Abstract
Autophagosomes are double-membrane vesicles characteristic of macroautophagy, a degradative pathway for cytoplasmic material and organelles terminating in the lysosomal or vacuole compartment for mammals and yeast, respectively. This highly dynamic, multi-step process requires significant membrane reorganization events at different stages of the macroautophagic process. Such events include exchange and flow of lipids and proteins between membranes and vesicles (e.g., during initiation and growth of the phagophore), vesicular positioning and trafficking within the cell (e.g., autophagosome location and movement) and fusion of autophagosomes with the boundary membranes of the degradative compartment. Here, we review current knowledge on the contribution of different organelles to the formation of autophagosomes, their trafficking and fate within the cell. We will consider some of the unresolved questions related to the molecular mechanisms that regulate the “life and death” of the autophagosome.
Keywords: autophagosome, degradation, lysosome, macroautophagy, mammals, membrane, organelle, yeast
1. Autophagy
Autophagy refers to a set of cellular homeostasis processes conserved across all eukaryotes that collectively serve as a tightly regulated intracellular surveillance mechanism, which is indispensable for maintenance of cell health. Induced by cellular stress, such as nutrient limitation, autophagy is the means by which lysosomes in mammalian cells and the vacuole in yeast contribute to the turnover of cellular components (autophagic cargo), including long-lived proteins, macromolecules, whole organelles and even pathogens. By degrading a plethora of (intra)cellular constituents for recycling, autophagy enables cells to survive periods of stress, whether initiated by events intrinsic or extrinsic to the cell. In multi-cellular organisms, impaired autophagic function seems to underlie a range of pathological conditions [1–5].
In mammalian cells, there are three distinct forms of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). CMA is apparently absent from lower eukaryotes, including yeast. The relative contribution of each form of autophagy under different physiological or pathological settings is poorly understood. In CMA, soluble proteins bearing a KFERQ pentapeptide motif are recognized, unfolded and transported directly across the limiting membrane of the lysosome by specific protein machinery [6]. Microautophagy involves the direct engulfment of cargo at the lysosome/vacuole surface by invagination, or protrusion and septation of the lysosome/vacuole membrane [7–9] or internalization by late endosomes (endosomal microautophagy) [10,11]. By contrast, during macroautophagy, cargo is sequestered into a double-membrane vesicle, termed the autophagosome (AP) [12–18]. It is this transient vesicle, the morphological hallmark of macroautophagy, which is the subject of this review. In particular, we will focus on membrane events, including the exchange and flow of lipids and proteins at different stages of the macroautophagic process.
2. Macroautophagy
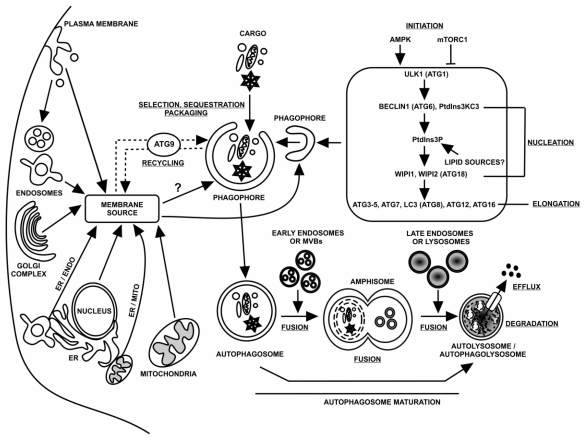
This pathway consists of a number of distinct steps and includes: (1) formation of the phagophore (i.e., the membrane that will become the AP boundary membrane, sometimes referred to as the isolation membrane); (2) phagophore expansion; (3) cargo selection and packaging; (4) AP formation; (5) AP maturation (which may involve AP fusion with endosomes or multivesicular bodies (MVBs)); (6) cargo delivery by fusion of the “mature” AP with the lysosome/vacuole; (7) breakdown of the sequestered cargo to metabolic “building blocks” and (8) efflux of breakdown products [1]. Upon induction of macroautophagy by nutrient deprivation, or some other signal, the formation of the phagophore is initiated. As expansion takes place, membrane curvature is induced to facilitate eventual sequestration of the cargo. A capacity for variable phagophore expansion allows the cell to adjust the size of APs in order to sequester cellular constituents over a very wide range in size, including organelles, such as a mitochondrion [19,20]. Eventually, the two opposing ends of the elongating phagophore membrane fuse to form an AP, thereby sequestering the cargo from the rest of the cell. The outer-membrane of a newly formed AP may then fuse with the boundary membrane of lysosomes (mammalian cells) or the vacuole (yeast cells). Such a fusion event allows the mixing of contents carried by the respective membrane-bound compartments as well as the delivery of the AP sequestered cargo into the acidic lumen (pH ~ 4.5–6). Through the action of resident lysosomal/vacuolar hydrolases, cargo is degraded to basic metabolic building blocks (e.g., nucleotides, amino acids, sugars, fatty acids), which are exported to the cytosol for reuse by the cell (Figure 1) [21].
