Abstract
The CAPRICE (CPC)-like MYB gene family encodes R3-type MYB transcription factors in Arabidopsis. There are six additional CPC-like MYB sequences in the Arabidopsis genome, including TRYPTICHON (TRY), ENHANCER OF TRY AND CPC1 and 2 (ETC1 and ETC2), ENHANCER OF TRY AND CPC3/CPC-LIKE MYB3 (ETC3/CPL3), and TRICHOMELESS1 and 2 (TCL1 and TCL2). We independently identified CPC-LIKE MYB4 (CPL4), which was found to be identical to TCL2. RT-PCR analysis showed that CPL4 is strongly expressed in shoots, including true leaves, but not in roots. Promoter-GUS analyses indicated that CPL4 is specifically expressed in leaf blades. Although CPC expression was repressed in 35S::ETC1, 35S::ETC2 and 35S::CPL3 backgrounds, CPL4 expression was not affected by ETC1, ETC2 or CPL3 over-expression. Notably, several chimeric transcripts may result from inter-genic alternative splicing of CPL4 and ETC2, two tandemly repeated genes on chromosome II. At least two chimeric transcripts named CPL4-α and CPL4-β are expected to encode complete CPC-like MYB proteins.
Keywords: Arabidopsis, epidermal cell differentiation, MYB, transcription factor, trichome
1. Introduction
Epidermal cell differentiation in Arabidopsis, including root-hair and trichome cell formation, has been used as a model system to analyze plant cell fate determination. Several regulatory factors are known to be involved in epidermal cell differentiation events. The glabra 2 (gl2) and werewolf (wer) mutants induce an increased number of root-hair cells [1,2]. The GL2 gene encodes a homeodomain leucine-zipper protein, and the WER gene encodes an R2R3-type MYB transcription factor that activates GL2 expression [1–4]. The GL1 and MYB23 genes encode R2R3-type MYB genes that are closely related to WER and are also involved in epidermal cell fate determination in Arabidopsis. The GL1 gene promotes trichome formation, and the gl1 mutant phenotype shows a reduced number of leaf trichomes [5]. GL1 is expressed in developing trichomes [6]. Constitutive expression of the MYB23 gene under the control of the CaMV 35S promoter induces ectopic trichome formation [7]. MYB23 and WER are preferentially expressed in non-hair cells in Arabidopsis roots [8,9]. GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) encode basic helix-loop-helix (bHLH) transcription factors that affect non-hair cell differentiation in a redundant manner [10]. There are two other bHLH genes, AtMYC1 [11] and TRANSPARENT TESTA8 (TT8) [12], that are in the same gene family as GL3 and EGL3 [13]. The TRANSPARENT TESTA GLABRA1 (TTG1) gene encodes a WD40-repeat protein that regulates non-hair cell formation and trichome differentiation [14,15]. Using a yeast two-hybrid system, GL3 and EGL3 were shown to interact with WER [10] and with a WD40 protein (TTG1) [16–18].
The CAPRICE (CPC) gene encodes an R3-type small MYB protein, and the cpc mutation strongly reduces the formation of root hairs [19]. Previously, we reported that the CPC protein moves from non-hair cells to root-hair cells and represses GL2 expression [20,21]. In addition, we proposed a model in which the CPC gene could have arisen by evolution from the WER gene [22]. A protein complex including WER, GL3/EGL3 and TTG1 induces GL2 expression [2,3,14,19,23]. The CPC protein disrupts this protein complex by competitively binding with WER, leading to repression of GL2 expression [24,25]. Arabidopsis has several additional CPC-like MYB sequences in its genome, including TRYPTICHON (TRY), ENHANCER OF TRY AND CPC1 and 2 (ETC1 and ETC2), ENHANCER OF TRY AND CPC3/CPC-LIKE MYB3 (ETC3/CPL3), and TRICHOMELESS1 and 2 (TCL1 and TCL2) [26–33]. Trichome cluster formation on leaves of the try mutant indicates that the TRY protein has a regulatory role in trichome differentiation [26,34]. ETC1 and ETC2 have been identified and their function with CPC and TRY genetically examined [27–29]. TCL1 and TCL2 negatively regulate trichome formation on the inflorescence stems and pedicels [32,33]. We have recently identified a seventh CPC-like MYB gene between At2g30430 and ETC2 (At2g30420) independently of Gan et al. [33], and have named it CPC-LIKE MYB4 (CPL4). In this paper, we examine the expression of the CPL4 gene in Arabidopsis. Notably, between CPL4 and ETC2, there were several chimeric transcripts generated through alternative splicing. Around 14% of the protein coding genes of Arabidopsis are annotated as producing multiple transcript variants through alternative splicing [35]. Our study proposes that inter-genic alterative splicing also characterizes the CPC-like MYB gene family.
