Abstract
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is a major global health threat. During infection, bacteria are believed to encounter adverse conditions such as iron depletion. Mycobacteria synthesize iron-sequestering mycobactins, which are essential for survival in the host, via the intermediate salicylate. Salicylate is a ubiquitous compound which is known to induce a mild antibiotic resistance phenotype. In M. tuberculosis salicylate highly induces the expression of Rv0560c, a putative methyltransferase. We identified and characterized the promoter and regulatory elements of Rv0560c. PRv0560c activity was highly inducible by salicylate in a dose-dependent manner. The induction kinetics of PRv0560c were slow, taking several days to reach maximal activity, which was sustained over several weeks. Promoter activity could also be induced by compounds structurally related to salicylate, such as aspirin or para-aminosalicylic acid, but not by benzoate, indicating that induction is specific to a structural motif. The −10 and −35 promoter elements were identified and residues involved in regulation of promoter activity were identified in close proximity to an inverted repeat spanning the −35 promoter element. We conclude that Rv0560c expression is controlled by a yet unknown repressor via a highly-inducible promoter.
Introduction
Tuberculosis (TB) is a major global health threat. According to the WHO, TB causes nearly 2 million deaths each year, and one third of the world's population is believed to be latently infected [1]. TB can be caused by any member of the Mycobacterium tuberculosis complex, however, M. tuberculosis is the major causative agent of TB in humans [1]. Despite an increased amount of research effort in the past decade, many processes underlying M. tuberculosis physiology and pathogenesis are still poorly understood. M. tuberculosis is an intracellular pathogen, and during infection the mycobacteria are believed to be exposed to adverse conditions such as hypoxia, nitric oxide and iron starvation [2]–[5].
Iron is an indispensable component of many prokaryotic and eukaryotic enzymes. When bacteria encounter conditions of low iron, for example during macrophage infection, they produce iron-sequestering siderophores in order to maintain cellular functions [6], [7]. M. tuberculosis produces two types of siderophores (mycobactins), whose production is essential for infection and survival in macrophages [8]–[10]. Expression of the genes required for mycobactin synthesis is controlled by the regulator of iron homeostasis IdeR [11]–[13]. Mycobactin biosynthesis involves the conversion of isochorismate into salicylate by the enzyme MbtI [14]–[16]. As a result of this, mycobacteria accumulate salicylate under iron-depleted conditions [14], [17]–[19].
Given the natural accumulation of salicylate under conditions encountered by the bacteria during an infection, it is interesting to note exogenous salicylate is able to induce a multiple antibiotic resistant (“mar”) phenotype in M. tuberculosis [20]. This effect is also seen in both Gram negative and Gram positive bacteria, and is thought to be due to the induction of efflux pumps via transcriptional repressors such as MarR in Escherichia coli or CmeR in Campylobacter jejuni [21]–[24].
The mechanism of the salicylate-induced mar phenotype in M. tuberculosis is poorly understood. Salicylate exposure affects the expression of 58 genes, none of which are known efflux pumps, and results in a general reduction of transcriptional and translational activities, as well as changes in energy metabolism [25]. Interestingly, two genes, Rv0560c and Rv0559c, are upregulated to a degree much higher (30- and 8- fold respectively) than any other of the differentially expressed genes [25].
Rv0560c is a non-essential gene encoding a putative benzoquinone methyltransferase. There have been suggestions that Rv0560c plays a role related to the biosynthesis of the isoprenoid lipid menaquinone [26], [27]. The fact that this gene is not expressed during aerobic growth [26], but upregulated during hypoxia [28] and intraphagosomal growth in macrophages [12] is interesting, indicating that this gene might play a role during infection. The Rv0560c protein is also induced in response to para-aminosalicylic acid (PAS), naphthoquinones such as menadione, and plumbagin, as well as the peroxisome proliferator gemfibrozil, and its structural relatives fenofibrate and clofibrate [26], [27].
The range of conditions under which Rv0560c is induced and the huge extent of its induction in response to salicylate are intriguing. The aim of this study was to identify and characterise the promoter of Rv0560c to gain further insight into its expression. Here, we demonstrate for the first time that Rv0560c is expressed from a salicylate-inducible promoter (PRv0560c) which is highly active. We investigated the induction kinetics of this promoter, which took several days to reach maximal activity and remained highly induced for several weeks. We show that PRv0560c is also induced by structural analogues of salicylate as well as fenofibrates and is mildly induced under conditions of low iron. The −10 and −35 promoter elements, as well as residues involved in its regulation were identified. Our results suggest that this regulatory control is likely to be mediated via a repressor. The data presented here reveal PRv0560c to be a promoter with a high level of induction after salicylate treatment.
Results
Rv0561c and Rv0560c are expressed from separate promoters
We wanted to identify the promoter responsible for the salicylate-dependent induction of Rv0560c. Rv0560c has been suggested to be in an operon with its upstream gene Rv0561c and its downstream gene Rv0559c (Fig. 1A) [26]. Interestingly, the upstream gene Rv0561c has not been reported to be induced by salicylate [25].
