Abstract
Telomeric DNA and C-myc22 are DNA G-quadruplex (G4)-forming sequences associated with tumorigenesis. Ligands that can facilitate the formation and increase the stabilization of G4 can halt tumor cell proliferation and have been regarded as potential anti-cancer drugs. In the present study, we have investigated the interaction of 11 natural alkaloids with G4 formed by telomeric DNA and C-myc22 sequences. Our results indicated that sanguinarine (San), palmatine (Pal), and berberine (Beb) of the first series (S1) can induce the formation of G4 as well as increase the stabilization ability. Daurisoline (S2-1), O-methyldauricine (S2-2), O-diacetyldaurisoline (S2-3), daurinoline (S2-4), dauricinoline (S2-5), N,N′-dimethyldauricine iodide (S2-6), and N,N′-dimethyldaurisoline iodide (S2-7) of the second series (S2) showed similar stabilization ability. We found that unsaturated ring C, N+ positively charged centers, and conjugated aromatic rings are key factors to increase the stabilization ability of S1, and we gave some advice on structure modification to S2 through structure-activity study. Besides, we found San and Pal to be cell cycle blocker in G1. San was speculated to bind to G4 through intercalation or end stacking.
Introduction
DNA has been recognized to play a passive role in genetic information storage as well as an active role in biological processes. Specific regions of the genome can exist in forms other than the Watson-Crick duplex (Ren and Chaires, 1999). Millimolar concentrations of guanine alone were observed to form a gel in aqueous solution (BANG, 1910). The structure was determined to consist of π-stacked guanine quartets, each of which consisted of 4 guanines in a cyclic planar arrangement, known as G-quadruplex (G4, Table 2) (Gellert et al., 1962). G4 is regarded as an important drug-design target for the treatment of various human disorders. First, G4 forming sequences are prevalent in human genome, exist in many important regions of the eukaryotic genome, such as telomeres ends and the regulatory regions of many oncogenes (Eddy and Maizels, 2006), including proto-oncogene C-myc (Simonsson et al., 1998; Grand et al., 2002; Siddiqui-Jain et al., 2002) and oncogenes c-kit (Rankin et al., 2005; Fernando et al., 2006), V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRas) (Cogoi and Xodo, 2006), platelet-derived growth factor subunit A (PDGF-A) (Qin et al., 2007), vascular endothelial growth factor (VEGF) (Sun et al., 2005), and others (Kumar and Maiti, 2008; Wong et al., 2009). Second, G4 is stable in physiological conditions, believed to be a competitive structure compared with double stranded DNA (dsDNA) in vivo. G4 was observed to be stable enough to postpone or prevent the formation of dsDNA in an equimolar mixture of dAG3(T2AG3)3 and d(C3TA2) C3T at acidic pH (Manzini et al., 1994; Li et al., 2002). Additionally, B-form dsDNA was reported to interconvert to intermolecular G4 under certain cation concentrations (Miura and Thomas, 1994; Deng and Braunlin, 1995). Third, G4 has been shown experimentally to play various significant functional roles in cells (Wong et al., 2009). Finally, the conformations of G4 can provide some selective recognition sites for small molecules. It is clear that G4 is no longer just a biophysical oddity and must be given serious consideration as an important target for the treatment of various human disorders (Ou et al., 2008).
Table 2.
Abbreviation List
| Full Name | Abbreviation |
|---|---|
| G-quadruplex | G4 |
| Sanguinarine | San |
| Palmatine | Pal |
| Berberine | Beb |
| Tetrahydropalmatine | Tep |
| Daurisoline | S2-1 |
| O-methyldauricine | S2-2 |
| O-diacetyldaurisoline | S2-3 |
| Daurinoline | S2-4 |
| Dauricinoline | S2-5 |
| N,N′-dimethyldauricine iodide | S2-6 |
| N,N′-dimethyldaurisoline iodide | S2-7 |
| Triethylene tetraamine | TETA |
| The first series | S1 |
| The second series | S2 |
| The nuclease hypersensitivity element | NHE |
| Traditional Chinese medicine | TCM |
| Melting temperature | Tm |
| Change in Tm | ΔT |
| Induced circular dichroism | ICD |
| Flow cytometric assay | FCM |