2. Results and Discussion
2.1. CPC-LIKE MYB4 (CPL4) Gene in Arabidopsis
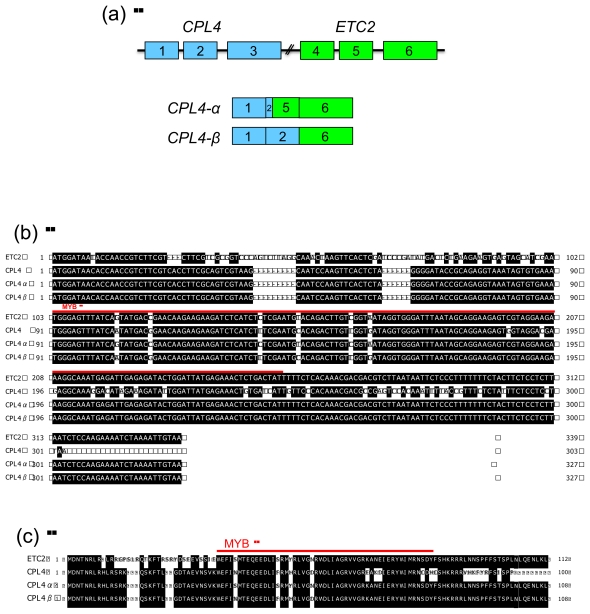
CPC encodes an R3-type MYB transcription factor and promotes root-hair cell differentiation [19]. A search of the Arabidopsis genome sequence revealed six MYB gene sequences with high homology to CPC: TRY, ETC1, ETC2, CPL3/ETC3 and TCL1 [26–32]. In addition to these six CPC-like MYB genes, we independently identified the CPC-LIKE MYB4 (CPL4) gene in the Arabidopsis genome. The CPL4 gene encodes a CPC-LIKE R3-type MYB sequence and is situated between TCL1 (At2g30432) and ETC2 (At2g30420) (Figure 1a). Amino acid sequence alignment showed that these two tandemly repeated genes, CPL4 and ETC2, share high homology (Figure 1a,b). CPL4 was identical with TRICHOMELESS2 (TCL2) [33]. The CPL4 protein shares 80% amino acid homology with TCL1, 70% with TRY, 68% with ETC2, 49% with CPL3/ETC3, 48% with CPC and 47% with ETC2. As expected from the amino acid sequence of CPL4, overexpression of CPL4 resulted in a glabrous phenotype similar to CPC, TRY, ETC1, ETC2, CPL3/ETC3 or TCL1 overexpressors [33].
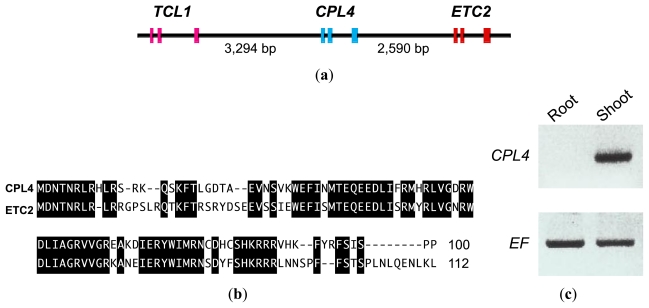
Figure 1.