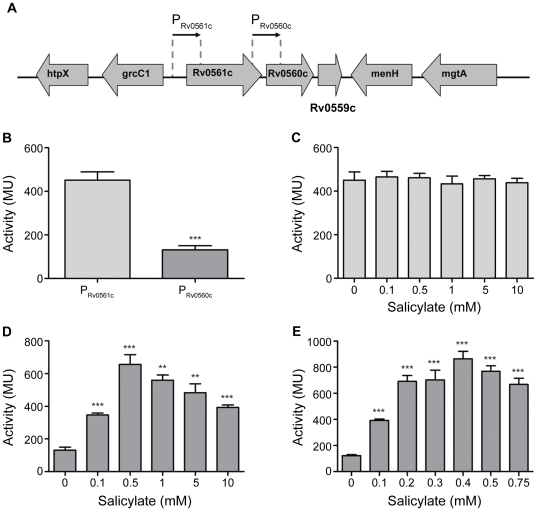
Figure 1. Identification of a salicylate-inducible promoter in M. tuberculosis.
A. The genetic organization of Rv0561c and Rv0560c in M. tuberculosis. The regions tested for promoter activity are indicated as PRv0561c, and PRv0560c. B–E. Promoter activity was measured in M. tuberculosis transformants grown under aerobic growth conditions. B Activity in the absence of salicylate. C, D and E: Promoter activity of PRv0561c (C), PRv0560c (D and E), after 2 h treatment with varying concentrations of salicylate. Results are the average and standard deviation of three independent transformants assayed in duplicate. Activity is given in Miller Units. A significant difference compared using Student's t-test to the untreated control is marked by an * for p<0.05) ** for p<0.01, *** for p<0.0001.
To determine whether these genes had their own promoters, the upstream regions of both genes were tested for promoter activity in M. tuberculosis by linking them to a lacZ reporter gene. The upstream regions of Rv0560c and Rv0561c are referred to as PRv0560c and PRv0561c respectively (Fig. 1A). Promoter activity of ∼400 MU was detected from PRv0561c, indicating that this upstream region contains an active promoter (Fig. 1B). Promoter activity was also detected from PRv0560c (130 MU) although lower than that of PRv0561c (Fig. 1B). Therefore each of the genes has its own promoter.
PRv0560c is salicylate-inducible in M. tuberculosis
To test if one or both promoter regions were salicylate-inducible, we assayed the effect of varying concentrations of salicylate on promoter activity (Fig. 1C–E). PRv0561c activity did not change in response to treatment with salicylate (Fig. 1C) and thus, PRv0561c is not salicylate-inducible. In contrast to this, PRv0560c activity increased up to 6-fold to over 800 MU (Fig. 1D–E). Promoter activity did not increase any further with concentrations higher than 0.5 mM salicylate. Thus this promoter is the one responsible for the induction of Rv0560c upon salicylate exposure. PRv0560c is not only salicylate-inducible, but also displays dose responsive behavior at concentrations <0.4 mM (Fig. 1D–E), with maximal expression being achieved at 0.4 mM salicylate.
PRv0560c induction kinetics
The induction kinetics of PRv0560c were investigated by monitoring promoter activity over time after treatment with salicylate. PRv0560c activity tripled from basal level to an activity of 301 MU after as little as 30 min of treatment with the inducer (p<0.05) and continued to increase over 4 hours (Fig. 2A). This trend persisted over the course of several days; a 100-fold increase (10315 MU) was observed after 3 d of treatment (Fig. 2B) and the promoter remained induced for at least 35 d (Fig. 2B). The level of induction is very high, and although the promoter responds in less than an hour to inducer, the full extent of the response takes several days and lasts for at least 5 weeks.
Figure 2. PRv0560c induction kinetics after exposure to salicylate in M. tuberculosis.
A and B. Promoter activity was measured in M. tuberculosis transformants grown under aerobic growth conditions exposed to 0.4 mM salicylate (A and B), or 0.2 mM or 0.4 mM salicylate (C) or with the use of LacZ tagged for degradation(D). Results are the average and standard deviation of three independent transformants assayed in duplicate. Activity is given in Miller Units. LacZ-ASV was tagged with AANDENYAASV; LacZ-LAA was tagged with AANDENYALAA.
Induction kinetics changed with the amount of inducer present (Fig. 2C). With lower amounts of inducer (0.2 mM), activity of the promoter was lower with only a 20-fold induction being observed; and that only after 7–14 d of treatment. This shows PRv0560c to have slower induction kinetics and to be of lower strength when exposed to less inducer. Thus PRv0560c activity varies depending on inducer concentration and length of treatment.
The slow induction kinetics in our system could be due to accumulation of LacZ in the cells which is not degraded and accumulates over time. However, LacZ has been used widely as a reporter of promoter activity including determination of kinetics making this seem unlikely [29]. In order to test this possibility we constructed an unstable variant of LacZ which incorporated a protein degradation tag previously shown to function in mycobacteria [30]. Two LacZ variants incorporating either AANDENYAASV or AANDENYALAA at the C-terminal end were engineered; the induction kinetics from PRv0560c was measured as before. The induction kinetics were slightly slower for both variants taking 14 d to reach maximal, but still reached the high levels seen with native LacZ (Fig. 2D). The maximal level of expression was also slightly lower, 8,000 Mu compared to 10,000, but this likely reflects a higher turnover of synthesized protein carrying the degradation tag rather than reduced expression levels. Since the steady state levels were similar for all, we discounted the possibility that LacZ stability was responsible for increased activity over time.
