Abstract
Purpose
Gastroenteropancreatic neuroendocrine tumors (GEP-NETs) are rare group of tumors with a wide spectrum of clinical behavior. However, there are no known clinically relevant biomarkers to predict metastasis.
Experimental Design
To investigate differential gene expression signatures of metastatic vs non-metastatic NETs, we studied cell cycle regulatory genes in 19 metastatic and 22 non-metastatic colorectal NETs by PCR arrays. Immunohistochemistry (IHC) and quantitative real-time RT-PCR were performed to verify the results and another set of 38 GEP-NETs were further studied for validation.
Results
We first delineated six candidate genes for metastasis including ATM, CCND2, RBL2, CDKN3, CCNB1, and GTSE1. ATM was negatively correlated with metastatic NETs (p<0.001) with more than 2-fold change compared to non-metastatic NETs. Overexpression of ATM protein by IHC was strongly correlated with high ATM mRNA levels and low Ki-67 labeling index. Patients with ATM-negativity by IHC showed significantly decreased overall survival than patients with ATM-positivity (median OS, metastatic vs non-metastatic NETs; 2.7 years vs not reached; p = 0.003) and 85.7% of metastatic NETs were ATM-negative. In another validation set of GEP-NETs, decreased mRNA of ATM gene was associated with metastasis and remained significant (p = 0.023).
Conclusions
ATM down-regulation was strongly associated with metastatic NETs when compared with non-metastatic NETs and ATM may be a potential predictive marker for metastasis as well as a novel target in metastatic GEP-NETs.
Introduction
Gastroenteropancreatic neuroendocrine tumors (GEP-NETs) are a heterogeneous group of malignancies derived from neuroendocrine cell compartments in various organs including pancreas, colorectum, biliary tract, and stomach. Data from recent population-based studies demonstrate a significant increase in the reported incidence of NETs over time and the incidence ranges from 2.5 to 5 cases per 100,000 in Caucasian population [1]–[4]. The increase in incidence is likely attributable, in part, to increasing awareness, improved diagnostic strategies for NETs, and possibly other undetermined environmental and genetic factors [5].
NETs are a broad family of tumors with a wide range of clinical presentations and outcomes. Given the biological diversity of GEP-NETs, the treatment of GEP-NETs is now becoming more optimized according to subgroups. Recently, WHO classified NETs as well-differentiated neuroendocrine tumor, well-differentiated neuroendocrine carcinoma, and poorly differentiated neuroendocrine carcinoma [6]–[8]. Nevertheless, there are no candidate biological markers to guide a novel targeted therapeutic approach, partially owing to the fact that little is known about the underlying mechanisms that regulate development and progression of NETs. Several clinicopathological factors have been reported to predict poor clinical outcome in NETs. These include histologic grade, disease stage, sex, race, age, year of diagnosis, tumor size, and proliferation indices including Ki-67 labeling index and mitotic count [1], [9]–[11]. In addition to tumor size, proliferation index are one of the most important factors associated with metastasis. These findings suggest that certain defects in cell cycle regulatory mechanism are associated with cell proliferation and may predict malignant behavior of NETs. In this study, we studied gene expression signatures of cell cycle regulatory genes in metastatic and non-metastatic colorectal NETs to delineate candidate genes which may predict metastatic behavior.
Materials and Methods
Patient samples
Cases of colonic (n = 4) and rectal (n = 15) metastatic NETs (n = 19) were selected from July 1, 1996 to December 31, 2008 who were treated at Samsung Medical Center. This retrospective study was approved by the institutional review board at Samsung Medical Center and was waived for written informed consent form. Case selection criteria for metastatic and non-metastatic NETs were as following: complete demographic data, treatment record (Appendix S1), availability of fresh frozen tissues and paraffin-embedded tissue blocks. For control cases, colorectal NETs (n = 22) which were localized initially at diagnosis and during the median follow-up period of 6.8 years (range, 3.9–10.4 years) were selected from the database. To verify results and further investigate, another set of 38 metastatic (n = 20) and non-metastatic (n = 18) GEP-NETs from pancreas (n = 32), stomach (n = 3), ampulla of Vater (n = 1) and rectum (n = 2) were also analyzed.
A histopathologic review was conducted by two pathologists (C.O.S and K.M.K) to verify the diagnosis and histopathology of all cases. All study materials were obtained under the protocol approved by the Institutional Review Board of Samsung Medical Center. Histological grade was categorized as following; carcinoid tumor or well differentiated NET was classified as G1 tumor, atypical carcinoid or well differentiated neuroendocrine carcinoma was classified as G2 tumor, and poorly differentiated neuroendocrine carcinoma was classified as G3 tumor [12]. Among 41 NETs studied, 36 were G1 and 5 were G2 tumors. G3 tumors were excluded from case selection. The demographic characteristics of cases and controls are outlined in Table 1.