| d(T2AG3) | Hum6 |
| d((T2AG3)4) | Hum24 |
| d(TGAGGGTGGGGAGGGTGGGGAA) | C-myc22 |
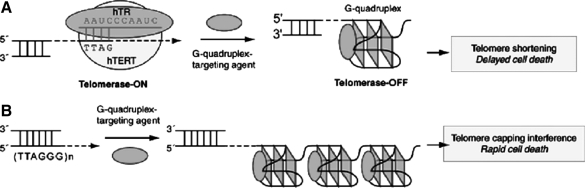
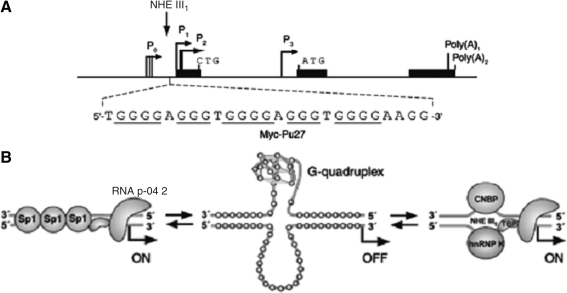
Considering the important role of G4, specific ligands that can facilitate the formation and stabilize G4 structures are regarded as potential drugs, in particular as inhibitors of telomerase activity and oncogene expression (Zahler et al., 1991; FRANCESCHIN, 2009). Telomerase activity has been identified in most human tumors (85%∼90%) but is almost undetectable in the majority of somatic cells, suggesting its close relationship with the degree of malignancy and the likelihood of tumor progression (Naasani et al., 1999). G4-targeting compounds have been shown to inhibit telomerase activity, thereby making the telomeric DNA G4 an attractive target for cancer therapeutic intervention (Fig. 1. A). Moreover, G4-targeting compounds have been shown to disrupt telomere capping and induce rapid apoptosis (Fig. 1. B) (Yang and Okamoto, 2010). Furthermore, the formation of G4 can also suppress the expression of oncogenes, such as C-myc. The aberrant overexpression of proto-oncogene C-myc is related to the increasing of cellular proliferation in a variety of different malignant tumors (Tian et al., 2010). C-myc contains parallel-stranded G4 in the nuclease hypersensitivity element III1 upstream of the P1 and P2 promoters (Wang et al., 2008) which can regulate transcription (Fig. 2) (Seenisamy et al., 2004). G4-ligands can regulate C-myc transcription and are speculated to halt tumor cell proliferation potentially.
FIG. 1.
The binding of ligands with G-quadruplex (G4) inhibit the formation of cancer. (A) Mechanism of telomerase inhibition by G4 ligands. (B) Mechanism of drug-mediated interference of telomere capping by G4 ligands (Yang and Okamoto, 2010). hTR, RNA component of human telomerase; hTERT, telomerase reverse transcriptase.
FIG. 2.
The formation of G4 can prevent the expression of c-myc. (A) The promoter structure of the human c-myc gene. (B) Alternative forms of the nuclease hypersensitivity element III1 (NHE III1) (Table 2) of the c-myc promoter associated with transcriptional activation or silencing (Yang and Okamoto, 2010).
Since the first report on the interaction between small molecules and G4 DNA in 1997 (Wang et al., 2010), many leading compounds that target G4 have been found, including anthraquinones (Sun et al., 1997; Perry et al., 1998a, 1998b), cationic porphyrins (Anantha et al., 1998; Han et al., 2001), perylene (Rossetti et al., 2002), benzoindoloquinolines (Alberti et al., 2002), ethidium derivatives (Koeppel et al., 2001), acridine derivatives (Harrison et al., 1999), piperazines (Riou et al., 2001), pentacyclicacridinium salts (Gowan et al., 2001), fluoroquinophenoxazines (Duan et al., 2001), and others. Most of these compounds have the same features: (1) a π-delocalized system that is able to stack on the face of a guanine quartet, (2) a partial positive charge that is able to lay in the center of the quartet, increasing stabilization by substituting the cationic charge of the potassium or sodium that would normally occupy that site, and (3) positively charged substituents to interact with the grooves and loops of G4 and the negatively charged backbone phosphates (Reed et al., 2006; Yang et al., 2011). In accordance with these desirable features, there are 3 interaction modes of G4-ligands and G4: (1) external stacking: ligands stack on the terminal G-quarter through π-π accumulation; (2) intercalating: ligands insert into the space of two G-quarters; and (3) groove binding: ligands bind to the grooves or loops of the G4. Notably, a few of the ligands have already entered preclinical or clinical trials, among which quarfloxin (CX-3543) has entered phase 2 clinical trials (Bates et al., 2007). Considering the importance of G4 and its ligands, our team has carried out a series of research in this area (Sun et al., 2006, 2007, 2009; Zhou et al., 2008b, 2009; Li et al., 2009; Yang et al., 2009, 2010a, 2010b, 2010c; Tian et al., 2010; Yang and Okamoto, 2010; Zhang et al., 2010; Ji et al., 2011a, 2011b).