Genomic structure, amino acid sequence and expression of the CPL4 gene. (a) Structure of tandemly arranged genes, ETC2, CPL4 and TCL1, in the genome. The exons are represented by pink (ETC2), blue (CPL4) and red (TCL1). The lengths of DNA sequences between genes are shown in base pairs; (b) Amino acid sequence comparison of CPL4 and ETC2 proteins generated using Genetyx ver.16.0.2 software [36]. Identical amino acids are shaded in black; (c) Expression of CPL4 in shoots of Arabidopsis seedlings. Tissues from Arabidopsis roots or shoots were collected, RNA was isolated, and RT-PCR was performed to check for the expression of CPL4. The PCR product size using primer pair (RT341/RT342) was 241 bp. The expression of Elongation Factor1α (EF) was used as a control.
CPL4 expression was examined in Arabidopsis roots and shoots using semi-quantitative RT-PCR analysis (Figure 1c). CPL4 was strongly expressed in shoots (including a few small true leaves). We did not detect CPL4 expression in Arabidopsis seedling roots (Figure 1c). Through this assay, we confirmed the expression of a seventh CPC-like MYB gene that had not been previously recognized.
2.2. Promoter-GUS Analysis
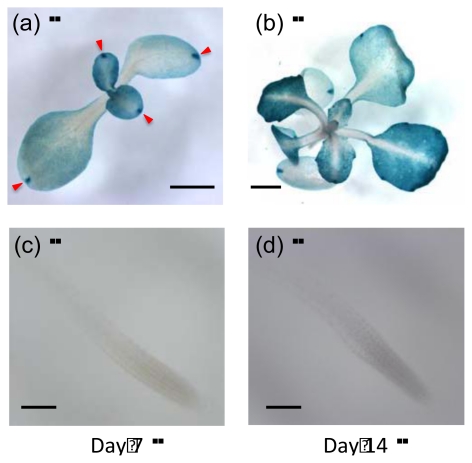
To analyze CPL4 expression at the tissue level, we made CPL4 promoter-GUS fusions. CPL4p::GUS was expressed in young true leaves and cotyledons of 7-day-old seedlings (Figure 2a). CPL4p::GUS expression was also observed in two-week-old rosette leaves (Figure 2b). Previously, we showed that the expression patterns of CPC-like MYB genes could be roughly classified into two groups [31]. CPCp::GUS, TRYp::GUS and ETC1p::GUS are expressed mainly in roots and trichomes, and ETC2p::GUS and CPL3p::GUS are expressed in young leaves and mainly in guard cells [31]. Thus, GUS expression by the CPC-like MYB family is found in tissues throughout the entire plant body. We did not detect CPL4p::GUS expression in trichomes. Unexpected strong CPL4p::GUS expression was observed in hydathodes (Figure 2a). Consistent with the results of RT-PCR (Figure 1b), we did not detect CPL4p::GUS expression in 7-day- and two-week-old Arabidopsis roots (Figure 2c,d).
Figure 2.
Expression of CPL4p::GUS in Arabidopsis leaves and roots. (a) CPL4p::GUS expression in cotyledons and true leaves of 7-day-old seedlings. Arrowheads indicate hydathodes; (b) CPL4p::GUS expression in two-week-old rosette leaves; (c) CPL4p::GUS expression in roots of 7-day-old seedlings; (d) CPL4p::GUS expression in roots of two-week-old plants. Scale bars, 1 mm ([a] and [b]), 100 μm ([c] and [d]).
2.3. CPL4 Expression in 35S::ETC1, 35S::ETC2 and 35S::CPL3/ETC3
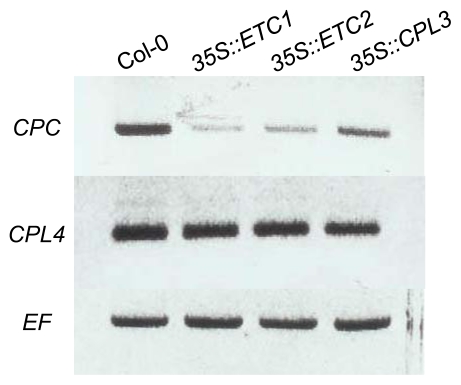
In addition to the spatial expression patterns, CPC expression was regulated by the CPC MYB protein itself as shown in 35S::CPC transgenic lines [24]. The mechanism for this repression may be by negative feedback, thereby contributing to the root-hair and non-hair cell differences. Because CPC protein shares high amino acid sequence homology with the other CPC-like MYBs, we checked the effect of ETC1, ETC2 or CPL3 overexpression on CPC expression (Figure 3). Semi-quantitative RT-PCR analyses showed that CPC expression was clearly repressed in the 35S::ETC1, 35S::ETC2 and 35S::CPL3 backgrounds (Figure 3). This result suggests the existence of a similar feedback loop resulting in the proper trichome distribution on leaves. To compare the regulation of gene expression, we also performed RT-PCR analyses using CPL4-specific primers. In plants harboring 35S::ETC1, 35S::ETC2 and 35S::CPL3, CPL4 was expressed at almost the same level as wild-type (Col-0) (Figure 3). These results suggest that CPL4 is not involved in a feedback regulatory cascade controlled by CPC-like MYBs. The CPC-like MYB family members are thought to have evolved by gene duplication [31] and have diverged to different regulatory functions in the course of evolution. The CPC-like genes retain some functional redundancy that may represent intermediate stages of regulatory specification [37,38]. Thus, CPL4 may have acquired specific functions different from that of CPC.