Table 1. Characteristics of the cases used in the analysis.
| Characteristic | Metastatic NET (n = 19) | non-metastatic NET (n = 22) |
| Age, years | ||
| Median | 62 | 53 |
| Range | 34–79 | 30–72 |
| Sex | ||
| Male | 11 (41%) | 16 (59%) |
| Female | 8 (57%) | 6 (43%) |
| Location | ||
| Rectum | 15 (41%) | 22 (59%) |
| Colon | 4 (100%) | 0 (0%) |
| Histology grade | ||
| G1 | 14 (39%) | 22 (61%) |
| G2 | 5 (100%) | 0 (0%) |
| Treatment | ||
| Biopsy | 11 (39%) | 7 (61%) |
| Local excision | 3 (17%) | 15 (83%) |
| Radical resection | 5 (100%) | 0 (0%) |
NET, neuroendocrine tumor.
Expression of cell cycle regulatory genes by PCR Arrays
All quantitative RT-PCR experiments were conducted with a Light Cycler 480 real-time PCR system (Roche Applied Science, Indianapolis, IN). For PCR array experiments, a RT2 Profiler Custom PCR Array was used to simultaneously examine the mRNA levels of the genes of 84 genes key to cell cycle regulation and five reference genes including 18SrRNA, HPRT, RPL13A, GAPDH, and β-actin, according to the protocol of the manufacturer (SuperArray Bioscience, Frederick, MD). Total RNAs from formalin-fixed paraffin-embedded (FFPE) tumor tissue was extracted using RT2 FFPE RNA extraction kit according to the manufacturer's instructions (SuperArray). Between 0.3 and 2.8 µg of total RNA was added to each well of the ReactionReady First Strand cDNA Synthesis Kit (SuperArray no. C-01; SuperArray). 5 ul of the first-strand cDNA synthesis reaction were pre-amplified by the RT2 PreAMP PCR Master Mix and RT2 FFPE PreAMP Primer Mix at 95°C for 10 min; 8 cycles of 95°C for 15 s, and 60°C for 2 min. The equivalent of 0.8 µl cDNA was added to each well of the PCR array. A total volume of 25 µl of PCR mixture, which included 12.5 µl of RT2 Real-Time SYBR Green/Fluorescein PCR master mix from SuperArray Bioscience, 11.5 µl of milliQ, and 1 µl of template cDNA, was loaded in each well of the PCR array. Arrays were run with the following parameters: cycle 1, 10 min at 95°C; and cycle 2, 15 sec at 95°C, followed by 1 min at 60°C (40 repeats), with optical data collection at 60°C for each repeat. This experiment was repeated three times. Data were analyzed using the comparative cycle threshold method with normalization of the raw data to 5 reference genes. Functional gene grouping of 84 genes is shown in Table 2.
Table 2. Functional grouping of 84 cell cycle regulatory genes used in PCR array analysis.
| Functional group | Gene |
| G1 phase and G1/S transition | ANAPC2, CCND1, CCNE1, CDC34, CDK4, CDK6, CDKN1B, CDKN3, CUL1, CUL2, CUL3, SKP2 |
| S phase and DNA replication | ABL1, MCM2, MCM3, MCM4, MCM5, PCNA, RPA3, SUMO1, UBE1 |
| G2 phase and G2/M transition | ANAPC2, ANAPC4, DIRAS3, BCCIP, BIRC5, CCNB1, CCNG1, CCNH, CCNT1, CCNT2, CDK5R1, CDK5RAP1, CDK7, CDKN3, CKS1B, CKS2, DDX11, DNM2, GTF2H1, GTSE1, HERC5, KPNA2, MNAT1, SERTAD1 |
| M phase | CCNB2, CCNF, CDC2, CDC16, CDC20, MRE11A, RAD51 |
| Cell cycle checkpoint and cell cycle arrest | ATM, ATR, BRCA1, BRCA2, CCNG2, CDC2, CDC34, CDK2, CDKN1A, CDKN1B, CDKN2A, CDKN2B, CDKN3, CHEK1, CHEK2, CUL1, CUL2, CUL3, GADD45A, HUS1, KNTC1, MAD2L1, MAD2L2, NBN, RAD1, RAD17, RAD9A, RB1, RBBP8, TP53 |
| Regulation of cell cycle | ABL1, ANAPC2, ANAPC4, DIRAS3, ATM, ATR, BCCIP, BCL2, BRCA2, CCNB1, CCNB2, CCNC, CCND1, CCND2, CCNE1, CCNF, CCNH, CCNT1, CCNT2, CDC16, CDC2, CDC20, CDK2, CDK4, CDK5R1, CDK6, CDK7, CDK8, CDKN1A, CDKN1B, CKS1B, DDX11, E2F4, GADD45A, KNTC1, MKI67, PCNA, RAD9A, RB1, SKP2, TFDP1, TFDP2 |
| Negative regulation of cell cycle | ATM, BAX, BRCA1, CDKN2B, RBL1, RBL2, TP53 |
Bold indicate significant genes in this study.