Figure 1.
Schematic model of dynamic membrane events contributing to autophagosome life and death. The life and death of autophagosomes, which is evolutionarily conserved, involves the following stages (underlined): initiation, phagophore nucleation, phagophore elongation through two conjugation cascades, cargo selection, sequestration and packaging, membrane recycling regulated by Atg9, fusion with lysosomes, degradation and efflux. Key proteins that act at these stages during such processes are listed. Potential sources for the phagophore include the endoplasmic reticulum (ER), nuclear membranes, mitochondria, plasma membrane, Golgi complex and endosomes. The phagophore expands, sequestering and packaging the selected autophagy cargo, and finally closes, forming an immature autophagosome. During the maturation process, an amphisome is formed from the fusion of an autophagosome with early endosomes or multivesicular bodies, whereas an autolysosome/autophagolysosome is formed as a product of fusion of amphisomes events with late endosomes or lysosomes. Once in the lysosomes, the autophagic cargo is degraded into essential building blocks that are transported back into the cytoplasm. Abbreviations: AMPK, AMP-activated protein kinase; Atg, autophagy-related; BECLIN 1, BCL-2 interacting myosin/moesin-like coiled-coil protein 1; ER, endoplasmic reticulum; ER/ENDO, endoplasmic reticulum/endosomes contact; ER/MITO, endoplasmic reticulum/mitochondria contact; LC3, light chain 3; mTORC1, mammalian target of rapamycin complex 1; MVBs, multivesicular bodies; PM, plasma membrane; PtdIns3P, phosphatidylinositol 3-phosphate; PtdIns3KC3; phosphatidylinositol 3-kinase class III; ULK1, UNC51-like kinase 1; WIPI1/WIPI2, WD repeat domain phosphoinositide-interacting 1/2.
Macroautophagy utilizes a set of core gene products (many are designated as autophagy-related: Atg), but may require cargo type-specific components also. Although macroautophagy was initially described as a non-selective process in terms of cargo selection, it is now well recognized that particular cell components, structures and organelles can be selectively targeted (Table 1) [22–54].
Table 1.
Selective types of autophagy. For further information on different types of selective autophagy, we refer the reader to the references listed.
| Selective type of autophagy | Cargo | Organism | References |
|---|---|---|---|
| Aggrephagy | Protein aggregates | Mammals | [22–25] |
| Cytoplasm-to-vacuole targeting (Cvt) pathway | Pro-aminopeptidase 1 (prApe1), pro-α mannosidase 1 (prAms1) and aspartyl aminopeptidase (Ape4) | Yeast | [22,26–28] |
| ER-phagy/reticulophagy | ER | Yeast and Mammals | [22,28–32] |
| Lipophagy | Lipids | Mammals | [33–35] |
| Lysophagy/Lysosomophagy | Vacuole/Lysosomal membrane | Yeast and Mammals | [22] |
| Mitophagy | Mitochondria | Yeast and Mammals | [20,22,36–39] |
| Nucleophagy | Nucleus | Yeast and Mammals | [22,40,41] |
| Pexophagy | Peroxisomes/peroxisome cluster | Yeast and Mammals | [22,42–46] |
| Piecemeal-microautophagy of the nucleus (PMN) | Portions of the nucleus | Yeast | [22,47–49] |
| Ribophagy | Ribosomes | Yeast | [28,50,51] |
| Secretophagy | Atg15 protein | Yeast | [52] |
| Vacuole import and degradation (Vid) pathway | Fructose-1,6-bisphosphatase (FBPase) | Yeast | [22,53] |
| Xenophagy | Pathogens (bacteria and viruses) | Plants and Mammals | [22,23,54] |
3. Autophagosomes (APs)
As the defining structure of the macroautophagy pathway, the AP is characterized by a number of unique properties: site(s) of initiation, structure, source of membrane lipids, trafficking and fusion events.
3.1. Site(s) of Initiation
The first crucial, but least understood, event in AP formation is the induction and nucleation of the phagophore membrane that will grow into the limiting membrane that sequesters cargo. In mammalian cells, multiple sites of AP formation can be detected throughout the cytoplasm [55,56]. In yeast, APs are generated at, or around the pre-autophagosomal structure (PAS), a single functional site situated close to the vacuole membrane [56]. It is not known why the PAS should be located close to the vacuole and how it is actually formed. It has been suggested that it may be physically linked to the ER, which is consistent with the suggestion that it is usually localized at a nuclear-vacuolar junction [14,57,58]. In serving as the phagophore initiation and assembly site the PAS is considered to be a dynamic structure to which many Atg proteins are recruited in an ordered manner and rapidly disassembled from the PAS once they fulfill their purpose [14,57,58].