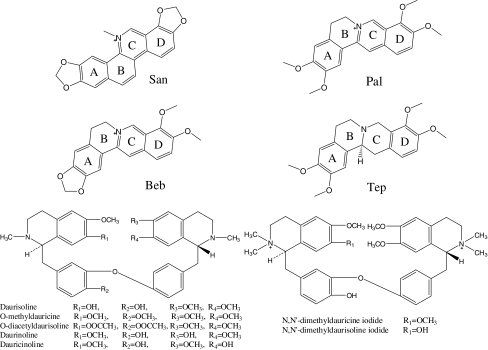
In the long term, natural products from traditional Chinese medicine (TCM) (Table 2) can contribute to the development of molecular target-guided therapies and individualized treatment strategies (Efferth et al., 2007) tested as a molecular library for seeking new drugs (Liu et al., 2010). With rapidly increasing export rates of TCM products to Europe and the United States, scientists in the western world show tremendous interest in TCM. Alkaloids, which represent one important class of active compounds in TCM, have various biological activities. A large number of natural alkaloids can form molecular complexes with nucleic acid structures (Maiti and Kumar, 2007). Studies on the interactions between alkaloids and DNA are necessary, since such interactions may not only provide the molecular basis for better understanding their bioactivity mechanisms, but also guide the rational design of more efficient DNA-binding molecules for cancer therapy (Wang et al., 2008). In this paper, we have investigated the interactions of 11 alkaloids originating from Chinese herbal medicine with G4 formed by human telomeric DNA and C-myc22. The first series (S1) (Table 2) contains 4 alkaloids with similar structures, Sanguinarine (San), Palmatine (Pal), Berberine (Beb), and tetrahydropalmatine (Tep) (Fig. 3, Table 2). Alkaloids of S1 exhibit a wide range of pharmacological effects (Ghosh et al., 1985; Schmeller et al., 1997; Wu et al., 1999; Adhami et al., 2004), including anti-cancer activity (Zhao et al., 1991; Ahmad et al., 2000), which is believed to be related to wide biological activities; their interactions with G4 is believed to be one of the most important activities. San (Bai et al., 2008), Beb (Ren and Chaires, 1999; Zhou et al., 2008a), and Pal (Zhou et al., 2008a) are known to bind to G4 structure. In addition, Beb can inhibit telomere elongation (Naasani et al., 1999). Tep has a similar structure to these alkaloids and is speculated to have similar function. To our knowledge, there is no report to give a comparison of their stabilization ability. We arranged them in a series, trying to find some regularities in structure activity. The second series (S2) (Table 2) contains 7 alkaloids: daurisoline (S2-1), O-methyldauricine (S2-2), O-diacetyldaurisoline (S2-3), daurinoline (S2-4), dauricinoline (S2-5), N,N′-dimethyldauricine iodide (S2-6), and N,N′-dimethyldaurisolineiodide (S2-7) (Fig. 3, Table 2). S2 has a more complex structure than S1, similar to bis-benzyltetrahydroisoquinoline alkaloids, which we reported to be G4-ligands (Ji et al., 2011a). So far, there is no report on the interaction of S2 with G4.
FIG. 3.
The structure of 11 natural alkaloids. Alkaloids in the first series (S1) and second series (S2) have similar structure with some tiny differences.
In this article we have investigated the interactions of 11 alkaloids with telomeric DNA G4 and C-myc22 G4, trying to get some regularities on structure activity through comparing the stabilization ability of S1 and S2, to find the potential binding mode as well as the biological function. Our results demonstrated that all of the alkaloids except Tep can stabilize G4. We found that unsaturated ring C and N+ positively charged centers connecting ring B and C help to build better planar structure and larger a conjugated system and to increase the stabilization ability. We also gave some advice on structure modification to S2. Besides Beb, San and Pal can induce the formation of C-myc22 G4 and Hum24 G4. San and Pal proved to be cell cycle blockers in gap 1 phase (G1) in flow cytometric assay (FCM) (Table 2). In addition, the binding mode of San was speculated to be intercalation or end stacking.
Materials and Methods
Materials
DNA samples d(T2AG3) (Hum6), d((T2AG3)4) (Hum24), and d(TGAGGGTGGGGAGGGTGGGGAA) (C-myc22) (Table 2) were purchased from Invitrogen (Beijing, China), purified by polyacrylamide gel electrophoresis. All of the alkaloids were purchased from National Institutes for Food and Drug Control. PI and ribonuclease were purchased from Sigma-Aldrich. Tris(hydroxymethyl)aminomethane (Tris) (D11, 98%) was purchased from Cambridge Isotope Laboratories, Inc. Analytical grade inorganic salts were purchased from Sinopharm Chemical Reagent Beijing Co. Trypsinase was purchased from AMRESCD.
Cell line
The adenocarcinomic human alveolar basal epithelial cells (A549) were provided by College of Life Sciences, Beijing Normal University. Cell lines were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (Beijing YuanHeng ShengMa Biology Technology Research Institute, China) in the presence of 5% CO2 at 37°C.
Sample preparation
DNA Samples (Hum24, Hum6, and C-myc22) were prepared according to references (Lu et al., 1993; Phan and Patel 2003). The buffer solution for circular dichroism (CD) spectroscopy was Tris-HCl; for melting-CD Tris-HCl-KCl (150mM K+, 1mM EDTA) (pH=7.4) was used. The alkaloids were dissolved in dimethyl sulfoxide/water mixture. The measured samples were prepared by mixing a quantity of alkaloids and DNA. For the CD spectroscopy experiment, the concentrations of Hum24 and C-myc22 were 2.5μM and 3μM, respectively. For melting-CD, the concentrations of Hum6 and Hum24 were 35μM and 3.5μM, respectively. The sample solutions were equilibrated at 4°C for more than 12 hours before measurements.