PRv0560c can be induced by structural analogues of salicylate
PAS, an antimycobacterial drug, and aspirin, a common painkiller, are structural analogues of salicylate; these compounds, as well as benzoate and the structurally-related compounds menadione, fenofibrate and gemfibrozil, have been shown to induce Rv0560c protein expression [26], [27]. We were interested in the effect of these compounds on PRv0560c activity.
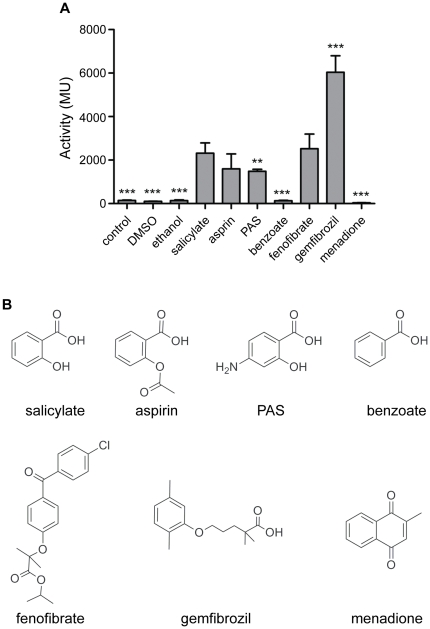
We determined the effect of each compound on promoter activity at a fixed concentration (0.4 mM) (Fig. 3A). Salicylate resulted in a 64-fold induction (∼10000 MU), whereas its structural analogues aspirin and PAS induced a comparatively moderate 10-fold increase in PRv0560c (∼1500 MU; Fig. 3A). Surprisingly, the structural analogue benzoate did not induce PRv0560c activity at all (Fig. 3A). These results show that PRv0560c induction is specific to a certain structure present in salicylate, and to a certain extent in aspirin and PAS, but not benzoate (Fig. 3B).
Figure 3. PRv0560c induction by structural analogs of salicylate in M. tuberculosis.
Promoter activity was measured in M. tuberculosis transformants grown under aerobic growth conditions. A. Promoter activity of PRv0560c after treatment with 0.4 mM of compound for 3 d. Results are the average and standard deviation of three independent transformants assayed in duplicate. Activity is given in Miller Units. B. Chemical structures of compounds of interest. A significant difference compared using Student's t-test to the untreated control is marked by an * for p<0.05) ** for p<0.01, *** for p<0.0001.
Amongst the fibrates tested, fenofibrate evoked a 17-fold induction (2524 MU; Fig. 3A), and gemfibrozil evoked a 41-fold induction (6038 MU), which is closer to the levels achieved by salicylate exposure (Fig. 3B). Under the conditions tested here, menadione repressed PRv0560c activity 4-fold. These results show that other compounds that are not direct structural analogues of salicylate, are able to modulate PRv0560c activity.
PRv0560c activity remains high after removal of exogenous inducer
We determined the off kinetics of the promoter. PRv0560c activity was induced to maximal level by growing M. tuberculosis in the presence of salicylate and promoter activity was monitored after removal of salicylate from the growth medium by washing in salicylate-free medium.
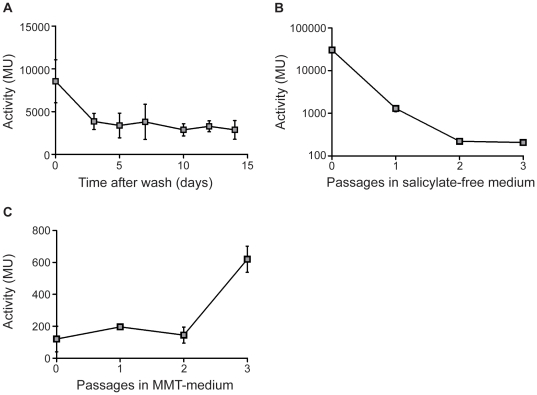
PRv0560c activity remained high immediately after the wash in salicylate-free medium (Fig. 4A), but then decreased over 3 d by approximately 2.5-fold (10000 to 3849 MU). Even after 2 weeks, activity did not decrease further, and was still 20-fold higher than the basal level of activity (∼150 MU). This shows that PRv0560c has slow off kinetics and that the remaining promoter activity could be due to the presence of residual salicylate in the medium. Increasing the number of washes carried out to remove salicylate from the growth medium up to four times did not allow for promoter activity to return to basal level over the course of 1 week (data not shown). Therefore, residual activity of PRv0560c could be due to high intracellular levels of salicylate which would not be removed merely by washing cells. To test this possibility, PRv0560c off kinetics were monitored during growth in salicylate-free medium during several passages; a salicylate-exposed culture was used to inoculate salicylate-free medium and cultured to late log phase before further passaging in salicylate-free medium. Promoter activity decreased with each passage (Fig. 4B), returning to basal level after 3 passages, suggesting that residual salicylate was gradually being consumed in the cells during growth.