Cell cycle regulatory gene expression signature analysis using bioinformatic methods
Volcano plot was used to identify significant differential expression genes between metastatic NETs and non-metastatic NETs. Briefly, the differences (log2fold change) between the two groups were plotted on X-axis. The –log10 (P value) were plotted on the y-axis. A cutoff value of P value<0.01 and fold change >1.5 were used to determine the differential gene expression signature. Principal component analysis was performed using R program (version 2.12.0: http://www.r-project.org) with prcomp function.
Immunohistochemistry
Immunohistochemistry (IHC) was performed for Ki-67 (clone MIB-1, DAKO M7240, dilution 1∶70) and Ataxia Telangiectasia Mutated (ATM) (mouse monoclonal, ab78, Abcam, Cambridge, MA, Cat No. ab78, dilution 2 ul/ml) on 3 µm sections from FFPE tissues and mounted on positively charged slides. Tissue sections on glass slides were deparaffinized in xylene, hydrated in descending concentrations of alcohol, and then washed in distilled water. Antigen retrieval was performed with a microwave for 5 minutes 2 times with 10 mM citrate buffer (pH 6.0) for Ki-67, and with 97°C for 20 minutes with Tris/EDTA buffer (pH 8.0) for ATM. The slides were washed in phosphate buffer for 3 times. The slides were incubated with primary antibodies for 60 minutes, and incubated with biotinylated antibodies (Envision plus, Dako, Carpinteria, CA, USA) for 25 minutes and 60 minutes, respectively. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in distilled water for 10 minutes. After washing, the slides were incubated with peroxidase-labeled streptavidin complex for 25 minutes. The slides were incubated in a solution of 3% diaminobenzidine for 20 minutes and counterstained with hematoxylin. To generate Ki-67 labeling proliferation index, image analysis was performed with the Applied Imaging Ariol SL-50 (Genetix, San Jose, USA). The immunostained slides were scanned under KiSight protocol for Ki-67 analysis. A first-scan pass at 1.25× objective magnification was performed to acquire a low-resolution image to recognize the tissue. Selection of regions of interest was performed by a pathologist. A second scan-pass was performed at 20× objective magnification to obtain a high resolution image. After training color and shape classifiers, the regions were selected for analysis. For Ki-67, more than 1,000 tumor cells were analyzed. On the basis of the intensity of staining and the nuclear morphology, the machine calculated the results as a percentage of positive cells. The ATM positivity was defined as greater than 5% ATM (+) tumor cells in whole tumor section area.
RNA Extraction and Real-Time Quantitative PCR of ATM
RNAs from FFPE tumor tissue was extracted using the RNeasy FFPE Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. ATM mRNA quantification was measured by real-time PCR based on TaqMan Gene Expression Assays (Applied Biosystems) as previously described [13]. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) mRNA was used as an internal control to normalize ATM mRNA level. Real-time PCR quantitation of transcripts was expressed as ATM/GAPDH ratios.
Statistical analysis
Differences between metastatic NET and non-metastatic NET samples were calculated using t-test or Mann-Whitney U test as appropriate. Correlations were examined Pearson's χ2, Fisher's exact test, Pearson's or Spearman's test as appropriate. Overall survival (OS) was determined using the Kaplan-Meier method and survival curves were compared using the long-rank test. Survival was calculated from the date of diagnosis and all patients were followed through March 23, 2011. All tests were two-sided and statistical significance was set at a threshold of p<0.05. Statistical analyses were performed using R program or SPSS (SPSS 18, Chicago, IL, USA).
Results
Metastatic NETs show different expression patterns of cell cycle regulatory genes
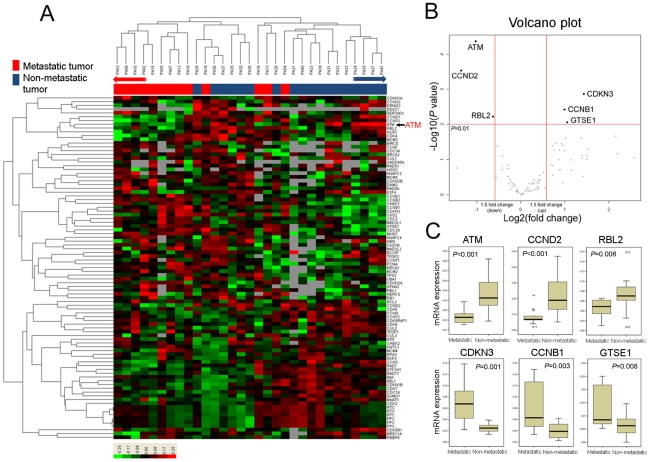
Of 19 metastatic NETs, 13 samples passed the initial quality control for RNA and 18 of 22 non-metastatic NETs passed the quality check and thus included for cell cycle regulatory gene expression profile analysis. The experiment was repeated three times with the same result. Hierarchical clustering analysis of gene expression data (filtering ≥2 standard deviation) from all tissues revealed that metastatic NETs and non-metastatic NETs were clustered with distinctive separation at the end of both sides with few exceptions (Figure 1A). Thus, gene expression pattern of cell cycle regulatory genes in metastatic NETs was apparently distinct from that observed in non-metastatic NETs. Next step, to identify significantly different set of genes expressed between the two groups, we applied volcano plot considering both p value (by two-sample t test) and fold-changes (metastatic vs non-metastatic NETs). Using this method, we selected six genes including ATM, CCND2, RBL2, CDKN3, CCNB1, and GTSE1 which are significantly associated with metastasis (Figure 1B). Among them, ATM, CCND2, and RBL2 were significantly down-regulated in metastatic NETs, whereas CDKN3, CCNB1, and GTSE1 were significantly up-regulated in metastatic NETs (all six genes, p<0.05, Figure 1C). Of note, ATM showed the lowest p value (p<0.001) with more than 2-fold change between non-metastatic vs metastatic NETs.