3.2. Structure
The AP can be considerably larger than most other vesicles in the cell and has a membrane that is apparently enriched for a relatively small number of different proteins. This seemingly limited repertoire of proteins suggests that the AP membranes contain only the minimal components necessary to load the cargo destined for eventual degradation and to facilitate subsequent fusion with the limiting membrane of the degradative compartment [59–61]. The compositions of the outer and inner AP membranes seem to be quite different [62], and this may reflect their different roles: cargo sequestration by the inner membrane cargo and fusion with the degradative compartment by the outer membrane. APs in yeast range from 0.4–0.9 μm in diameter [63], whereas mammalian APs are usually larger, being 0.5–1.5 μm in diameter [64–66]. The size of APs may be determined in relation to a specific cargo, which can range from proteins to intracellular bacteria [4]. While the volume of each AP represents less than 0.1% of the total cellular volume in mammalian cells, because the half-life of an AP is considered to be very short (5–10 min), the total degradative capacity of macroautophagy can be large [64–67].
3.3. Source of Membrane Lipids
The origin of the AP membranes, whether from pre-existing membranes or formed by de novo synthesis, has long been a controversial but fundamental question in the field of autophagy. Recently, a number of studies have suggested the ER, nucleus, mitochondria, plasma membrane, endosomes and Golgi complex may each serve as a source of lipids (Figure 1) [14,15]. The relative contribution of each of these sites to formation of the AP is not presently known. It is possible that different membrane sources are utilized, dependent on the cell type, stress and intended cargo.
3.4. Trafficking and Fusion Events
The completed APs loaded with cargo must traffic to and fuse with lysosomes or the vacuole in order to acquire degradative capacity. Moreover, APs can either fuse homotypically with other APs, or receive inputs from the endocytic pathway (Figure 1) by fusing heterotypically with early endosomes or multivesicular bodies (MVBs) to form amphisomes. In turn, amphisomes can fuse with late endosomes. Using time-lapse fluorescence microscopy, Kimura and colleagues [68] have shown that, in mammalian cells, APs do not move far from their site of formation until they are completed. After completion, mammalian APs exhibit rapid vectorial, dynein- and microtubule-dependent movement in the direction of lysosomes; the average velocity of AP movement being 5 μm/s [68]. However, the detailed mechanism by which this process occurs is still far from fully understood.
4. Autophagosome Origin and Birth
In this section of the review, we discuss and analyze current understanding of the origin of APs (Figure 1) and the mechanism(s) that lead to AP formation. However, a major challenge in understanding these processes arises from the fact that the different potential membrane sources and mode of transport of lipids from them are only now beginning to be probed rigorously.
4.1. ER and Mitochondrial Membranes
Ktistakis and colleagues [69] have reported that in mammalian cells subjected to amino acid starvation, PI3P-enriched structures named omegasomes form in close proximity to ER membranes and Vps34-positive endosomes. An omegasome marker, DFCP1 (a phospholipid binding protein), colocalizes with the autophagy-specific proteins, Atg5 and LC3 (mammalian counterpart of yeast Atg8), which are recruited to sites of AP formation by upstream factors such as the ULK1 complex, the PI3 kinase complex and at a later stage Atg9 (Figure 1), promoting the formation of the curved, cradle-like phagophore by membrane invagination at the centre of the omegasome. Once formed, an autophagic structure seems to exit the omegasome [69,70].
Electron tomography studies [71,72] have delineated the 3D architecture of the relationship between the ER and the phagophore. These studies confirmed that a portion of the ER forms a cradle-like structure surrounding the phagophore such that the phagophore is sandwiched between two ER cisternae. A narrow membrane extension connects the phagophore and ER, giving a rise to the ER-isolation membrane (ER-IM) complex. Immuno-electron microscopy revealed that GFP-DFCP1 localizes to the ER-IM complex, indicating that the cradle is possibly related to the omegasome, at least in that both contain DFCP1 [71,72]. It is possible that the phagophore grows and expands inside the cradle with the associated ER membranes acting as a lipid donor for membrane expansion [14].
Lippincott-Schwartz and colleagues [73] have suggested an alternative model in which DFCP1 may be located at sites where the ER and mitochondria make contact. Rapid formation of APs was proposed to drive lipid transfer from the ER to mitochondria where lipids are modified and then utilized in the formation of APs, which subsequently bud from the outer membrane (OM) of the mitochondrion [73]. The proteins anchored in the outer leaflet of the mitochondrial OM, but not transmembrane proteins of the inner membrane or the matrix proteins, colocalized with the AP markers, Atg5 and LC3. The mitochondrial OM proteins did not label the autophagosomal lumen, but rather appeared in the form of ring-shaped structures. Of possible relevance is the report that phosphatidylethanolamine (PE), which is a membrane component of APs, is produced in mitochondria from phosphatidylserine, which is also abundant in the ER [73–75].