Figure 3.
Semi-quantitative RT-PCR analyses of CPL4 expression in Arabidopsis. Tissues from 12-day-old rosette leaves of Arabidopsis seedlings were collected, RNA was isolated, and RT-PCR was performed to investigate the expression of CPC and CPL4 in 35S::ETC1, 35S::ETC2 or 35S::CPL3 overexpressing backgrounds. The expression of EF was used as a control.
2.4. CPL4-Related Gene
2.4.1. CPL4-Related Gene Expression
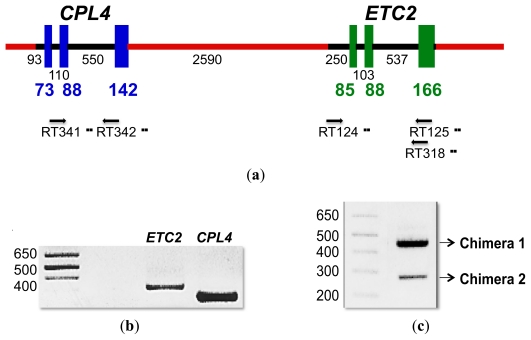
CPL4 is 2590 bp upstream of the other CPC-like MYB gene, ETC2, on chromosome II and lies in a head-to-tail orientation (Figures 1a and 4a). Both genes are composed of three exons (Figures 1a and 4a). As shown in Figure 1b, CPL4 shares high amino acid sequence homology with ETC2. Therefore, the forward primer beginning with the predicted start codon of ETC2 can also act as the forward primer beginning with the start codon of CPL4. Therefore, it was necessary to carefully design primer pairs specific for ETC2 and CPL4. Gene-specific RT-PCR was performed for ETC2 and CPL4 using rosette leaves from 12-day-old Arabidopsis seedlings. Both primer pairs designed for ETC2 (RT124/RT125) and for CPL4 (RT341/RT342) amplified specific, single gene products of the expected sizes (320 bp and 240 bp) (Figure 4b). Using the CPL4-specific forward primer (RT341) and ETC2-specific reverse primer (RT125) as a primer pair, two DNA fragments named CPL4-related Chimera 1 and Chimera 2 were amplified (Figure 4c). An abundantly expressed band (Chimera 1) was estimated to be approximately 450 bp, and a less expressed band (Chimera 2) was approximately 280 bp in size (Figure 4c). To confirm the existence of CPL4-related chimeric gene expression, we repeated the RT-PCR experiment using new RNA samples and another primer pair (RT341/RT318). As a result, the existence of Chimera 1 and 2 cDNA products was confirmed.
Figure 4.
Existence of CPL4-related chimeric transcripts. (a) Diagram illustrating the CPL4 and ETC2 genes located on Arabidopsis chromosome II. The blue boxes represent CPL4 exons. The green boxes represent ETC2 exons. The black lines represent introns and UTR. The red lines represent intergenic DNA. The lengths of DNA sequences are noted in base pairs. The positions of the PCR primers used in the RT-PCR are indicated with arrows; (b) Expression of ETC2 and CPL4 transcripts in Arabidopsis leaves. RNA was isolated from 12-day-old rosette leaves, and RT-PCR was performed to examine the expression of ETC2 and CPL4 transcripts. The size of the marker in base pairs is shown on the left; (c) Expression of ETC2 and CPL4 chimeric transcripts in Arabidopsis leaves. RNA was isolated from 12-day-old rosette leaves, and RT-PCR was used to examine the expression of chimeric transcripts. The size of the marker in base pairs is shown on the left.