Figure 1. Hierarchical clustering (filtering ≥2 standard deviation) (A) and volcano plot (B) of expression data on cell cycle regulatory genes.
In metastatic neuroendocrine tumors, ATM, CCND2, and RBL2 were significantly down-regulated whereas CDKN3, CCNB1, and GTSE1 were significantly up-regulated compared to non-metastatic neuroendocrine tumors (C).
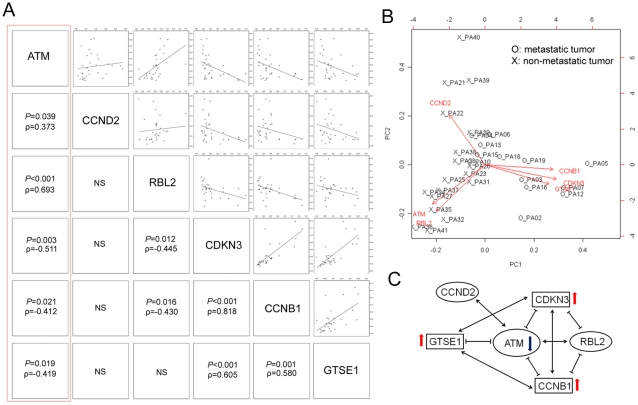
Correlation analysis of six significant genes reveals that ATM plays key role as a tumor suppressor in NETs
In the correlation analysis of six genes, only ATM expression was consistently correlated with expressions of all other 5 candidate genes with statistical significance (Figure 2A). ATM expression demonstrated strong positive correlations with CCND2 and RBL2, and negative correlations with CDKN3, CCNB1, and GTSE1. Using these six genes, principal component analysis was performed which revealed two distinct groups of metastatic and non-metastatic NETs (Figure 2B). Furthermore, principal component analysis suggested that CCNB1, CDKN3, and GTSE1 were important genes associated with metastatic NETs, whereas ATM and RBL2 were highly expressed in non-metastatic NET with CCND2 expressed in minor group of non-metastatic NETS. ATM and RBL2 are already known as negative regulators of cell cycle which support their role as tumor suppressor genes [14], [15]. In line with these findings, a network interaction diagram constructed based on correlation analyses clearly showed that an ATM down-regulation leads to increased expressions of CDKN3, CCNB1, and GTSE1 (Figure 2C). Based on these results, ATM seems to be one of the key regulators for cell cycle in NETs.
Figure 2. Correlation analysis of six genes reveals relationship with each other.
Expression of ATM was only significantly associated with all other 5 genes (A). Principal component analysis showed separation of metastatic and non-metastatic neuroendocrine tumors with contributing genes (B). Network interaction diagram constructed based on correlation of six genes (C).
