCGAGAT GGTTGGATCTTTGGAATCTG-3′) and L126GFP-BamHI-R (5′-AATGGGGATCCCGCTTGT GAGCTGGACAGAT-3′). The amplified AtSDC ORF was subcloned into pEZTNL between the XhoI and BamHI sites, which resulted in pEZTNL-GFP-AtSDC. pCAM1200-AtSDCg and pCAM1381-AtSDCp-GUS were transferred to Agrobacterium GV3101 (pMP90) and pEZTNL-GFP-AtSDC to Agrobacterium LBA4404 by electroporation at 1250V with capacitance of 25 F and resistance of 400. After appropriate antibiotic selection and PCR confirmation, selected Agrobacterium was grown at 28 C in LB medium (Luria-Bertani, bacto-tryptone 1% (w/v), bacto-yeast extract 0.5% (w/v), NaCl 1% (w/v) pH 7.0) overnight and then used for in planta transformation by floral infiltration.
4.5. Microscopic Analysis
Glufosinate-ammonium resistant GFP-AtSDC transgenic seedlings, selected in soil by spraying 30 mg/L Finale (AgrEvo Environmental Health, Montvale, NJ, USA), were mounted on glass slides, and green fluorescence images were taken under a BioRad MRC-1024 confocal laser-scanning microscope with 488 nm laser excitation and an emission filter of 522/DF35. For GUS staining, the buffer 3 mM X-Gluc, 0.1 M Na-phosphate buffer, pH 7, 0.1% Triton X-100, and 8 mM β-mercaptoethanol was added to the AtSDCp-GUS transgenic seedlings. The samples were incubated overnight at 37 °C and subjected to treatment with 70% ethanol at 70 °C to remove chlorophyll. GUS pictures were taken under a dissecting microscope.
5. Conclusions
In this study, we have characterized the Arabidopsis atsdc-1 mutant defective in serine decarboxylase and analyzed the expression of the AtSDC gene. The atsdc-1 mutants showed leaf necrosis, multiple inflorescence development, floral sterility, and early flowering in short day conditions. These defects were rescued by ethanolamine or choline application to atsdc-1, implicating that ethanolamine and ethanolamine-derived metabolites are important in these developmental processes. Gene expression analysis of the AtSDC gene suggests that Arabidopsis serine decarboxylase is cytosol-localized and expressed in all tissue.
Acknowledgments
This work was supported by the Sogang University Research Grants (201010056 and 201114003) and the Korea Polar Research Institute Grant (PE11060).
References
- 1.Gibellini F., Smith T.K. The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 2.Mudd S.H., Datko A.H. Synthesis of ethanolamine and its regulation in Lemna paucicostata. Plant Physiol. 1989;91:587–597. doi: 10.1104/pp.91.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz C.R. Molecular genetics of membrane phospholipid synthesis. Annu. Rev. Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes D., Hanson A.D. Quaternary ammonium and tertiary sulfonium compouns in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:357–384. [Google Scholar]
- 5.Zinser E., Sperkagottlieb C.D.M., Fasch E.V., Kohlwein S.D., Paltauf F., Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- 7.Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W.I., Voelker D.R., Basset G., Hanson A.D. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 2001;276:35523–35529. doi: 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- 8.Rontein D., Rhodes D., Hanson A.D. Evidence from engineering that decarboxylation of free serine is the major source of ethanolamine moieties in plants. Plant Cell Physiol. 2003;44:1185–1191. doi: 10.1093/pcp/pcg144. [DOI] [PubMed] [Google Scholar]
- 9.Voelker D.R. Phosphatidylserine decarboxylase. Biochim. Biophys. Acta-Lipids Lipid Metab. 1997;1348:236–244. doi: 10.1016/s0005-2760(97)00101-x. [DOI] [PubMed] [Google Scholar]
- 10.Vance J.E., Steenbergen R. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 2005;44:207–234. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Elabbadi N., Ancelin M.L., Vial H.J. Phospholipid metabolism of serine in Plasmodium-infected erythrocytes involves phosphatidylserine and direct serine decarboxylation. Biochem. J. 1997;324:435–445. doi: 10.1042/bj3240435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lykidis A. Comparative genomics and evolution of eukaryotic phospholipid biosynthesis. Prog. Lipid Res. 2007;46:171–199. doi: 10.1016/j.plipres.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy E.P., Weiss S.B. The function of cytidine coenzymes in the biosynthesis of phospholipids. J. Biol. Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 14.Wallis J.G., Browse J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 2002;41:254–278. doi: 10.1016/s0163-7827(01)00027-3. [DOI] [PubMed] [Google Scholar]
- 15.Mou Z.L., Wang X.Q., Fu Z.M., Dai Y., Han C., Ouyang J., Bao F., Hu Y.X., Li J.Y. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell. 2002;14:2031–2043. doi: 10.1105/tpc.001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada K., Lim J., Dale J.M., Chen H.M., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 17.Sandmeier E., Hale T.I., Christen P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 1994;221:997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x. [DOI] [PubMed] [Google Scholar]
- 18.The Arabidopsis Information Resource Home Page. [accessed on 14 September 2007]. Available online: http://www.arabidopsis.org.
- 19.Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H.M., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 20.SeedGenes Project Home Page. [accessed on 7 November 2008]. Available online: http://www.seedgenes.org.
- 21.The Arabidopsis Information Resource Germplasm Datapage. [accessed on 10 January 2009]. Available online: http://www.arabidopsis.org/servlets/TairObject?type=germplasm&id=1005161765.
- 22.Tian G.W., Mohanty A., Chary S.N., Li S.J., Paap B., Drakakaki G., Kopec C.D., Li J.X., Ehrhardt D., Jackson D., et al. High-throughput fluorescent tagging of full-length arabidopsis gene products in planta. Plant Physiol. 2004;135:25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizoi J., Nakamura M., Nishida I. Defects in CTP: PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell. 2006;18:3370–3385. doi: 10.1105/tpc.106.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaoka Y., Yu Y.B., Mizoi J., Fujiki Y., Saito K., Nishijima M., Lee Y., Nishida I. Phosphatidylserine Synthase1 is required for microspore development in Arabidopsis thaliana. Plant J. 2011;67:648–661. doi: 10.1111/j.1365-313X.2011.04624.x. [DOI] [PubMed] [Google Scholar]
- 25.Lightner J., James D.W., Dooner H.K., Browse J. Altered Body Morphology Is Caused by Increased Stearate Levels in a Mutant of Arabidopsis. Plant J. 1994;6:401–412. [Google Scholar]
- 26.Masclaux-Daubresse C., Valadier M.H., Carrayol E., Reisdorf-Cren M., Hirel B. Diurnal changes in the expression of glutamate dehydrogenase and nitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ. 2002;25:1451–1462. [Google Scholar]
- 27.Schaffer R., Landgraf J., Accerbi M., Simon V., Larson M., Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matt P., Schurr U., Klein D., Krapp A., Stitt M. Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the Nia transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta. 1998;207:27–41. doi: 10.1007/s004250050452. [DOI] [PubMed] [Google Scholar]
- 29.Liu J., Zhu J.K. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell C.J., Ecker J.R. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 31.Monsanto Arabidopsis Polymorphism and Ler Sequence Collections Page. [accessed on 6 March 2007]. Available online: http://www.arabidopsis.org/Cereon/index.html.