2.4.2. CPL4-Related Chimera Sequences
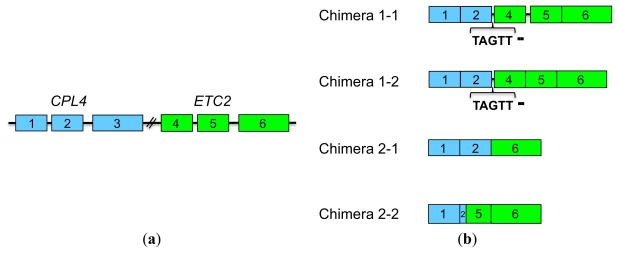
To determine the precise structure of CPL4-related chimeric transcripts (Chimera 1 and 2), we cloned these RT-PCR products and subsequently sequenced the constructs. Chimera 1 included two different amplicons, named Chimera 1-1 and Chimera 1-2 (Figures 4c and 5b). Chimera 2 also included two different amplicons, named Chimera 2-1 and Chimera 2-2 (Figures 4c and 5b). As shown in Figure 5a, CPL4 and ETC2 lie in a tandem orientation (Figure 5a). All chimeric transcripts may result from alternative splicing of CPL4 and ETC2. Chimera 1-1 and Chimera 1-2 contained the first two exons of CPL4 and all three exons of ETC2 (1, 2, 4, 5 and 6 exons) (Figure 5b). Both chimeric transcripts (Chimera 1-1 and Chimera 1-2) contain a TAGTT additional sequence between the second exon of CPL4 and the first exon of ETC2. This linker sequence ‘TAGTT’ was just upstream of the 5′-UTR region of the first exon of ETC2. Although Chimera 1-1 included the first intron of ETC2, Chimera 1-2 did not (Figure 5b). Chimera 2-1 contained the first and second exons of CPL4 and the third exon of ETC2 (1, 2 and 6 exons) (Figure 5b). The “TAGTT” linker insertion of Chimera 1-1 and 1-2 was produced by GT-AG splicing between the second intron of CPL4 and an upstream region of the first exon of ETC2. This linker sequence included a “TAG” sequence that could function as a stop codon. Therefore, Chimera 1-1 and 1-2 are expected to produce a truncated MYB protein that includes only the first two exons of CPL4. On the other hand, transcripts of Chimera 2-1 and 2-2 are expected to encode complete CPC-like MYB proteins. Chimera 2-1 contained the first two exons of CPL4 and the third exon of ETC2 (exons 1, 2 and 6) (Figure 5b). Chimera 2-2 contained the first CPL4 exon, the first 16 bp of the second exon of CPL4, the second exon of ETC2 lacking the first 16 bp, and the third exon of ETC2 (exons 1, 2, 5 and 6) (Figure 5b). Thus, we renamed Chimera 2-1 and 2-2 to CPL4-α and CPL4-β, respectively (Figure 6a). CPL4-α shares 90% identity with ETC2 and 83% identity with CPL4 at the nucleotide level. CPL4-β shares 89% identity with ETC2 and 84% identity with CPL4 at the nucleotide level (Figure 6b). CPL4-α shares 86% identity with ETC2 and 76% identity with CPL4 at the amino acid level. CPL4-β shares 82% identity with ETC2 and 80% identity with CPL4 at the amino acid level (Figure 6c). Both CPL4-α and CPL4-β contain the conserved amino acid signature [D/E]Lx2[R/K]x3Lx6Lx3R that is required for the interaction with R/B-like bHLH transcription factors [39]. Their results strongly suggest that CPL4-α and CPL4-β serve as CPC-like MYB proteins harboring similar functions of ETC2 and/or CPL4.
Figure 5.