CD spectroscopy
In melting-CD spectroscopy, CD spectra were collected from 220 to 350 nm. Each spectrum was the average of two scans. In CD Spectroscopy of room temperature, CD spectra were collected from 200 to 350 nm on a Jasco-815 automatic recording spectropolarimeter with a 1-cm pathlength quartz cell at 25°C. Each spectrum was the average of three scans. A buffer blank correction was made for all spectra. The temperature of the cell holder was regulated by a Jasco PTC-423S temperature controller, and the cuvette-holding chamber was flushed with a constant stream of dry N2 gas to avoid water condensation on the cuvette exterior. Melting curves of the G4 were obtained by recording the CD intensity at 262 nm. The heating rate was 2.0°C per minute. The condition of induced CD was the same as CD spectroscopy.
Molecular modeling simulations
Molecular modeling simulations were performed according to a method described previously (Luu et al., 2006; Ji et al., 2011a). All the molecular modeling works and simulations were performed using the Insight II 2005 software package on a Dell Precision T5400 workstation under CHARMM force field.
Flow cytometric assay
The effects of San, Pal, S2-1, S2-4, S2-5, S2-6, and S2-7 on the cell cycle were assessed by FCM by measuring the percentage of cells in G1, synthesis (S), and gap 2 (G2) phases, with and without treatment with the investigational compounds. The FCM evaluation of the cell cycle status and apoptosis was performed according to a method described previously (Wang et al., 2007; Ji et al., 2011a).
Results and Discussion
G4 stabilization ability
The stability of G4 is quite an important property for alkaloids to exert telomerase inhibition ability. We applied melting-CD to monitor the melting temperature (Tm) (Table 2) of induced G4 formed by Hum6 and Hum24 at 262 nm to discuss the stabilities. The change in Tm (ΔT) (Table 2) in the folded and unfolded G4 structures upon interacting with the ligands provides evidence of thermal stabilization of the DNA structure (Yang et al., 2011). We found an obvious increase in Tm with the presence of S1 except Tep (Supplementary Fig. 1; Supplementary Data are available online at www.liebertonline.com/nat).
The change in temperature (ΔT) of alkaloids in S1 turns out to be in an order as San > Pal > Beb > Tep to both Hum6 and Hum24 (Table 1). Higher ΔT stands for better stabilization ability. This may be the result of the slight difference in their structure (Fig. 3). We analyzed the structure–activity relationship and reached some conclusions. First, unsaturation of ring C is critical for the DNA-binding property. The adjacent aromatic rings C and D conjugate π electrons to form a planar structure, which is necessary for the intercalative binding to the DNA (Wang et al., 2008). We can see that all of the alkaloids of S1 showing stabilization ability have an unsaturated ring C. Second, the conjugated aromatic rings and the positively charged centers N+ connecting rings B and C may be critical for the DNA-binding property. Tep, which showed no stabilization ability, has no conjugated aromatic rings. The N of Tep has a different electron structure from the N+-containing alkaloids. Third, the existing of chiral atom may decrease the planarity and in turn decrease stabilization ability. Tep has a chiral carbon that connects rings B and C with 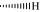 , this conformation can decrease the planarity and increase the steric hindrance. Fourth, previous study revealed that a larger number of benzo[1,3]dioxole groups of ligands increase the ability to interact with G4 (Yang et al., 2011). According to our results, it is not the number of the benzo[1,3]dioxole groups that affect the interaction with G4, but whether the group can help built a good planar structure (Supplementary Fig. 2). In our experiments, Pal, which has no benzo[1,3]dioxole group, showed a better stabilization ability than Beb, which has a benzo[1,3]dioxole group. Pal is also a stronger G4 stabilizer than Beb to d(TTGGGTT)4 (Zhou et al., 2008a). All in all, the factors that can help build a larger conjugated system and a better planar structure can increase the binding ability. Besides, the stabilization ability of S1 to Hum6 and Hum24 was in a similar order. It seemed that S1 might bind to sites that have a similar steric hindrance in Hum6 and Hum24, such as Plane2 or Site6, instead of loops or groups.
, this conformation can decrease the planarity and increase the steric hindrance. Fourth, previous study revealed that a larger number of benzo[1,3]dioxole groups of ligands increase the ability to interact with G4 (Yang et al., 2011). According to our results, it is not the number of the benzo[1,3]dioxole groups that affect the interaction with G4, but whether the group can help built a good planar structure (Supplementary Fig. 2). In our experiments, Pal, which has no benzo[1,3]dioxole group, showed a better stabilization ability than Beb, which has a benzo[1,3]dioxole group. Pal is also a stronger G4 stabilizer than Beb to d(TTGGGTT)4 (Zhou et al., 2008a). All in all, the factors that can help build a larger conjugated system and a better planar structure can increase the binding ability. Besides, the stabilization ability of S1 to Hum6 and Hum24 was in a similar order. It seemed that S1 might bind to sites that have a similar steric hindrance in Hum6 and Hum24, such as Plane2 or Site6, instead of loops or groups.