4.2. ER and Nuclear Membranes
As the nuclear membrane is continuous with the ER membrane network, the nuclear envelope (inner and outer nuclear membranes) could serve as a source of AP membranes. In this context, a coiling of the nuclear membrane has been observed at locations where viral proteins are enriched during a late phase of Herpes simplex type 1 (HSV-1) virus infection of murine macrophages. The formation of LC3-positive AP-like structures then occurs as follows: (1) viral capsids assemble and accumulate in the nucleus of infected cells; (2) during the egress process, HSV-1 capsids fuse with the inner nuclear membrane and acquire their envelope in the perinuclear space; (3) a second step of fusion releases a naked HSV-1 capsid into the cytoplasm; and (4) during this process, some of these capsids are trapped by the emerging 4-layer nuclear-derived APs [76,77]. It is not clear whether viruses other than HSV-1 can promote AP formation by a similar mechanism.
4.3. Plasma Membrane
Formation of APs from the plasma membrane (PM) in mammalian cells requires components of the endocytic pathway (Figure 1). Recent data suggest that the PM can contribute to early autophagic precursors, a phenomenon that is dependent on the association of Atg16L1-positive vesicles with the PM through Atg16L1–AP2 (clathrin adaptor protein at the Golgi)/clathrin heavy-chain interactions [78,79]. Subsequent scission of the Atg16L1/clathrin/AP2-associated structures, leading to the formation of early endosomal-like intermediates, is a crucial step that enables the liberation and maturation of these Atg16L1 vesicles into APs. These autophagic precursors subsequently mature to form phagophores and are proposed to represent an earlier stage in AP assembly. The ability of PM to contribute to AP formation in mammalian cells (as described above) may be particularly important during periods of increased autophagy, because the large surface area of the PM may serve as a large membrane reservoir that allows cells periods of AP synthesis at levels many-fold higher than under basal conditions, without compromising other processes [78–80].
4.4. Golgi Complex and Endosomes
In yeast, tubulo-vesicular compartments often found adjacent to mitochondria and that originate from the secretory pathway are believed to serve as Atg9 reservoirs. The suggested role of Atg9 reservoirs is to deliver and exchange material, including lipids and possibly proteins with the endocytic system and mitochondria. It is believed that one or more of the Atg9 reservoirs, together with other Atg proteins, act in close proximity to the vacuole as a signal for generation of the PAS and formation of an AP [81–83].
Mammalian Atg9 (mAtg9) is found both on the trans-Golgi network and endosomes in nutrient-rich cells, and LC3-positive APs in nutrient-starved cells. It is not known from which compartment mAtg9 traffics to reach the developing AP, but the process does require other Atg proteins, namely, the ULK1 (yeast Atg1 counterpart)/Atg13 complex and Beclin 1 (yeast Atg6 counterpart) as well as lipid kinases to be delivered continuously to the phagophore [84]. Wang and colleagues [85] demonstrate that nutrient starvation induces the tubulation and fragmentation of Atg9-positive Golgi membranes in a manner that is dependent on the membrane curvature-driving protein Bif-1/Endophilin B1 and the class III phosphatidylinositol-3-kinase (PI3KC3) complex II. Starvation-induced Atg9 foci colocalized not only with Bif-1, but also with an early endosome marker, Rab5, and an AP precursor marker, Atg16L. Knockout or knockdown of Bif-1 as well as inhibition of the PI3KC3 complex II, by PI3K inhibitor or knockdown of Beclin 1 or UVRAG, impaired Golgi fission, Atg9 trafficking and LC3 foci formation. Hence, these authors proposed that Bif-1-mediated fragmentation of the Golgi complex during nutrient starvation plays a crucial role in Atg9 trafficking and AP biogenesis in mammalian cells [85].
5. Phagophore Expansion and Autophagosome Development
The process of membrane expansion is regulated by a number of different Atg proteins that includes ubiquitin-like (Ubl) proteins, which participate in the two conjugation reactions. Both systems include proteins displaying three different enzymatic activities: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-protein ligase (E3) [56,86]. The first Ubl system essential for autophagy and membrane expansion is responsible for generating an Atg12-Atg5 protein conjugate [56,86,87]. In this system, the C-terminal glycine of Atg12 is covalently attached to Atg5 through an internal lysine residue. This process requires the action of Atg7 (E1 enzyme) [88] and Atg10 (E2 enzyme) ([89]. A third protein, Atg16, binds to the Atg5 component of the Atg12-Atg5 conjugate and dimerizes to link a pair of Atg12-Atg5 conjugates. This multi-protein Ubl system is constitutively active and crucial for phagophore expansion and AP formation, but these proteins dissociate from the expanding phagophore before its completion and subsequent fusion with lysosomes [1,90].