Gene structure of CPL4-related chimeric transcripts. (a) Structure of CPL4 and ETC2 genes. The blue boxes 1, 2 and 3 represent CPL4 exons. The green boxes 4, 5 and 6 represent ETC2 exons. The black lines represent introns; (b) Structure of CPL4-related chimeric transcripts, Chimera 1-1, 1-2, 2-1 and 2-2, produced from CPL4 and ETC2 by intergenic alternative splicing. The blue boxes 1, 2 and 3 represent CPL4 exons. The green boxes 4, 5 and 6 represent ETC2 exons. The black lines represent introns. The linker sequences have been inserted between CPL4 and ETC2 in Chimera 1-1 and Chimera 1-2.
Figure 6.
Gene structure and amino acid sequences of CPL4-α and CPL4-β. (a) Structure of CPL4 and ETC2 genes. The blue boxes 1, 2 and 3 represent CPL4 exons. The green boxes 4, 5 and 6 represent ETC2 exons. The black lines represent introns; (b) cDNA sequence alignment of ETC2, CPL4, CPL4-α and CPL4-β transcripts. Identical nucleotides are shaded in black. The red line indicates the MYB region; (c) Amino acid sequence alignment of ETC2, CPL4 and deduced CPL4-α and CPL4-β proteins. Identical amino acids are shaded in black. The red line indicates the MYB region.
Our study raises the possibility that there are unexpected alternative splicing sites spanning two homologous genes. In Arabidopsis, there are many examples of genes where alternative splicing provides a regulatory mechanism controlling aspects of development and other processes including flowering [40]. Although many genes produce alternatively spliced transcripts, the critical role of alternative splicing is poorly understood. This work provides additional evidence for the function of alternative splicing.
3. Experimental Section
3.1. Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild type in this study. Seeds were surface-sterilized and sown on 1.5% agar plates as described previously [41]. Construction of the 35S::CPL3 transgenic line was described previously [31]. Seeded plants were kept at 4 °C for 2 days and then incubated at 22 °C under constant white light (50–100 μmol·m−2·s−1).
3.2. Gene Constructs
Sequences of all primers used in this paper are listed in Table 1. All PCR-generated constructs were completely sequenced following isolation of the clones to check for amplification-induced errors.
Table 1.
Primer sequences used in this study.
| Primer Name | Sequence |
|---|---|
| RT124 | 5′-GATAATACCAACCGTCTTCGTCTTC-3′ |
| RT125 | 5′-TTCTTGGAGATTAAGAGGAGAAGTAG-3′ |
| RT128 | 5′-CTTCTTGTTTCTCGAGATTTATTCTC-3′ |
| RT129 | 5′-AATAGTAATTCAAGGACAGGTACATTTC-3′ |
| RT318 | 5′-GAATTATTAAGACGTCGTCGTTTGTGAG-3′ |
| RT341 | 5′-AAGCAATCCAAGTTCACTCTAGGG-3′ |
| RT342 | 5′-CGGTAAATTTGTGGACTCGG-3′ |
| NEKO45 | 5′-ATATGTCGACTACCAAAATCACTCCACCATTTTC-3′ |
| NEKO47 | 5′-ATATGGATCCGTTGGTGTTATCCATTGGTATTTG-3′ |
| TW1165 | 5′-ATATGGTACCAATAAAAAATAAATCAC-3′ |
| TW1166 | 5′-TGCTTGTCGACTGTATACACTAA-3′ |
| TW1169 | 5′-ATATGGTACCACTTCATGTTCTTCCCTT-3′ |
| TW1170 | 5′-ATATGTCGACAAGCCAATACATATCCA-3′ |
3.2.1. Promoter::GUS Constructs
A 2.7 kb PCR-amplified promoter region of CPL4 (primers NEKO45/NEKO47) was digested with SalI and BamHI and subcloned into pBluescript SK+ (Stratagene, La Jolla, CA, USA) to create pBS-CPL4p. The SalI and BamHI digested fragment of pBS-CPL4p was ligated into the SalI and BamHI sites of binary vector pBI101 (Clontech Laboratories, Inc., Mountair View, CA, USA) to create the CPL4p::GUS constructs.