Table 1.
Change in Temperature of Hum6 and Hum24 Guanine-quadruplexes with First Series Alkaloids by Melting-Circular Dichroism
| Alkaloids | Hum6 (°C) | Hum24 (°C) |
|---|---|---|
| San | 12 | 20 |
| Pal | 10 | 11 |
| Beb | 8 | 6 |
| Tep | 0 | 0 |
Beb, berberine; Pal, palmatine; San, sanguinarine; Tep, tetrahydropalmatine.
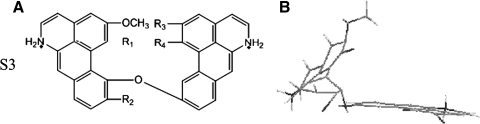
The alkaloids in S2 have a similar parent structure with some tiny difference in the side chains (Fig. 3). In our experiments, alkaloids in S2 had similar ΔT of ∼6°C, regardless of the difference in the side chains (Supplementary Fig. 3). It seemed that the functional part of S2 was their parent structure. To small molecules with π-aromatic surfaces of fewer than 2 rings, neither the position of the positively charged centers nor the substitutions could adjust their ability to interact with G4 (Yang et al., 2011). From the structure of S2, we can see that they have no more than 2 consecutive rings, but linked by some loops; this may be the reason that the substituents do not affect the ability to interact with G4 so much. If we change the parent structure of S2 to a structure with more than 2 consecutive rings, like third series (S3) (Fig. 4), from the 3-dimensional structure of thrid series (S3) we can see that it is an L-shaped structure with a larger conjugated system, which may result in a better G4 stabilization ability. However, S3 is not synthesized and our hypothesis needs further experiment to test and verify. Moreover, some substituents such as -OCH3 can still show influence on the stabilization ability, 2 alkaloids in S2 with 4 or 5 -OCH3 groups had relatively higher stabilization ability. The -OOCH3 group may form hydrogen bonds with hydroxyl or amine groups of G4, making alkaloids bind to G4 more tightly.
FIG. 4.
The chemical structure (A) and the 3-dimensional structure (B) of third series alkaloid (S3).
G4-inducing ability
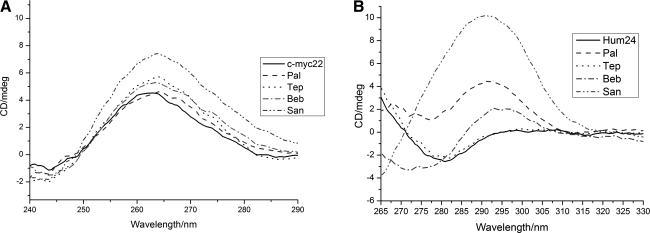
CD Spectroscopy has been extensively applied to study secondary structures of DNA. Different DNA structures have distinct CD spectral features, which can be used to recognize different DNA motifs conveniently (Yang et al., 2010c, 2011). We applied CD to investigate the G4-inducing ability of S1. The DNA sequences we used were C-myc22 and Hum24. To C-myc22, we can see a band shift from ∼262nm to ∼264nm (Fig. 5 A), suggesting S1 induces C-myc22 to form parallel G4. To Hum24, there was an obvious positive peak at ∼295nm after adding San, Pal, and Beb, suggesting the formation of antiparallel G4 (Fig. 5 B), while Tep cannot induce the formation of Hum24 G4. Considering that C-myc22 is known to be very stable, with the Tm as high as 85°C (Ambrus et al., 2005) and parallel G4 seems to be a preponderant structure in adverse situation (Ji et al., 2011a), we speculated that C-myc22 G4 may be easier to form than other G4, like Hum24 G4.
FIG. 5.
(A) The CD spectra of c-myc22 with Pal, Tep, Beb, and San. The concentration of c-myc22 and the alkaloids are 3μm and 10μm, respectively. (B) The CD spectra of Hum24 with Pal, Tep, Beb, and San. The concentration of Hum24 and the alkaloids are 2.5μm and 10μm, respectively. CD, circular dichroism.
Biological function
It is known that the in vivo environment is complex, and that G4 DNA is a dynamic structure, making it necessary and a particular challenge to study the biological functions. Previous studies showed that telomerase activity varies during the cell cycle (Zhu et al., 1996; Izbicka et al., 1999). Cells actively undergoing the cell cycle are targeted in cancer therapy, as the DNA is relatively exposed during cell division and hence susceptible to damage by drugs or radiation (THOMPSON, 1995; Schmitt and Lowe, 1999). We applied FCM to determine the effects of the alkaloids on the cell cycle.