A second Ubl system involves the processing and subsequent conjugation of Atg8 (LC3) to PE by sequential action of three Atg proteins: Atg4, Atg7 and Atg3 [1,86,91]. LC3 is proteolytically processed by the protease Atg4, forming an intermediate, cytosolically localized LC3-I. Then, the E1 enzyme, Atg7 activates the processed LC3 and transfers it to Atg3 (functions like E2 enzyme). Atg3 finally conjugates LC3 to PE (lipidation process), resulting in a tight association of LC3-PE to the membrane (membrane-bound LC3-II) where it functions in the formation of the AP [56]. Although LC3-II is inserted into both sides of the phagophore membrane, after fusion of the membrane tips to form an AP, the molecules on the outer face are eventually delipidated by Atg4 and recycled [92]. As an aside, LC3-II levels generally correlate with the number of APs present in the cell, making it the basis for autophagy assays [93].
The two Ubl systems do not act independently. The Atg16-Atg12-Atg5 complex can bring LC3 to the site of lipidation and act as an E3 for LC3-II conjugation [94]. Meanwhile, Atg10, the E2 enzyme in Atg12-Atg5 conjugation, also facilitates the conversion of LC3 to the lipidated form, although LC3 is not a substrate of Atg10 [95,96].
In yeast and mammals, Atg8/LC3 has been found to promote membrane tethering and hemi-fusion, suggesting that it enables the growth and expansion of the forming phagophore/AP in vitro [97] and in vivo [15,96]. Membrane tethering mediated by LC3-PE leads to membrane hemi-fusion, which is normally a transient intermediate in membrane fusion reactions. Hemi-fusion involves lipid mixing only between the outer and proximal leaflets of the membranes, but not between the inner leaflets [97,98].
Mammalian cells contain a number of Atg8 orthologs that can be divided into two subgroups based on their amino acid sequence homology, where LC3A-C (including two variants of LC3A originating from an alternative splicing event) constitute the LC3 subfamily and GABARAP, GABARAPL1, GATE-16 (also known as GABARAPL2), and GABARAP-L3 constitute the GABARAP/GATE-16 subfamily [99,100]. Of these eight Atg8 orthologs identified in mammals, only LC3B has been extensively studied. LC3B is known to decorate APs and recruit adaptor proteins such as p62 and NBR1 [101,102]. Other Atg8 orthologs such as GATE-16 and GABARAP were initially characterized as intra-cellular trafficking factors [103,104] and later shown to be localized to starvation-induced APs [105]. The occurrence of several Atg8 orthologs in the mammalian system raises the question whether each has a distinct and crucial role in autophagy. Elazar and colleagues [106] have shown that both the LC3 and GABARAP/GATE-16 subfamilies are indispensable for the autophagic process, acting differentially at early stages of AP biogenesis. Thus, the LC3 subfamily is required for elongation of the phagophore membrane, whereas the GABARAP/GATE-16 subfamily is required for a later stage in AP maturation.
6. Autophagosome Fusion Events
In yeast cells, APs are formed at the single PAS, adjacent to the vacuole. By contrast, mammalian APs are formed at many sites upon nutrient-deprivation or rapamycin (a pharmacological autophagy inducer) treatment [4]. Furthermore, once completed, APs are transported to endosomes and lysosomes (Figure 1). The mechanisms employed for this directed movement are not well understood. However, it seems that cytoskeletal elements, such as microtubules and actin microfilaments, may play a crucial role. Interestingly, in yeast it seems that neither type of cytoskeletal element is required for bulk autophagy, but that actin microfilaments are essential for selective types of autophagy. In mammalian cells, it has been shown that AP movement and transport to lysosomes depends on microtubules, whereas the role of actin cables in such events remains unclear [107].
In yeast, the AP is transported to the vacuole and the outer membrane of the AP vesicle docks and fuses with the vacuolar membrane, in a process that is dependent upon two proteins, Ccz1 and Mon1, which form a complex that facilitates homotypic vacuole fusion. Other components involved in AP-vacuole fusion are the SNARE proteins Vam3 and Vti1 (found on the vacuolar membrane), Vam7 and Ykt6 (found on the AP), NSF (Sec18), SNAP (Sec17), Sec19, the Rab protein Ypt7, and members of the class C Vps/HOPS complex [21]. After fusion, the AP inner single-membrane vesicle is released into the vacuole lumen and is now termed the autophagic body. Subsequently, the membrane of the autophagic body is broken down and complete degradation of the autophagic body is dependent on resident vacuolar proteases and acidification of the vacuole [21,63,108]. The Atg15 lipase is required for this degradation process [109,110]. Subsequent recycling of the products of controlled degradation occurs via the action of a group of partially redundant vacuolar effluxers, Atg22, Avt3, and Avt4, which mediate the efflux of leucine and other amino acids resulting from autophagic degradation. The release of autophagy-derived and recycled molecules allows the maintenance of cellular metabolism protein synthesis and viability during starvation conditions [111,112].