3.2.2. 35S::ETC1 and 35S::ETC2 Constructs
A 0.5 kb PCR-amplified linear ETC1 genome fragment (primers TW1169/TW1170), and a 1.0 kb PCR-amplified linear ETC2 genome fragment (primers TW1165/TW1166) were subcloned into pBS using Pyrobest DNA polymerase (Takara, Tokyo, Japan) to make pBS-ETC1 and pBS-ETC2. Next, Acc65I to SalI fragments were ligated into the Acc65I to SalI sites of the pCHF3 binary vector [42] to create 35S::ETC1 and 35S::ETC2.
3.3. Transgenic Plants
Plant transformation was performed by a floral dip method [43], and transformants were selected on 0.5 × MS agar plates containing 50 mg/L kanamycin. Homozygous transgenic lines were selected for kanamycin resistance. At least twelve T1 lines were isolated for each construct and at least six T2 and three T3 lines were selected on the basis of their segregation ratios for kanamycin resistance.
3.4. RNA Isolation and Semi-Quantitative RT-PCR
Total RNA was extracted using RNeasy Plant Mini Kits (Qiagen, Valencia, CA, USA). On-column DNase I digestion was performed during RNA purification following the protocol described in the RNeasy Mini Kit handbook. First-strand cDNA was synthesized from 1 μg total RNA in a 20 μL reaction mixture using the Prime Script RT Regent Kit (Takara).
Semi-quantitative RT-PCR reactions were conducted as described by Kurata et al. [44]. The CPL4 fragment was amplified with RT341/RT342 primer pairs. The CPC fragment was amplified with RT128/RT129 primer pairs. The ETC2 fragment was amplified with RT124/RT125 primer pairs. EF was amplified with the EF1α-F/EF1α-R primer pair as described by Kurata et al. [21]. To determine the sequence of the PCR products, we cloned them into the pT7 blue T-vector (Novagen) and subsequently sequenced the constructs.
3.5. Histology
Promoter::GUS plants were excised from the growth medium and immersed in X-Gluc solution containing 1.0 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-glucuronide), 1.0 mM K3Fe(CN)6, 1.0 mM K4Fe(CN)6, 100 mM NaPi (pH 7.0), 100 mM EDTA and 0.1% Triton X-100. Primary roots of 7-day- or two-week-old seedlings were incubated at 37 °C overnight. Cotyledons of 7-day-old seedlings and two-week-old rosette leaves were incubated at 37 °C for 3 h.
3.6. Microscopy
Root GUS activity was observed using an Olympus Provis AX70 microscope. At least five individual primary roots of 7-day- and two-week-old seedlings were analyzed for root GUS activity. To observe leaf GUS activity, an Olympus SZH binocular microscope was used. At least five cotyledons of 7-day-old seedlings and two-week-old rosette leaves were analyzed for leaf GUS activity.
3.7. Accession Numbers
CPC (At2g46410), TRY (At5g53200), ETC1 (At1g01380), ETC2 (At2g30420), CPL3/ETC3 (At4g01060), CPL4 (At2g30424) and TCL1 (At2g30432).
4. Conclusions
In this paper, we investigated the expression of CPL4 and CPL4-related chimeric transcripts in Arabidopsis using promoter-GUS and RT-PCR analyses. The results showed that CPL4 is specifically expressed in leaf blades, and CPL4 expression was not affected by ETC1, ETC2 or CPL3 overexpression. We hypothesize that the proteins created by inter-genic alternative splicing between CPL4 and ETC2 act in a redundant manner with other CPC-like MYB family proteins. Much work remains to determine the function of CPC-like MYB gene family members. By overexpressing 35S::CPL4-α and 35S::CPL4-β and investigating the localization of CPL4::CPL4-α:GFP and CPL4::CPL4-β:GFP in transgenic plants, we should be in a better position to elucidate the roles of these CPC-like MYB genes in Arabidopsis epidermal cell differentiation.
Acknowledgments
We thank Tetsuya Ishida, Ryosuke Sano, Tetsuya Kurata and Takuji Wada for useful suggestions, and Mineko Iwata for technical supports. This work was financially supported by the Program to Disseminate Tenure Tracking System from the Ministry of Education, Culture, Sports, Science and Technology, a grant for Scientific Research on Priority Areas from the University of Miyazaki, Grant-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science (No. 23570057) and Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (No. 23012035).