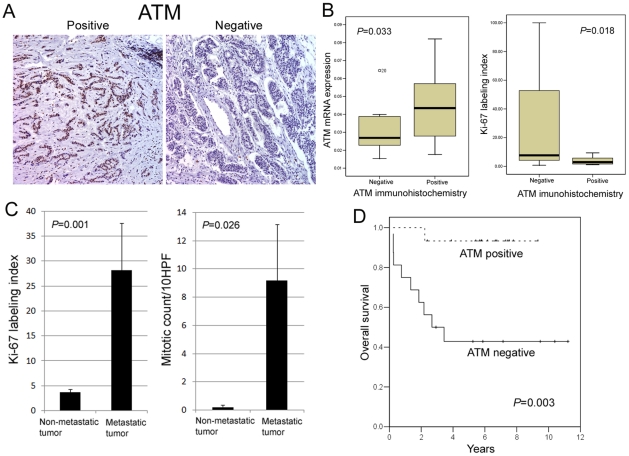
ATM gene down-regulation is strongly associated with metastatic NET and proliferation of tumor cells
To validate ATM protein expression, we performed ATM IHC from 14 of 19 metastatic NETs, and 17 of 22 non-metastatic NETs due to availability of FFPE tissue specimens. ATM IHC revealed nuclear staining of tumor cells (Figure 3A), and 15 (48.4%) of 31 cases were positive for ATM staining. The ATM positivity was defined as greater than 30% ATM (+) tumor cells in whole tumor areas. Moreover, ATM overexpression was strongly correlated with high ATM mRNA levels (Figure 3B) and low Ki-67 labeling index (Figure 3B). In line with these results, we identified that Ki-67 labeling index and mean mitotic counts were considerably increased in metastatic NETs (Figure 3C). Of 14 metastatic NETs, 12 cases (85.7%) were ATM-negative, which indicated that ATM down-regulation is strongly associated with metastatic NETs. However, there was no significant relationship between ATM expression and various clinicopathologic parameters including tumor site, grade, gender and age (Table 3). Accordingly, patients with ATM-negativity showed significantly decreased overall survival than patients with ATM-positivity (p = 0.003, Figure 3D), which correlated well with their metastatic behaviors.
Figure 3. ATM immunohistochemistry in colorectal neuroendocrine tumors.
ATM protein expression was significantly correlated with high ATM mRNA expression and low Ki-67 labeling index (A, B). Ki-67 labeling index and mean mitotic counts were increased in metastatic neuroendocrine tumors (C). Patients with negative ATM protein expression showed significantly shorter overall survival than patients with positive ATM protein expression (D).
Table 3. Relationships between ATM protein expression and various clinicopathologic factors in colorectal neuroendocrine tumors.
| ATM expression | ||||
| Characteristics | n | Negative | Positive | P value |
| Tumor site | ||||
| Colon | 4 | 4 | 0 | 0.101 |
| Rectum | 27 | 12 | 15 | |
| Grade | ||||
| G1 | 27 | 12 | 15 | 0.101 |
| G2 | 4 | 4 | 0 | |
| Sex | ||||
| Female | 8 | 6 | 2 | 0.22 |
| Male | 23 | 10 | 13 | |
| Age | ||||
| <60 | 18 | 9 | 9 | 0.833 |
| ≥60 | 13 | 7 | 6 | |
| Tumor behavior | ||||
| Metastatic tumor | 14 | 12 | 2 | 0.001 |
| Non-metastatic tumor | 17 | 4 | 13 | |
ATM RNA expression levels in validation set
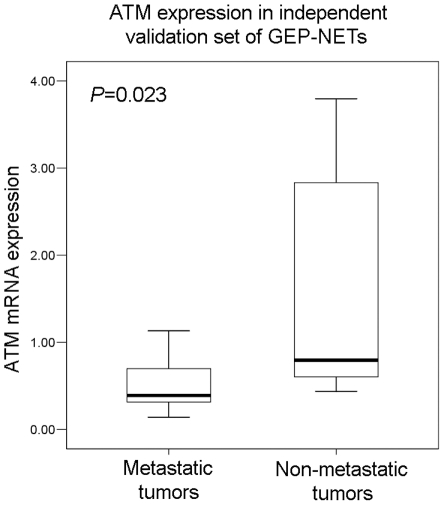
In the validation set of 20 metastatic and 18 non-metastatic GEP-NETs from various organs including pancreas, stomach, ampulla of Vater and rectum, mean mRNA levels of ATM in metastatic NETs were significantly low in metastatic NETs compared to non-metastatic NETs (p = 0.023) (Figure 4).
Figure 4. Comparison of mRNA expression levels of ATM in metastatic (mean = 0.5370±0.33263, median = 0.3912, minimum 0.14, maximum 1.13) and non-metastatic GEP-NETs (mean = 1.5075±1.22467, median = 0.7964, minimum 0.44, maximum 3.80).
Discussion
We investigated differential gene expression signatures of metastatic vs non-metastatic colorectal NETs focusing on cell cycle regulatory gene set, and delineated six candidate genes for metastasis including ATM, CCND2, RBL2, CDKN3, CCNB1, and GTSE1. Among them, we have focused on ATM since ATM has been known as a tumor suppressor gene which encodes a principal DNA damage-signaling protein. In this study, decreased expression of ATM protein was strongly correlated with low ATM mRNA levels and high Ki-67 labeling index. Tumor cells with ATM dysfunction exhibit increased loss of cell cycle check points and p53 dysfunction in selected cancer types [16]–[18]. For the first time, we demonstrated here that ATM down-regulation was strongly associated with metastatic NETs when compared with non-metastatic NETs and that >80% of metastatic NETs do not express ATM protein.