Figure 4. PRv0560c off kinetics in salicylate-free or iron-free medium in M. tuberculosis.
M. tuberculosis transformants were grown under aerobic growth conditions in the presence of 0.4 mM salicylate. Cultures were washed and inoculated into salicylate-free medium A. Washed cells. Transformants were washed and resuspended in salicylate-free medium. B. Cells were washed and inoculated into salicylate-free medium; cultures were passaged into fresh salicylate-free medium at a dilution of 1/10. C. Cells were washed and inoculated in low iron minimal medium (MMT); cultures were passaged into fresh MMT medium at a dilution of 1/100. Results are the average and standard deviation of three independent transformants assayed in duplicate. Activity is given in Miller Units.
PRv0560c is induced in iron-depleted M. tuberculosis
Salicylate is known to accumulate in iron-depleted mycobacteria as an intermediate during the biosynthesis of the iron-sequestering mycobactin [14], [18], [19]. Hence, one would expect Rv0560c to be upregulated under this condition. However, existing data on whether Rv0560c is upregulated under iron-limiting conditions are conflicting [11], [26]. We tested if PRv0560c activity increased in M. tuberculosis grown in iron-free medium. After a state of iron-depletion had been achieved (three passages in iron-free medium), PRv0560c activity increased by 3-fold to 620 MU (Fig. 4C). Thus, as expected the promoter is induced when intracellular levels of iron are depleted. Interestingly induction was not as pronounced as in salicylate-exposed cells, suggesting that the normal intracellular concentrations of salicylate are low.
PRv0560c is a negatively regulated, sigA-dependent promoter
To characterise PRv0560c and its regulator(s) further, an attempt was made to map its promoter elements. According to the current annotation on TubercuList (http://tuberculist.epfl.ch/), there is a short intergenic region of 24 bp between the stop codon of Rv0561c and the start codon of Rv0560c. A search for putative −10 elements within the upstream region of Rv0560c was carried out. Three putative −10 elements (PM1: TGTGTT, PM2: TATATC, PM3: TATATAT) were found, all located downstream of the annotated translational start site of Rv0560c, but immediately upstream of a putative alternative translational start site (Fig. 5A). To establish if and which one of these putative −10 elements was part of the Rv0560c promoter, residues in each motif were mutated.
Figure 5. Identification of the promoter and regulatory elements.
A. DNA sequence of the PRv0560c region. The predicted translation start site of Rv0560c according to TubercuList is marked with **. Protein sequences of Rv0561c and Rv0560c are shown. Potential −10 promoter elements (PM1, PM2, PM3) are underlined. The −35 and extended −10 element are in bold. A palindromic moitif is indicated by grey shading. B. Promoter activity following mutation of the promoter region. M. tuberculosis transformants were grown under aerobic growth conditions in the absence/presence of 0.4 mM salicylate. Results are the average and standard deviation of three independent transformants assayed in duplicate. Activity is given in Miller Units. A significant difference compared to the wild type is marked by an * (p<0.05).
Mutation of PM3 abolished promoter activity to a level seen in the empty vector control (11 MU) confirming that it is the most probable −10 element of the Rv0560c promoter. Interestingly, mutations in PM1 and PM2 both resulted in constitutive expression, albeit to different levels (Fig. 5B). Mutation of PM1 had minimal impact on the basal activity (260 MU), but resulted in complete lack of induction by salicylate (Fig. 5B). In contrast, mutation of PM2 resulted in high level constitutive activity (15000–17000 MU). Closer examination of the sequence revealed a putative −35 region with the consensus sequence TTGACA located 17 bp upstream of the confirmed −10 element; this region was mutated in PM1 (to TGGACA) suggesting that it is the −35 element and is involved in regulation of promoter activity.
The sequence of the promoter elements suggests that PRv0560c is highly likely to be a SigA–dependent promoter. The SigA consensus sequence is TTGACW-N16–21-TATAMT [31]. The promoter region identified has a perfect match in the −35 region of TTGACA, with a spacing of 17 bp to the −10 element TATAta (matching bases in capital). Furthermore, PRv0560c looks to be an extended promoter due to the presence of a TGN motif (in this case TGA) directly in front of the −10 element [32].
Mutations in the −10 element affect promoter strength
We decided to mutate some of the residues in the −10 element to test their effect on promoter strength and regulation (Fig. 5B). Mutation of the second residue (PM4) severely weakened promoter strength. Basal activity under uninduced conditions was abolished. However, the promoter retained inducibility, albeit to a lower level (133 MU). Similar effects were seen when mutating the fourth residue (PM5), although the promoter was slightly more active (54 MU and 1239 MU in the uninduced and induced states respectively). These results are as expected since mutating residues away from the sigma-factor consensus would weaken recognition and binding of the factor, resulting in less transcription.