In mammalian cells, once delivered to lysosomes, APs tether, dock and then fuse with lysosomal membranes in separately regulated events, independent of lysosomal acidification. However, changes in the intracellular lysosomal and AP lipid content (occasioned by metabolic disorders) and/or protein composition may have pronounced effects on the fusion step and thereby affect the overall degradative activity of macroautophagy. Two kinds of fusion can occur between APs and lysosomes: (1) complete fusion that creates a hybrid compartment (the autolysosome/autophagolysosome); and (2) kiss-and-run fusion during which transfer of some content occurs while still maintaining the separateness of the contributing vesicles [4,113–116]. The relative contribution of each form of fusion and whether there is any related physiological significance is currently unclear.
As stated earlier, en route to fusion with lysosomes in mammalian cells, APs can fuse with early endosomes or MVBs to form amphisomes (Figure 1). The fusion step involves proteins such as the ESCRT complex, SNAREs, Rab7 and the class C Vps proteins [4,116–118]. Fusion of APs with lysosomes versus MVBs may be differentially regulated, as fusion of MVBs with APs is a calcium-dependent event involving Rab11 [119]. UVRAG, a Beclin 1 interacting protein, is involved in the maturation step by recruiting the class C Vps proteins and activating Rab7, which in turn promotes fusion with late endosomes and lysosomes [120]. Furthermore, another Beclin 1 interacting protein, Rubicon, also functions in the maturation of APs. Rubicon is thought to be a part of a distinct Beclin 1 complex containing Vps34, Vps15 and UVRAG that suppresses AP maturation. However, more work is required to definitively characterize the different Beclin 1 complexes and their roles in the autophagy pathway [4,121].
7. Outstanding Questions
As described above, APs are responsible for the sequestration of autophagic cargo and its delivery to lysosomes or the vacuole for degradation and recycling. However, a number of questions remain to be addressed.
7.1. AP Origin
Although we have improved understanding of the possible sources of lipid for the AP membrane, the relative contribution of each source under any one set of conditions remains to be determined. In this context, what determines whether the membrane comes from any particular source remains unknown. Do the relative numbers of APs formed from any one source change under autophagy-induction or pathological conditions? Do non-selective and selective types of macroautophagy use the same, or different membrane sources for AP formation? How is mobilization of membrane from different sources achieved and how is the supply of various lipids and proteins to the AP membrane regulated?
7.2. AP Development
What is the driving force for phagophore curvature and expansion? In addition to known Atg proteins, what are other (co)factors or proteins are necessary for phagophore curvature and AP formation? What are the interactions between these proteins and how such events are regulated? What is the precise molecular mechanism required for sealing of the phagophore membrane to from the AP membrane? What factors regulate AP size and number, and how do these parameters vary under different physiological or pathophysiological conditions? What is the driving force for AP transport/trafficking and how are cytoskeletal elements connected to APs?
7.3. AP Maturation and Death
Are different conditions and/or machineries required for APs to undergo fusion with early/late endosomes or MVBs? Are different fusion components required to act on AP membranes having distinct lipid compositions?
Answering these questions will ensure that understanding the “life and death” of autophagosomes remains a vibrant and active area of investigation of both membrane biogenesis and function in the broader context of autophagy for some years to come.
References
- 1.Legakis J.E., Klionsky D.J. Overview of Autophagy. In: Deretic V., editor. Autophagy in Immunity and Infection. Vol. 1. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germnay: 2006. p. 3. Chapter 1. [Google Scholar]
- 2.Deretic V., Klionsky D.J. How cells clean house. Sci. Am. 2008;298:74–81. doi: 10.1038/scientificamerican0508-74. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 5.Sridhar S., Botbol Y., Macian F., Cuervo A.M. Autophagy and disease: Always two sides to a problem. J. Pathol. 2011;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., Orenstein S.J., Wong E., Cuervo A.M. Chaperone-mediated autophagy at a glance. J. Cell Sci. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uttenweiler A., Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 2008;445:245–259. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- 8.Mijaljica D., Prescott M., Devenish R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 9.Li W.W., Li J., Bao J.K. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2011 doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santambrogio L., Cuervo A.M. Chasing the elusive mammalian microautophagy. Autophagy. 2011;7:652–654. doi: 10.4161/auto.7.6.15287. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z., Klionsky D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tooze S.A., Yoshimori T. The origin of autophagosomal membrane. Nat. Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N., Yoshimori T., Ohsumi Y. The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 15.Weidberg H., Shvets E., Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 16.Mari M., Tooze S.A., Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol. Rep. 2011;3 doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Codogno P., Mehrpour M., Proikas-Cezanne T. Canonical and non-canonical autophagy: Variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 18.Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012;22:R229–R234. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 19.