In our study, low ATM mRNA expression was significantly associated with increased expression of CDKN3, GTSE1, and CCNB1 as well as high Ki-67 proliferation index. These findings suggest that ATM down-regulation can lead to increased cell cycle regulatory genes such as CDKN3 and CCNB1, and then increased CDKN3 and CCNB1 leading to increased cell division and cell proliferation. CDKN3, also known as kinase-associated phosphatase (KAP), is another molecule involved in cell cycle checkpoint and cell cycle arrest. It has been reported that KAP is over-expressed in breast and prostate tumors, and is associated with proliferation of tumor cells [19]. We also found that RBL2, a key regulator of entry into cell division was significantly down-regulated in metastatic neuroendocrine tumors, suggesting growth-suppressive properties of RBL2 was decreased and tumor cells proloferate [20].
ATM gene has been described to be causal gene of Ataxia-telangiectasia, and inactivation of ATM is associated with increased risk of carcinogenesis and radiosensitivity [21]–[24]. Bartkova et al. [25] suggested that tumorigenic events early in the progression of major human cancer types activate the ATM-regulated checkpoint through deregulated DNA replication and DNA damage, and thereby activate an inducible barrier against tumor progression and genetic instability. Loss-of-function events in the ATM-Chk2-p53 tumor-suppressive cascade facilitates survival and proliferation of genetically unstable cancer cells [26]. Main mechanism of ATM inactivation in ataxia-telangiectasia associated patient is mutation such as truncating mutation and missense mutation, and missense mutation might be more prone to carcinogenesis [27]. Mutational inactivation of the ATM gene has been demonstrated in mantle cell lymphoma, T-cell prolymphocytic leukemias, rhabdomyosarcomas, and gastric cancer [28]–[31], and ATM heterozygous state is more common in breast cancer patients than in the general population [32], [33]. On the other hand, promoter methylation or deletion of ATM seems to be important mechanism for ATM inactivation in some sporadic cancers. Indra et al. [34] reported that 36% of cervical cancer samples showed ATM promoter methylation and 31% of cervical cancer samples showed ATM deletion. However, mutation in ATM gene was rare [35]. Ai et al. [36] also reported that frequent aberrant methylation of ATM promoter (25% of 100 samples) was found in head and neck squamous cell carcinomas. Both two mechanisms including methylation and deletion were well correlated with reduced mRNA and protein expression of ATM [34], [37]. Methylation or deletion of ATM may be a mechanism for reduced expression in NET, however, in our study, because of further study need to confirm.
Cell lines derived from Ataxia-telangiectasia patients exhibit abnormalities related to DNA damage and repair such as chromosomal instability, cell-cycle checkpoint defects in G1/S and G2/M, sensitivity to ionizing radiation and telomere end-to-end fusions [38]. There is a growing evidence that oral poly (ADP-ribose) polymerase inhibitor, olaparib impedes the growth of ATM null or ATM mutant lymphoma cells in vitro and in vivo by instigating the accumulation of intolerable levels of DNA damage in cycling tumor cells [39], [40]. Consistent with these reports, ATM knockdown breast cancer cells, like BRCA1/2 null cells, exhibited selective sensitivity to PARP inhibition. Given the recent positive results from phase III trial comparing conventional chemotherapy to chemotherapy plus olaparib or iniparib in breast cancer, [39], [40]. Currently, there is no standard treatment for metastatic NETs although recent trials support the use of everolimus or sunitinib as one of the treatment options [41]. In contrast to the NETs in the West, where pancreatic NET is the most prevalent NETs, colorectum is the most common anatomic tumor location in Asia [1], [42]. Hence, a clinical research with PARP inhibitor in ATM-negative colorectal NETs should be actively implemented in Asian countries.
One of the major limitations of this study was small number of tumor specimens owing to the rarity of metastatic NETs with limited availability of fresh frozen tissues for gene expression profiling. Distant metastases in rectal NETs are associated with 1.7 to 8.1% [43], and approximately 16 to 40% of colonic NETs [44]. The median survival of metastatic colorectal NETs ranges from 6 to 18 months. We showed that ATM-negativity was not significantly correlated with clinicopathologic parameters such as tumor site, histology grade, sex, and age. However, there was a tendency towards ATM negativity in colon NETs and G2 NETs when compared with rectal NETs and G1 NETS, respectively, although statistical significance was not reached (p>0.05).
In conclusion, our study identified six significant cell cycle regulating genes associated with metastatic colorectal NETs, and down-regulation of ATM and ATM-mediated cell cycle signaling is indicated as a key regulatory molecule leading to increased cell proliferation and involves metastasis in colorectal NETs. The anti-tumor effect of PARP inhibitor in ATM-negative, metastatic NETs should be studied in the context of clinical trials. Further studies are warranted to explain underlying biological and molecular mechanisms and develop new therapeutic targets.