Identification of further residues involved in repressor binding
Since mutation in the −35 region resulted in loss of induction, we predicted that there is a regulatory element in this region; such operator regions are often located between the −35 and 10 elements. We carried out further mutations to identify other residues involved in regulation of promoter activity located between the −10 and −35 region. Mutation of the two residues immediately downstream of the −35 element (PM6) resulted in a much higher basal level of activity (1232 MU). Interestingly, mutation of two bases further downstream (PM7) increased the basal activity even further (5959 MU). In both cases, induction was retained, with promoter activity of over 10000 MU in the presence of salicylate (Fig. 5B). This confirms that this region contains a regulatory site and suggests it is the binding site for a repressor.
Discussion
We were interested in the M. tuberculosis gene Rv0560c due to its strikingly high induction in response to salicylate, and its upregulation under conditions mimicking the in vivo environment the bacteria encounter during an infection [12], [28]. Although Rv0560c has been suggested to be in an operon with its upstream gene, our results show that each of the genes does have its own promoter, both of which are active during aerobic growth. Furthermore, we demonstrate that PRv0560c, but not PRv0561c is inducible by salicylate. This is in accordance with a previous transcriptome study showing that Rv0560c, but not Rv0561c is upregulated after salicylate treatment [25].
We found PRv0560c to be a strong promoter as compared to other M. tuberculosis promoters and with slow induction kinetics, taking several days to reach peak activity and remaining stably induced over a course of several weeks. The majority of M. tuberculosis promoters measured using LacZ as a reporter have activity in the range of 100–1000 MU; examples include the promoters of recA, pknH, embA, mbtB and higBA [33]–[37]. There are few examples of promoters with activity in the 10,000 range; for example PrpfA, which is reported as one of the strongest constitutive promoters in M. tuberculosis has an activity of 4500 MU [38], [39], whereas PwhiB1 has comparable activity to PRv0560c at 800–15,000 MU [40].
The high level of promoter activity after induction could be due to presence of a perfect match to the canonical −35 element TTGACA (which is not always present in most mycobacterial promoters), the extended promoter motif, and the high sequence similarity of the −10 element to the consensus sequence TATAAT [31], [32]. Indeed, these attributes are also present in the PwhiB1 promoter with comparable activity [41].
The slow induction kinetics could be due to a tight interaction of the repressor with its binding motif, taking a long time to alleviate and allow full access to the polymerase. Alternatively the mechanism leading to alleviation of repression (whether it occurs through salicylate binding to the repressor or via a relay of signal through other regulators) might be slow. It is interesting to note that there are other inducible promoters with slow kinetics, although most of these are not native M. tuberculosis systems, for example the ATc and pristinamcyin systems [42]–[44]. However, the kinetics of induction of PRv0560c appear to be particularly slow, since the ATc-inducible systems are fully induced with 24 h [36], [43], [45], [46] and even the “slow” induction of RecA expression previously noted in M. tuberculosis took 18–36 h, rather than 72 h [33]. These kinetics are not a general phenomenon since other promoters can be induced to maximal expression much more rapidly, for example induction of heat shock proteins takes less than an hour.
The fact that some structural analogues of salicylate (PAS and aspirin), but not others (benzoate) induce promoter activity confirms that induction is specific to a certain chemical structure present in salicylate, PAS and aspirin, but not benzoate. These findings are in accordance with a previous study on Rv0560crotein expression, except for aspirin (of which salicylate is a breakdown product) which was reported not to induce Rv0560c [26].
Compounds that can interfere with isoprenoid quinone action and are structurally related to salicylate (such as fenofibrate or gemfibrozil) also induced PRv0560c activity. In our study, menadione did not induce PRv0560c, but actually repressed it. Both results are in accordance with previous findings of a protein study [27]. It would be interesting to determine whether this induction is due to an indirect effect or due to a structural motif common to all these compounds.
The function of Rv0560c is unknown, but it has homology with a benzoquinone methyltransferase (UbiG) involved in ubiquinone biosynthesis in E. coli [26]. Quinones are lipid-soluble electron carriers involved in the electron transport chain, a process essential for growth. Rv0560c could be involved in a ubiquinone biosynthetic process, as M. tuberculosis does contain homologs of some of the genes present in the E. coli ubiquinone pathway [47], although there is currently no evidence that mycobacteria produce ubiquinones [48], [49].
Rv0560c has also been suggested to be involved in menaquinone biosynthesis, due to its genomic proximity to menH (Rv0558), menD (Rv0555) and menC (Rv0553) [26], [27]. Menaquinone biosynthesis is essential for mycobacterial viability and this synthetic pathway has been proposed as an attractive target for novel antimycobacterial drugs [50], [51]. Furthermore, a recent study linked menaquinones to the induction of the DosR regulon, which is implicated in the adaptation to hypoxia and the establishment of a dormant state [52], [53]. One could speculate Rv0560c is a methyltransferase carrying out functions equivalent of the methyltransferases MenH or MenG.
Alternatively, Rv0560c (which is not expressed during aerobic in vitro growth) could be involved in the synthesis of novel menaquinones such as the recently identified sulphated menaquinone [54] and/or menaquinone biosynthesis under certain stress conditions such as iron starvation. Our results show PRv0560c to be induced during iron starvation, when intracellular levels of salicylate are naturally elevated. The salicylate-dependent induction of PRv0560c accounts for the upregulation of Rv0560c during iron depletion, despite the absence of binding motifs for the main regulator of iron responsive genes (IdeR) upstream of Rv0560c [11], [13].