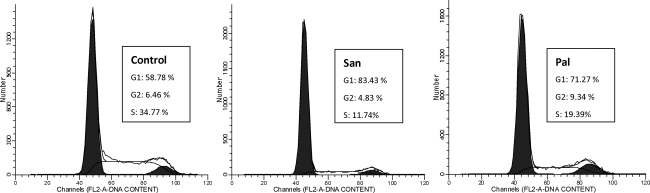
Our results showed that San and Pal can terminate the cell cycle in G1 (Fig. 6). Previous study on telomerase reported that as the cell progresses through the cell cycle, maximum telomerase activity was detected in S phase (Zhu et al., 1996). In our experiments, San and Pal seemed to inhibit the telomerase activity effectively by terminating the cell cycle enter S phase. We speculated that the 3 alkaloids induce differentiation or quiescence in A549 cells by interacting with G4 and in turn resulted in the telomerase repression instead of killing cells (Bestilny et al., 1996; Yin et al., 2004), as they functioned in a similar way compared to triethylene tetraamine (Table 2) (Bestilny et al., 1996; Yin et al., 2004) and bis-benzyltetrahydroisoquinoline alkaloids we reported in another paper (Ji et al., 2011a). Moreover, S2-1, S2-4, and S2-5 cannot terminate the cell cycle or only had weak ability to terminate it. S2-6 and S2-7 seemed to terminate cell cycle in S, with the percentage of G2 much lower than the control (Supplementary Fig. 4). In addition, the alkaloids of S2 may function through a different process instead of inhibiting the activation of telomerase. Compared with S2, San and Pal were more effective G4-ligands as well as potent anti-cancer lead compounds.
FIG. 6.
Results of flow cytometric assay. Cells in the control group (left) were not treated with drugs; cells in the experimental group were treated with San (middle) and Pal (right).
Binding mode
We have proved that San, Pal, and Beb can interact with G4. In order to confirm the action patterns of the three alkaloids with G4, we applied computer simulation and induced CD. In the molecular docking, the receptor model was G4 formed by the d[TTGGG (TTAGGG)3A] sequence, and according to the external character of telomere G4, it can be divided into 3 planes, 3 loops, and a site (Luu et al., 2006). Combining the results of melting-CD and molecular docking, we inferred the binding site to be Plane2 or Site6, which have a similar steric hindrance in Hum6 and Hum24. San showed the highest score, in accordance with its high stabilization ability (Supplementary Table 1; Supplementary Data are available online at www.liebertonline.com/nat), appeared to be a potentially effective G4-ligand. So we carried out induced circular dichroism (ICD), (Table 2) to further investigate the binding mode of San and G4.
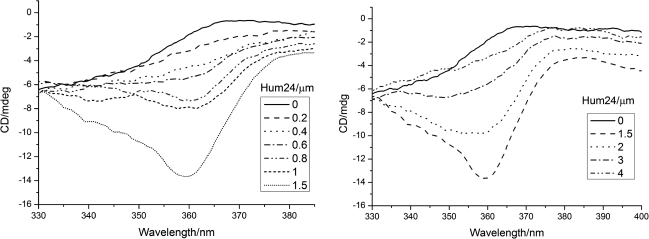
As far as we know, ligands generally interact with G4 structure through 3 modes: intercalation, groove binding, and end stacking. ICD can be used to detect the binding mode, since the molecular frame of ligands would be twisted and give rise to CD signals when the molecule bound to the G4 structure (Yang et al., 2010c). We applied ICD to investigate the signal of San to Hum24-G4 in 300∼600nm, since both Hum24 and San in a free state did not have CD signal in the region >300nm. We observed a weak negative peak at around 360nm with the presence of San and Hum24 (Fig. 7). It is reported that the binding mode could be intercalation or end stacking if there is a weak ICD signal (Sehlstedt et al., 1994). The result suggested San to bind to G4 through intercalation or end stacking. Moreover, we found that when the concentration of hum24 [Hum24] was under 1.5μm, the negative peak increased with the increase of [Hum24], while above 1.5μm, the negative peak began to decrease with the increase of [Hum24] (Fig. 7). It seemed that the saturation concentration for San(8μm) was ∼1.5μm. Besides, San is also similar to the shape of a G-quartet and has the central cationic aromatic cores, which suggest the ability to interact with G4 might be attributable to π-π stacking.
FIG. 7.
The result of induced CD. The concentration of San used was 8μm. The concentration of Hum24 was 0, 0.2, 0.4, 0.6, 0.8, 1, and 1.5 and 1, 1.5, 2, 3, and 4μm, respectively.
Conclusion
In the present study, we have proved San, Pal, and Beb to have the ability to induce the formation and increase the stabilization of G4. Factors like unsaturated ring C and positively charged centers N+ can enlarge the conjugated system and increase the ability to interact with G4. Subsequently, FCM results suggested that San and Pal function through interacting with G4 to induce cell differentiation or quiescence. In addition, San showed the highest stabilization ability, got the highest score in molecule docking, and displayed a better ability to terminate the cell cycle than did Pal. Through ICD, the binding mode of San was speculated to be intercalation or end stacking. Alkaloids in S2 showed similar stabilization ability to G4, suggesting the function part of S2 to be their parent structure. We also gave some advice on structure modification for building a larger conjugated system. However, further study is needed to test whether the new structures have better ability to interact with G4.