Supporting Information
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26:3063–3072. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- 2.Hauso O, Gustafsson BI, Kidd M, Waldum HL, Drozdov I, et al. Neuroendocrine tumor epidemiology: contrasting Norway and North America. Cancer. 2008;113:2655–2664. doi: 10.1002/cncr.23883. [DOI] [PubMed] [Google Scholar]
- 3.Hemminki K, Li X. Incidence trends and risk factors of carcinoid tumors: a nationwide epidemiologic study from Sweden. Cancer. 2001;92:2204–2210. doi: 10.1002/1097-0142(20011015)92:8<2204::aid-cncr1564>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 4.Lepage C, Rachet B, Coleman MP. Survival from malignant digestive endocrine tumors in England and Wales: a population-based study. Gastroenterology. 2007;132:899–904. doi: 10.1053/j.gastro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Chan JA, Kulke MH. New Treatment Options for Patients with Advanced Neuroendocrine Tumors. Curr Treat Options Oncol. 2011 doi: 10.1007/s11864-011-0148-2. [DOI] [PubMed] [Google Scholar]
- 6.Kloppel G, Perren A, Heitz PU. The gastroenteropancreatic neuroendocrine cell system and its tumors: the WHO classification. Ann N Y Acad Sci. 2004;1014:13–27. doi: 10.1196/annals.1294.002. [DOI] [PubMed] [Google Scholar]
- 7.Solcia EKG, Sobin LH. World Health Organization International Histological Classification of Tumours. World Health Organization International Histological Classification of Tumours: 2nd ed. New York, NY: Springer; 2000. [PMC free article] [PubMed] [Google Scholar]
- 8.Williams GT. Endocrine tumours of the gastrointestinal tract-selected topics. Histopathology. 2007;50:30–41. doi: 10.1111/j.1365-2559.2006.02570.x. [DOI] [PubMed] [Google Scholar]
- 9.Fahy BN, Tang LH, Klimstra D, Wong WD, Guillem JG, et al. Carcinoid of the rectum risk stratification (CaRRs): a strategy for preoperative outcome assessment. Ann Surg Oncol. 2007;14:1735–1743. doi: 10.1245/s10434-006-9311-6. [DOI] [PubMed] [Google Scholar]
- 10.Koura AN, Giacco GG, Curley SA, Skibber JM, Feig BW, et al. Carcinoid tumors of the rectum: effect of size, histopathology, and surgical treatment on metastasis free survival. Cancer. 1997;79:1294–1298. doi: 10.1002/(sici)1097-0142(19970401)79:7<1294::aid-cncr4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Shields CJ, Tiret E, Winter DC. Carcinoid tumors of the rectum: a multi-institutional international collaboration. Ann Surg. 2010;252:750–755. doi: 10.1097/SLA.0b013e3181fb8df6. [DOI] [PubMed] [Google Scholar]
- 12.Klimstra DS, Modlin IR, Adsay NV, Chetty R, Deshpande V, et al. Pathology reporting of neuroendocrine tumors: application of the Delphic consensus process to the development of a minimum pathology data set. Am J Surg Pathol. 2010;34:300–313. doi: 10.1097/PAS.0b013e3181ce1447. [DOI] [PubMed] [Google Scholar]
- 13.Mitui M, Nahas SA, Du LT, Yang Z, Lai CH, et al. Functional and computational assessment of missense variants in the ataxia-telangiectasia mutated (ATM) gene: mutations with increased cancer risk. Hum Mutat. 2009;30:12–21. doi: 10.1002/humu.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 15.Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 17.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 18.Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. J Clin Pathol. 2005;58:1009–1015. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SW, Reimer CL, Fang L, Iruela-Arispe ML, Aaronson SA. Overexpression of kinase-associated phosphatase (KAP) in breast and prostate cancer and inhibition of the transformed phenotype by antisense KAP expression. Mol Cell Biol. 2000;20:1723–1732. doi: 10.1128/mcb.20.5.1723-1732.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Falco G, Leucci E, Lenze D, Piccaluga PP, Claudio PP, et al. Gene-expression analysis identifies novel RBL2/p130 target genes in endemic Burkitt lymphoma cell lines and primary tumors. Blood. 2007;110:1301–1307. doi: 10.1182/blood-2006-12-064865. [DOI] [PubMed] [Google Scholar]
- 21.Iannuzzi CM, Atencio DP, Green S, Stock RG, Rosenstein BS. ATM mutations in female breast cancer patients predict for an increase in radiation-induced late effects. Int J Radiat Oncol Biol Phys. 2002;52:606–613. doi: 10.1016/s0360-3016(01)02684-0. [DOI] [PubMed] [Google Scholar]
- 22.Cole J, Arlett CF, Green MH, Harcourt SA, Priestley A, et al. Comparative human cellular radiosensitivity: II. The survival following gamma-irradiation of unstimulated (G0) T-lymphocytes, T-lymphocyte lines, lymphoblastoid cell lines and fibroblasts from normal donors, from ataxia-telangiectasia patients and from ataxia-telangiectasia heterozygotes. Int J Radiat Biol. 1988;54:929–943. doi: 10.1080/09553008814552331. [DOI] [PubMed] [Google Scholar]