The aim of our study was to identify and characterize the promoter of Rv0560c. Our results demonstrated PRv0560c to be a predicted SigA-dependent promoter and suggest that the translational start site of Rv0560c is currently misannotated. Furthermore, the expression of Rv0560c appears to be regulated by a repressor, which according to our results binds to residues close to the −35 element of PRv0560c, possibly to a palindromic motif that overlaps the −35 element. This is supported by the fact that when certain residues in this motif are mutated, PRv0560c is constitutively active at maximal level.
In Gram negative and Gram positive bacteria the salicylate-induced mar phenotype is mediated through salicylate binding directly to transcriptional repressors of the MarR family [21]–[23]. In E. coli, MarR binds to salicylate, relieves repression of its regulon and evokes induction of a mild antibiotic resistance phenotype through upregulation of efflux pumps [21]–[23], [55], [56]. The mar phenotype has been observed in M. tuberculosis although the mechanism of induction of multidrug resistance has not been determined [20]. Interestingly, M. tuberculosis possesses several MarR type regulators, with Rv1404 or Rv2887 showing the highest sequence similarity to the E. coli MarR, with some conservation of the salicylate binding sites. Thus either of these genes could make attractive candidates for the regulator of Rv0560c expression.
To conclude, we identified Rv0560c to have its own salicylate-inducible promoter. with a high level of induction. We present evidence that this promoter is controlled by an unknown repressor binding to a palindromic motif upstream of Rv0560c. Further studies are required to identify this repressor, which we speculate to be part of the MarR family. Whether Rv0560c is involved in menaquinone biosynthesis, if it is important for iron starvation; salicylate tolerance or even plays a role in the mar phenotype are interesting questions which would be worthy of further investigation.
Materials and Methods
Bacterial strains and culture conditions
M. tuberculosis H37Rv (ATCC 25618) was grown in Middlebrook 7H9 medium supplemented with 10% v/v oleic acid-albumin-dextrose-catalase (OADC) supplement and 0.05% w/v Tween 80, or on 7H10 agar supplemented with 10% v/v OADC. Cultures were grown without agitation in 50 mL conical tubes unless otherwise stated. Streptomycin was used at 20 mg L−1 and X-gal at 50 mg L−1 when required. Low iron minimal medium (MMT) was prepared as follows: 6 g L−1 Na2HPO4, 3 g L−1 KH2PO4, 0.5 g L−1 NaCl, 1 g L−1 NH4Cl and 0.0147 g L−1 CaCl2 supplemented with 0.05% w/v Tween 80 and 2% v/v glycerol and treated overnight with 5 g L−1 Chelex 100; 2 mM MgSO4 was added and the medium was filter-sterilised.
Plasmid construction
Primers were designed to amplify the upstream regions of Rv0561c and Rv0560c from M. tuberculosis genomic DNA using primer pairs UR561F CCCCCCGGGGGATC- GCGACGTTGTTAC and UR561R CCCCCCGGGCCGCCAGCCACTTAC for Rv0561c, and UR560F CCCCCCGGGGCGCCGGCTAGCGTTGTTAC and UR560R CCCCCCGGGCCTG- CCGTCATAGCCGGTAAACG for Rv0560c. Products were cloned as SmaI fragments (underlined) into the SacI site of pSM128 [57] upstream of the lacZ reporter gene. Protein tags were added to lacZ in pSM128 using SDM primer pairs TailLAAf GGTCTGGTGTCAAAAAGCAGCAAACGACGAAAACTACGCTTTAGC AGCTTAATAATAAC, TailLAAr GTTATTATTAAGCTGCTAAAGCGTAGTTTTCG TCGTTTGCTGCTTTTTGACACCAGAC or TailASVf GGTCTGGTGTCAAAAAGC AGCAAACGACGAAAACTACGCTGCATCAGTTTAATAATAAC, TailASVr GTTA TTATTAAACTGATGCAGCGTAGTTTTCGTCGTTTGCTGCTTTTTGACACCAGACC.
Site directed mutagenesis (SDM)
Amplification reactions were carried out in 50 µL total volume containing 2.5 units PfuUltra Hot Start high fidelity DNA polymerase (Stratagene), 1× buffer, 0.5 mM dNTPs, 10 pmol of each primer, 5 µL DMSO, and 10 ng template. The thermocycling programme used was: 94°C for 2 min, followed by 18 cycles of 94°C for 30 s, 56°C for 1 min and 68°C for 9 min, followed by 68°C for 10 min. Template was degraded using 10 units DpnI (Promega) at 37° for 2 h. 10 µL of each reaction were used to transform competent E. coli. Recombinant plasmids were isolated and sequence-verified.
Preparation of cell-free extracts for promoter activity assays
Electrocompetent mycobacteria were prepared as described previously [58], electroporated with 1 µg plasmid DNA and transformants selected on streptomycin. M. tuberculosis transformants were cultured to mid-log phase and exposed to compounds. Cells were harvested, washed and resuspended in 1 mL 10 mM Tris-Cl (pH 8) and added to 2 mL lysing matrix B tubes (MP Biomedicals) on ice. Cells were disrupted using a 30 s cycle at speed 6.0 using a FastPrep™ FP120 (MP Biomedicals). Extracts were centrifuged at 16000× g for 4 min and the supernatants recovered filter-sterilised through a 0.2 µm filter unit and recovered. Total protein concentration of the samples was determined using the BCA protein assay kit.