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanki T., Klionsky D.J. The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 2010;75:795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yorimitsu T., Klionsky D.J. Autophagy: Molecular machinery of self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky D.J., Cuervo A.M., Dunn W.A., Jr, Levine B., van der Klei I., Seglen P.O. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 23.Johansen T., Lamark T. Selective autophagy mediated by autophagy adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto A., Simonsen A. The elimination of accumulated and aggregated proteins: A role for aggrephagy in neurodegeneration. Neurobiol. Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamark T., Johansen T. Aggrephagy: Selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2011 doi: 10.1155/2012/736905. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch-Day M.A., Klionsky D.J. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umekawa M., Klionsky D.J. The cytoplasm-to-vacuole targeting pathway: A historical perspective. Int. J. Cell Biol. 2011;2012 doi: 10.1155/2012/142634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cebollero E., Reggiori F., Kraft C. Reticulophagy and ribophagy: Regulated degradation of protein production factories. Int. J. Cell Biol. 2011;2012 doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamasaki M., Noda T., Baba M., Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 30.Mijaljica D., Prescott M., Devenish R.J. Endoplasmic reticulum and Golgi complex: Contributions to, and turnover by, autophagy. Traffic. 2006;7:1590–1595. doi: 10.1111/j.1600-0854.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- 31.Bernales S., Schuck S., Walter P. ER-phagy: Selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 32.Yorimitsu T., Klionsky D.J. Eating the endoplasmic reticulum: Quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weidberg H., Shvets E., Elazar Z. Lipophagy: Selective catabolism designated for lipids. Dev. Cell. 2009;16:628–630. doi: 10.1016/j.devcel.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Navarro J.A., Cuervo A.M. Autophagy and lipids: Tightening the knot. Semin. Immunopathol. 2010;32:343–353. doi: 10.1007/s00281-010-0219-7. [DOI] [PubMed] [Google Scholar]
- 36.Tolkovsky A.M. Mitophagy. Biochim. Biophys. Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Kanki T., Klionsky D.J., Okamoto K. Mitochondria autophagy in yeast. Antioxid. Redox Signal. 2011;14:1989–2001. doi: 10.1089/ars.2010.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May A.I., Rodney J.D., Prescott M. The many faces of mitochondrial autophagy: Making sense of contrasting observations in recent research. Int. J. Cell Biol. 2011 doi: 10.1155/2012/431684. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park Y.E., Hayashi Y.K., Bonne G., Arimura T., Noguchi S., Nonaka I., Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 41.Mijaljica D., Prescott M., Devenish R.J. The intricacy of nuclear membrane dynamics during nucleophagy. Nucleus. 2010;1:213–223. doi: 10.4161/nucl.1.3.11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunn W.A., Jr, Cregg J.M., Kiel J.A., van der Klei I.J., Oku M., Sakai Y., Sibirny A.A., Stasyk O.V., Veenhuis M. Pexophagy: The selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 43.Sakai Y., Oku M., van der Klei I.J., Kiel J.A. Pexophagy: Autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 44.Manjithaya R., Nazarko T.Y., Farré J.C., Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oku M., Sakai Y. Peroxisomes as dynamic organelles: Autophagic degradation. FEBS J. 2010;277:3289–3294. doi: 10.1111/j.1742-4658.2010.07741.x. [DOI] [PubMed] [Google Scholar]
- 46.Till A., Lakhani R., Burnett S.F., Subramani S. Pexophagy—The selective degradation of peroxisomes. Int. J. Cell Biol. 2011 doi: 10.1155/2012/512721. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., Goldfarb D.S. Piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kvam E., Goldfarb D.S. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- 49.Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.L., Millen J., Goldfarb D.S., Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraft C., Deplazes A., Sohrmann M., Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 51.Kraft C., Peter M. Is the Rsp5 ubiquitin ligase involved in the regulation of ribophagy? Autophagy. 2008;4:838–840. doi: 10.4161/auto.6603. [DOI] [PubMed] [Google Scholar]
- 52.Tang F., Watkins J.W., Bermudez M., Gray R., Gaban A., Portie K., Grace S., Kleve M., Craciun G. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–886. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- 53.Brown C.R., Dunton D., Chiang H.L. The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J. Biol. Chem. 2010;285:1516–1528. doi: 10.1074/jbc.M109.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knodler L.A., Celli J. Eating the strangers within: Host control of intracellular bacteria via xenophagy. Cell. Microbiol. 2010;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itakura E., Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y., Klionsky D.J. The regulation of autophagy—Unanswered questions. J. Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki K., Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Baba M., Osumi M., Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell. Struct. Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 61.Øverbye A., Fengsrud M., Seglen P.O. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 62.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 63.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeifer U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. J. Cell Biol. 1978;78:152–167. doi: 10.1083/jcb.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schworer C.M., Shiffer K.A., Mortimore G.E. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J. Biol. Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- 66.Mizushima N., Klionsky D.J. Protein turnover via autophagy: Implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 67.Mortimore G.E., Pösö A.R. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 68.Kimura S., Noda T., Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell. Struct. Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 69.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simonsen A., Stenmark H. Self-eating from an ER-associated cup. J. Cell Biol. 2008;182:621–622. doi: 10.1083/jcb.200807061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 72.Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 73.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McEwan D.G., Dikic I. Not all autophagy membranes are created equal. Cell. 2010;141:564–566. doi: 10.1016/j.cell.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 75.Rambold A.S., Lippincott-Schwartz J. Starved cells use mitochondria for autophagosome biogenesis. Cell Cycle. 2010;9:3633–3634. doi: 10.4161/cc.9.18.13170. [DOI] [PubMed] [Google Scholar]