- 23.DahlBerg WK, Little JB. Response of dermal fibroblast cultures from patients with unusually severe responses to radiotherapy and from ataxia telangiectasia heterozygotes to fractionated radiation. Clin Cancer Res. 1995;1:785–790. [PubMed] [Google Scholar]
- 24.Gatti RA, Tward A, Concannon P. Cancer risk in ATM heterozygotes: a model of phenotypic and mechanistic differences between missense and truncating mutations. Mol Genet Metab. 1999;68:419–423. doi: 10.1006/mgme.1999.2942. [DOI] [PubMed] [Google Scholar]
- 25.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 26.Bartek J. DNA damage response, genetic instability and cancer: from mechanistic insights to personalized treatment. Mol Oncol. 2011;5:303–307. doi: 10.1016/j.molonc.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna KK. Cancer risk and the ATM gene: a continuing debate. J Natl Cancer Inst. 2000;92:795–802. doi: 10.1093/jnci/92.10.795. [DOI] [PubMed] [Google Scholar]
- 28.Schaffner C, Idler I, Stilgenbauer S, Dohner H, Lichter P. Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proc Natl Acad Sci U S A. 2000;97:2773–2778. doi: 10.1073/pnas.050400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoppa-Lyonnet D, Soulier J, Lauge A, Dastot H, Garand R, et al. Inactivation of the ATM gene in T-cell prolymphocytic leukemias. Blood. 1998;91:3920–3926. [PubMed] [Google Scholar]
- 30.Zhang P, Bhakta KS, Puri PL, Newbury RO, Feramisco JR, et al. Association of ataxia telangiectasia mutated (ATM) gene mutation/deletion with rhabdomyosarcoma. Cancer Biol Ther. 2003;2:87–91. doi: 10.4161/cbt.231. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Jia G, Li WM, Guo RF, Cui JT, et al. Alteration of the ATM gene occurs in gastric cancer cell lines and primary tumors associated with cellular response to DNA damage. Mutat Res. 2004;557:41–51. doi: 10.1016/j.mrgentox.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Atencio DP, Iannuzzi CM, Green S, Stock RG, Bernstein JL, et al. Screening breast cancer patients for ATM mutations and polymorphisms by using denaturing high-performance liquid chromatography. Environ Mol Mutagen. 2001;38:200–208. doi: 10.1002/em.1072. [DOI] [PubMed] [Google Scholar]
- 33.Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–1836. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 34.Mazumder Indra D, Mitra S, Roy A, Mondal RK, Basu PS, et al. Alterations of ATM and CADM1 in chromosomal 11q22.3–23.2 region are associated with the development of invasive cervical carcinoma. Hum Genet. 2011;130:735–748. doi: 10.1007/s00439-011-1015-8. [DOI] [PubMed] [Google Scholar]
- 35.Vorechovsky I, Rasio D, Luo L, Monaco C, Hammarstrom L, et al. The ATM gene and susceptibility to breast cancer: analysis of 38 breast tumors reveals no evidence for mutation. Cancer Res. 1996;56:2726–2732. [PubMed] [Google Scholar]
- 36.Ai L, Vo QN, Zuo C, Li L, Ling W, et al. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol Biomarkers Prev. 2004;13:150–156. doi: 10.1158/1055-9965.epi-082-3. [DOI] [PubMed] [Google Scholar]
- 37.Kim WJ, Vo QN, Shrivastav M, Lataxes TA, Brown KD. Aberrant methylation of the ATM promoter correlates with increased radiosensitivity in a human colorectal tumor cell line. Oncogene. 2002;21:3864–3871. doi: 10.1038/sj.onc.1205485. [DOI] [PubMed] [Google Scholar]
- 38.Xie P, Li L, Xing G, Tian C, Yin Y, et al. ATM-mediated NuSAP phosphorylation induces mitotic arrest. Biochem Biophys Res Commun. 2011;404:413–418. doi: 10.1016/j.bbrc.2010.11.135. [DOI] [PubMed] [Google Scholar]
- 39.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 40.Williamson CT, Muzik H, Turhan AG, Zamo A, O'Connor MJ, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito T, Sasano H, Tanaka M, Osamura RY, Sasaki I, et al. Epidemiological study of gastroenteropancreatic neuroendocrine tumors in Japan. J Gastroenterol. 2010;45:234–243. doi: 10.1007/s00535-009-0194-8. [DOI] [PubMed] [Google Scholar]
- 43.Modlin IM, Lye KD, Kidd M. A 5-decade analysis of 13,715 carcinoid tumors. Cancer. 2003;97:934–959. doi: 10.1002/cncr.11105. [DOI] [PubMed] [Google Scholar]
- 44.Soga J. Carcinoids of the colon and ileocecal region: a statistical evaluation of 363 cases collected from the literature. J Exp Clin Cancer Res. 1998;17:139–148. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)