Quantification of ß -galactosidase
Assays of ß-galactosidase activity were carried out as previously described [59]. To 100 µL of cell-free extract, 900 µL of Z-Buffer (60 mM Na2HPO4, 40 mM Na2PO4, 10 mM KCl, 1 mM MgSO4, pH 7) was added. Samples were pre-warmed to 37°C for 5 min and 200 µL of 4 mg mL−1 ONPG was added. Reaction mixtures were incubated at 37°C and reactions were stopped with 500 µL of 1 M NaHCO3 after 30–90 min. The OD420 was measured and ß-galactosidase activity was calculated as Miller units (MU) = amount of O-nitrophenol produced (nmol) per min per mg of total protein, using the following formula: Units = (OD420×1.7)/(time (min)×volume of cell-free extract (mL)×total protein concentration (mg mL−1)×0.0045).
Acknowledgments
We are grateful to Renan Goude and Amanda Brown for valuable discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the European Union Project LSHP-CT-2005-018923. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Global tuberculosis control: Epidemiology, strategy, financing. 2009. World Health Organization report.
- 2.Russell DG, Barry CE, Flynn JL. Tuberculosis: what we don't know can, and does, hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 4.Young M, Mukamolova GV, Kaprelyants AS. Parish T, editor. Mycobacterial dormancy and its relation to persistence. Mycobacterium: Molecular Microbiology: Horizon bioscience. 2005. pp. 265–320.
- 5.Stewart GR, Papatheodorou I, Young D. Parish T, editor. The stress response. Mycobacterium: Molecular Microbiology: Horizon Bioscience. 2005. pp. 245–265.
- 6.Weinberg ED. The role of iron in protozoan and fungal infectious diseases. J Eukaryot Microbiol. 1999;46:231–238. doi: 10.1111/j.1550-7408.1999.tb05119.x. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Barclay R, Ratledge C. Mycobactins and exochelins of Mycobacterium tuberculosis, M. bovis, M. africanum and other related species. J Gen Microbiol. 1988;134:771–776. doi: 10.1099/00221287-134-3-771. [DOI] [PubMed] [Google Scholar]
- 9.Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 10.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacon J, Dover LG, Hatch KA, Zhang Y, Gomes JM, et al. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 2007;153:1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 15.Harrison AJ, Yu M, Gardenborg T, Middleditch M, Ramsay RJ, et al. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J Bacteriol. 2006;188:6081–6091. doi: 10.1128/JB.00338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwahlen J, Kolappan S, Zhou R, Kisker C, Tonge PJ. Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2007;46:954–964. doi: 10.1021/bi060852x. [DOI] [PubMed] [Google Scholar]
- 17.Ratledge C, Marshall BJ. Iron transport in Mycobacterium smegmatis: the role of mycobactin. Biochim Biophys Acta. 1972;279:58–74. doi: 10.1016/0304-4165(72)90241-3. [DOI] [PubMed] [Google Scholar]
- 18.Ratledge C, Winder FG. The accumulation of salicylic acid by mycobacteria during growth on an iron-deficient medium. Biochem J. 1962;84:501–506. doi: 10.1042/bj0840501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Schaller A, Sun Z, Yang Y, Somoskovi A, Zhang Y. Salicylate reduces susceptibility of Mycobacterium tuberculosis to multiple antituberculosis drugs. Antimicrob Ag Chemother. 2002;46:2636–2639. doi: 10.1128/AAC.46.8.2636-2639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price CT, Lee IR, Gustafson JE. The effects of salicylate on bacteria. Int J Biochem Cell Biol. 2000;32:1029–1043. doi: 10.1016/s1357-2725(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 22.Rosner JL. Nonheritable resistance to chloramphenicol and other antibiotics induced by salicylates and other chemotactic repellents in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1985;82:8771–8774. doi: 10.1073/pnas.82.24.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafson JE, Candelaria PV, Fisher SA, Goodridge JP, Lichocik TM, et al. Growth in the presence of salicylate increases fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:990–992. doi: 10.1128/aac.43.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Z, Pu XY, Zhang Q. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant mutants in Campylobacter jejuni. Appl Environ Microbiol. 2011;77:7128–7133. doi: 10.1128/AEM.00763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denkin S, Byrne S, Jie C, Zhang Y. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch Microbiol. 2005;184:152–157. doi: 10.1007/s00203-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 26.Sun Z, Cheng SJ, Zhang H, Zhang Y. Salicylate uniquely induces a 27-kDa protein in tubercle bacillus. FEMS Microbiol Lett. 2001;203:211–216. doi: 10.1111/j.1574-6968.2001.tb10843.x. [DOI] [PubMed] [Google Scholar]
- 27.Garbe TR. Co-induction of methyltransferase Rv0560c by naphthoquinones and fibric acids suggests attenuation of isoprenoid quinone action in Mycobacterium tuberculosis. Can J Microbiol. 2004;50:771–778. doi: 10.1139/w04-067. [DOI] [PubMed] [Google Scholar]
- 28.Starck J, Kallenius G, Marklund BI, Andersson DI, Akerlund T. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology. 2004;150:3821–3829. doi: 10.1099/mic.0.27284-0. [DOI] [PubMed] [Google Scholar]