Supplementary Material
Acknowledgments
The work was support by the Chinese National Natural Science Foundation (No. 30973869) and the National Natural Science Foundation of China (Grant No. 81072576).
Author Disclosure Statement
No competing financial interests exist.
References
- ADHAMI V.M. AZIZ M.H. REAGAN-SHAW S.R. NIHAL M. MUKHTAR H. AHMAD N. Sanguinarine causes cell cycle blockade and apoptosis of human prostate carcinoma cells via modulation of cyclin kinase inhibitor-cyclin-cyclin-dependent kinase machinery. Mol. Cancer Ther. 2004;3:933–940. [PubMed] [Google Scholar]
- AHMAD N. GUPTA S. HUSAIN M.M. HEISKANEN K.M. MUKHTAR H. Differential antiproliferative and apoptotic response of sanguinarine for cancer cells versus normal cells. Clin. Cancer Res. 2000;6:1524–1528. [PubMed] [Google Scholar]
- ALBERTI P. SCHMITT P. NGUYEN C.H. RIVALLE C. HOARAU M. Benzoindoloquinolines interact with DNA tetraplexes and inhibit telomerase. Bioorg. Med. Chem. Lett. 2002;12:1071–1074. doi: 10.1016/s0960-894x(02)00080-x. [DOI] [PubMed] [Google Scholar]
- AMBRUS A. CHEN D. DAI J. JONES R.A. YANG D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- ANANTHA N.V. AZAM M. SHEARDY R.D. Porphyrin binding to quadruplexed T4G4. Biochemistry. 1998;37:2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- BAI L.P. HAGIHARA M. JIANG Z.H. NAKATANI K. Ligand binding to tandem G quadruplexes from human telomeric DNA. ChemBioChem. 2008;9:2583–2587. doi: 10.1002/cbic.200800256. [DOI] [PubMed] [Google Scholar]
- BANG I. Untersuchungen über die Guanylsäre. Biochemisce Zeitschrift. 1910;26:293–311. [Google Scholar]
- BATES P. MERGNY J.L. YANG D. Quartets in G-major. EMBO rep. 2007;8:1003–1010. doi: 10.1038/sj.embor.7401073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BESTILNY L.J. BROWN C.B. MIURA Y. ROBERTSON L.D. RIABOWOL K.T. Selective inhibition of telomerase activity during terminal differentiation of immortal cell lines. Cancer Res. 1996;56:3796–3802. [PubMed] [Google Scholar]
- COGOI S. XODO L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENG H. BRAUNLIN W.H. Duplex to quadruplex equilibrium of the self‐complementary oligonucleotide d (GGGGCCCC) Biopolymers. 1995;35:677–681. doi: 10.1002/bip.360350613. [DOI] [PubMed] [Google Scholar]
- DUAN W. RANGAN A. VANKAYALAPATI H. KIM M.Y. ZENG Q. SUN D. HAN H. FEDOROFF O.Y. NISHIOKA D. RHA S.Y. Design and Synthesis of Fluoroquinophenoxazines That Interact with Human Telomeric G-quadruplexes and their biological effects. Mol. Cancer Ther. 2001;1:103–120. [PubMed] [Google Scholar]
- EDDY J. MAIZELS N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFFERTH T. FU Y. ZU Y. SCHWARZ G. KONKIMALLA V.S.B. WINK M. Molecular target-guided tumor therapy with natural products derived from traditional Chinese medicine. Curr. Med. Chem. 2007;14:2024–2032. doi: 10.2174/092986707781368441. [DOI] [PubMed] [Google Scholar]
- FERNANDO H. RESZKA A.P. HUPPERT J. LADAME S. RANKIN S. VENKITARAMAN A.R. NEIDLE S. BALASUBRAMANIAN S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANCESCHIN M. G-quadruplex DNA structures and organic chemistry: more than one connection. European J. Org. Chem. 2009;2009:2225–2238. [Google Scholar]
- GELLERT M. LIPSETT M.N. DAVIES D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHOSH A.K. BHATTACHARYYA F.K. GHOSH D.K. Leishmania donovani: Amastigote inhibition and mode of actior of berberine. Exp. Par. 1985;60:404–413. doi: 10.1016/0014-4894(85)90047-5. [DOI] [PubMed] [Google Scholar]
- GOWAN S. BRUNTON L. VALENTI M. HEALD R. READ M. HARRISON J. STEVENS M. NEIDLE S. KELLAND L. Preclinical antitumor properties of G-quadruplex-interactive small molecule inhibitors of telomerase. Proc. Am. Assoc. Cancer Res. 2001;42 abstract 466. [Google Scholar]
- GRAND C.L. HAN H. MU OZ R.M. WEITMAN S. VON HOFF D.D. HURLEY L.H. BEARSS D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- HAN H. LANGLEY D.R. RANGAN A. HURLEY L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- HARRISON R.J. GOWAN S.M. KELLAND L.R. NEIDLE S. Human telomerase inhibition by substituted acridine derivatives. Bioorg. Med. Chem. Lett. 1999;9:2463–2468. doi: 10.1016/s0960-894x(99)00394-7. [DOI] [PubMed] [Google Scholar]