References
- 1.Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 2.Lee M.M., Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 3.Rerie W.G., Feldmann K.A., Marks M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 4.di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J.C., Oppenheimer D.G., Pollock S., Marks M.D. Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell. 1993;5:1739–1748. doi: 10.1105/tpc.5.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirik V., Schnittger A., Radchuk V., Adler K., Hulskamp M., Baumlein H. Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 2001;235:366–377. doi: 10.1006/dbio.2001.0287. [DOI] [PubMed] [Google Scholar]
- 8.Lee M.M., Schiefelbein J. Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development. 2001;128:1539–1546. doi: 10.1242/dev.128.9.1539. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell. 2009;21:1080–1094. doi: 10.1105/tpc.108.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- 11.Urao T., Yamaguchi-Shinozaki K., Mitsukawa N., Shibata D., Shinozaki K. Molecular cloning and characterization of a gene that encodes a MYC-related protein in Arabidopsis. Plant Mol. Biol. 1996;32:571–576. doi: 10.1007/BF00019112. [DOI] [PubMed] [Google Scholar]
- 12.Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 14.Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 15.Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne C.T., Zhang F., Lloyd A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esch J.J., Chen M., Sanders M., Hillestad M., Ndkium S., Idelkope B., Neizer J., Marks M.D. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 19.Wada T., Tachibana T., Shimura Y., Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1136. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 20.Wada T., Okada K. Development of root-hair and trichome in Arabidopsis. Tanpakushitsu Kakusan Koso. 2002;47:1599–1604. [PubMed] [Google Scholar]
- 21.Kurata T., Ishida T., Kawabata-Awai C., Noguchi M., Hattori S., Sano R., Nagasaka R., Tominaga R., Koshino-Kimura Y., Kato T., et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- 22.Tominaga R., Iwata M., Okada K., Wada T. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell. 2007;19:2264–2277. doi: 10.1105/tpc.106.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 24.Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., Marks M.D., Shimura Y., Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- 25.Koshino-Kimura Y., Wada T., Tachibana T., Tsugeki R., Ishiguro S., Okada K. Regulation of CAPRICE Transcription by MYB Proteins for Root Epidermis Differentiation in Arabidopsis. Plant Cell Physiol. 2005;46:817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- 26.Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jurgens G., Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirik V., Simon M., Huelskamp M., Schiefelbein J. The enhancer of try and CPC1 gene acts redundantly with triptychon and caprice in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 28.Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 2004;55:389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- 29.Esch J.J., Chen M.A., Hillestad M., Marks M.D. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004;40:860–869. doi: 10.1111/j.1365-313X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- 30.Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Tominaga R., Iwata M., Sano R., Inoue K., Okada K., Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135:1335–1345. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- 32.Wang S., Kwak S.H., Zeng Q., Ellis B.E., Chen X.Y., Schiefelbein J., Chen J.G. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134:3873–3882. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- 33.Gan L., Xia K., Chen J.G., Wang S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in. Arabidopsis. BMC Plant Biol. 2011;11 doi: 10.1186/1471-2229-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hulskamp M., Misra S., Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 35.English A.C., Patel K.S., Loraine A.E. Prevalence of alternative splicing choices in. Arabidopsis thaliana. BMC Plant Biol. 2010;10 doi: 10.1186/1471-2229-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genetyx Software, version 16.0.2. Genetyx; Tokyo, Japan: 2011. [Google Scholar]
- 37.Thomas J.H. Thinking about genetic redundancy. Trends Genet. 1993;9:395–399. doi: 10.1016/0168-9525(93)90140-d. [DOI] [PubMed] [Google Scholar]
- 38.Cooke J., Nowak M.A., Boerlijst M., Maynard-Smith J. Evolutionary origins and maintenance of redundant gene expression during metazoan development. Trends Genet. 1997;13:360–364. doi: 10.1016/s0168-9525(97)01233-x. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 40.Eckardt N.A. Alternative splicing and the control of flowering time. Plant Cell. 2002;14:743–747. doi: 10.1105/tpc.000000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada K., Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- 42.Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- 43.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Kurata T., Kawabata-Awai C., Sakuradani E., Shimizu S., Okada K., Wada T. The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 2003;36:55–66. doi: 10.1046/j.1365-313x.2003.01854.x. [DOI] [PubMed] [Google Scholar]