- 29.Carroll P, James J. Assaying promoter activity using LacZ and GFP as reporters. Methods in Molecular Biology. 2008:265–277. doi: 10.1007/978-1-59745-207-6_18. [DOI] [PubMed] [Google Scholar]
- 30.Blokpoel MC, O'Toole R, Smeulders MJ, Williams HD. Development and application of unstable GFP variants to kinetic studies of mycobacterial gene expression. J Microbiol Methods. 2003;54:203–211. doi: 10.1016/s0167-7012(03)00044-7. [DOI] [PubMed] [Google Scholar]
- 31.Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, et al. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bashyam MD, Tyagi AK. Identification and analysis of “extended −10” promoters from mycobacteria. J Bacteriol. 1998;180:2568–2573. doi: 10.1128/jb.180.9.2568-2573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papavinasasundaram KG, Anderson C, Brooks PC, Thomas NA, Movahedzadeh F, et al. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology. 2001;147:3271–3279. doi: 10.1099/00221287-147-12-3271. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal N, Tyagi AK. Role of 5′-TGN-3′ motif in the interaction of mycobacterial RNA polymerase with a promoter of ‘extended −10’ class. FEMS Microbiol Lett. 2003;225:75–83. doi: 10.1016/S0378-1097(03)00483-X. [DOI] [PubMed] [Google Scholar]
- 35.Amin AG, Goude R, Shi L, Zhang J, Chatterjee D, et al. EmbA is an essential arabinosyltransferase in Mycobacterium tuberculosis. Microbiology. 2008;154:240–248. doi: 10.1099/mic.0.2007/012153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll P, Schreuder LJ, Muwanguzi-Karugaba J, Wiles S, Robertson BD, et al. Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS ONE. 2010;5:e9823. doi: 10.1371/journal.pone.0009823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fivian-Hughes AS, Davis EO. Analyzing the regulatory role of the HigA antitoxin within Mycobacterium tuberculosis. J Bacteriol. 2010;192:4348–4356. doi: 10.1128/JB.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Triccas JA, Britton WJ, Gicquel B. Isolation of strong expression signals of Mycobacterium tuberculosis. Microbiology. 2001;147:1253–1258. doi: 10.1099/00221287-147-5-1253. [DOI] [PubMed] [Google Scholar]
- 40.Agarwal N, Raghunand TR, Bishai WR. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein. Microbiology. 2006;152:2749–2756. doi: 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- 41.Chauhan S, Sharma D, Singh A, Surolia A, Tyagi JS. Comprehensive insights into Mycobacterium tuberculosis DevR (DosR) regulon activation switch. Nucl Ac Res. 2011;39:7400–7414. doi: 10.1093/nar/gkr375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucl Ac Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blokpoel MC, Murphy HN, O'Toole R, Wiles S, Runn ES, et al. Tetracycline-inducible gene regulation in mycobacteria. Nucl Ac Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forti F, Crosta A, Ghisotti D. Pristinamycin-inducible gene regulation in mycobacteria. J Biotechnol. 2009;140:270–277. doi: 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Carroll P, Faray-Kele MC, Parish T. Identifying vulnerable pathways in Mycobacterium tuberculosis by using a knock-down approach. Appl Environ Microbiol. 2011;77:5040–5043. doi: 10.1128/AEM.02880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll P, Muttucumaru DGN, Parish T. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl Environ Microbiol. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 48.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230. doi: 10.1099/00221287-100-2-221. [DOI] [PubMed] [Google Scholar]
- 50.Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, et al. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A. 2005;102:4548–4553. doi: 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurosu M, Crick DC. MenA is a promising drug target for developing novel lead molecules to combat Mycobacterium tuberculosis. Med Chem. 2009;5:197–207. doi: 10.2174/157340609787582882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honaker RW, Dhiman RK, Narayanasamy P, Crick DC, Voskuil MI. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J Bacteriol. 2010;192:6447–6455. doi: 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balazsi G, Heath AP, Shi L, Gennaro ML. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol Syst Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holsclaw CM, Sogi KM, Gilmore SA, Schelle MW, Leavell MD, et al. Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem Biol. 2008;3:619–624. doi: 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Ag Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dussurget O, Timm J, Gomez M, Gold B, Yu S, et al. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J Bacteriol. 1999;181:3402–3408. doi: 10.1128/jb.181.11.3402-3408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parish T, Stoker NG. Electroporation of mycobacteria. Methods Mol Biol. 1998;101:129–144. doi: 10.1385/0-89603-471-2:129. [DOI] [PubMed] [Google Scholar]
- 59.Miller JH, editor. Experiments in Molecular Genetics. Cold Spring Harbor: Cold Spring Harbor Press; 1972. [Google Scholar]