- 76.English L., Chemali M., Duron J., Rondeau C., Laplante A., Gingras D., Alexander D., Leib D., Norbury C., Lippé R., et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.English L., Chemali M., Desjardins M. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy. 2009;5:1026–1029. doi: 10.4161/auto.5.7.9163. [DOI] [PubMed] [Google Scholar]
- 78.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ravikumar B., Moreau K., Rubinsztein D.C. Plasma membrane helps autophagosomes grow. Autophagy. 2010;6:1184–1186. doi: 10.4161/auto.6.8.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuervo A.M. The plasma membrane brings autophagosomes to life. Nat. Cell Biol. 2010;12:735–737. doi: 10.1038/ncb0810-735. [DOI] [PubMed] [Google Scholar]
- 81.Geng J., Klionsky D.J. The Golgi as a potential membrane source for autophagy. Autophagy. 2010;6:950–951. doi: 10.4161/auto.6.7.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mari M., Reggiori F. Atg9 reservoirs, a new organelle of the yeast endomembrane system? Autophagy. 2010;6:1221–1223. doi: 10.4161/auto.6.8.13792. [DOI] [PubMed] [Google Scholar]
- 83.Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tooze S.A., Jefferies H.B., Kalie E., Longatti A., McAlpine F.E., McKnight N.C., Orsi A., Polson H.E., Razi M., Robinson D.J., Webber J.L. Trafficking and signaling in mammalian autophagy. IUBMB Life. 2010;62:503–508. doi: 10.1002/iub.334. [DOI] [PubMed] [Google Scholar]
- 85.Takahashi Y., Meyerkord C.L., Hori T., Runkle K., Fox T.E., Kester M., Loughran T.P., Wang H.G. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2011;7:61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 87.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 88.Kim J., Dalton V.M., Eggerton K.P., Scott S.V., Klionsky D.J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mizushima N., Yoshimori T., Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2003;35:553–561. doi: 10.1016/s1357-2725(02)00343-6. [DOI] [PubMed] [Google Scholar]
- 91.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 92.Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nemoto T., Tanida I., Tanida-Miyake E., Minematsu-Ikeguchi N., Yokota M., Ohsumi M., Ueno T., Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- 96.Jimenez-Sanchez M., Thompson F., Zavodsky E., Rubinsztein D.C. Autophagy and polyglutamine diseases. Prog. Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.08.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 98.Subramani S., Farré J.C. A ubiquitin-like protein involved in membrane fusion. Cell. 2007;130:18–20. doi: 10.1016/j.cell.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 99.Xin Y., Yu L., Chen Z., Zheng L., Fu Q., Jiang J., Zhang P., Gong R., Zhao S. Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics. 2001;74:408–413. doi: 10.1006/geno.2001.6555. [DOI] [PubMed] [Google Scholar]
- 100.He H., Dang Y., Dai F., Guo Z., Wu J., She X., Pei Y., Chen Y., Ling W., Wu C., et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 101.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 103.Sagiv Y., Legesse-Miller A., Porat A., Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kittler J.T., Rostaing P., Schiavo G., Fritschy J.M., Olsen R., Triller A., Moss S.J. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol. Cell Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- 105.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 106.Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monastyrska I., Rieter E., Klionsky D.J., Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos. Soc. 2009;84:431–448. doi: 10.1111/j.1469-185X.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakamura N., Matsuura A., Wada Y., Ohsumi Y. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- 109.Epple U.D., Suriapranata I., Eskelinen E.L., Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Teter S.A., Eggerton K.P., Scott S.V., Kim J., Fischer A.M., Klionsky D.J. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Z., Huang J., Geng J., Nair U., Klionsky D.J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Z., Klionsky D.J. Permeases recycle amino acids resulting from autophagy. Autophagy. 2007;3:149–150. doi: 10.4161/auto.3631. [DOI] [PubMed] [Google Scholar]
- 113.Jahreiss L., Menzies F.M., Rubinsztein D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ravikumar B., Futter M., Jahreiss L., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Narayanan U., Renna M., et al. Mammalian macroautophagy at a glance. J. Cell Sci. 2009;122:1707–1711. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koga H., Kaushik S., Cuervo A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehrpour M., Esclatine A., Beau I., Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 117.Eskelinen E.L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 118.Noda T., Fujita N., Yoshimori T. The late stages of autophagy: How does the end begin? Cell Death Differ. 2009;16:984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- 119.Fader C.M., Sánchez D., Furlán M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in K562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 120.Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]