- IZBICKA E. WHEELHOUSE R.T. RAYMOND E. DAVIDSON K.K. LAWRENCE R.A. SUN D. WINDLE B.E. HURLEY L.H. VON HOFF D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- JI X. CHEN J. SUN H. ZHOU H. XIANG J. PENG A. TANG Y. ZHAO C. The interaction of telomere DNA G-quadruplex with three bis-benzyltetrahydroisoquinoline alkaloids. Nucleic Acid Ther. 2011a;21:415–422. doi: 10.1089/nat.2011.0311. [DOI] [PubMed] [Google Scholar]
- JI X. SUN H. ZHOU H. XIANG J. TANG Y. ZHAO C. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011b;21:185–200. doi: 10.1089/nat.2010.0272. [DOI] [PubMed] [Google Scholar]
- KOEPPEL F. RIOU J.F. LAOUI A. MAILLIET P. ARIMONDO P.B. LABIT D. PETITGENET O. HÉLÈNE C. MERGNY J.L. Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res. 2001;29:1087–1896. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUMAR N. MAITI S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 2008;36:5610–5622. doi: 10.1093/nar/gkn543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Q. XIANG J. LI X. CHEN L. XU X. TANG Y. ZHOU Q. LI L. ZHANG H. SUN H. Stabilizing parallel G-quadruplex DNA by a new class of ligands: two non-planar alkaloids through interaction in lateral grooves. Biochimie. 2009;91:811–819. doi: 10.1016/j.biochi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- LI W. WU P. OHMICHI T. SUGIMOTO N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- LIU Y. ZHENG B. XU X. YUAN G. Probing the binding affinity of small-molecule natural products to the G-quadruplex in C-myc oncogene by electrospray ionization mass spectrometry. Rapid Comm. Mass Spectrom. 2010;24:3072–3075. doi: 10.1002/rcm.4730. [DOI] [PubMed] [Google Scholar]
- LU M. GUO Q. KALLENBACH N.R. Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry. 1993;32:598–601. doi: 10.1021/bi00053a027. [DOI] [PubMed] [Google Scholar]
- LUU K.N. PHAN A.T. KURYAVYI V. LACROIX L. PATEL D.J. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAITI M. KUMAR G.S. Molecular aspects on the interaction of protoberberine, benzophenanthridine, and aristolochia group of alkaloids with nucleic acid structures and biological perspectives. Med. Res. Rev. 2007;27:649–695. doi: 10.1002/med.20087. [DOI] [PubMed] [Google Scholar]
- MANZINI G. YATHINDRA N. XODO L. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 1994;22:4634–4640. doi: 10.1093/nar/22.22.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIURA T. THOMAS G.J., JR Structural polymorphism of telomere DNA: interquadruplex and duplex-quadruplex conversions probed by Raman spectroscopy. Biochemistry. 1994;33:7848–7856. doi: 10.1021/bi00191a012. [DOI] [PubMed] [Google Scholar]
- NAASANI I. SEIMIYA H. YAMORI T. TSURUO T. FJ5002: a potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res. 1999;59:4004–4011. [PubMed] [Google Scholar]
- OU T. LU Y. TAN J. HUANG Z. WONG K.Y. GU L. G-quadruplexes: targets in anticancer drug design. ChemMedChem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- PERRY P.J. GOWAN S.M. RESZKA A.P. POLUCCI P. JENKINS T.C. KELLAND L.R. NEIDLE S. 1, 4-and 2, 6-disubstituted amidoanthracene-9, 10-dione derivatives as inhibitors of human telomerase. J. Med. Chem. 1998a;41:3253–3260. doi: 10.1021/jm9801105. [DOI] [PubMed] [Google Scholar]
- PERRY P.J. RESZKA A.P. WOOD A.A. READ M.A. GOWAN S.M. DOSANJH H.S. TRENT J.O. JENKINS T.C. KELLAND L.R. NEIDLE S. Human telomerase inhibition by regioisomeric disubstituted amidoanthracene-9, 10-diones. J. Med. Chem. 1998b;41:4873–4884. doi: 10.1021/jm981067o. [DOI] [PubMed] [Google Scholar]
- PHAN A.T. PATEL D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIN Y. REZLER E.M. GOKHALE V. SUN D. HURLEY L.H. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RANKIN S. RESZKA A.P. HUPPERT J. ZLOH M. GARY N. TODD A.K. LADAME S. BALASUBRAMANIAN S. NEIDLE S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REED J.E. ARNAL A.A. NEIDLE S. VILAR R. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel (II) complexes. J. Am. Chem. Soc. 2006;128:5992–5993. doi: 10.1021/ja058509n. [DOI] [PubMed] [Google Scholar]
- REN J. CHAIRES J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- RIOU J. MAILLIET P. LAOUI A. RENOU E. PETIGENET O. GUITTAT L. MERGNY J. Apoptosis, cell senescence, and telomere shortening induced by a new series of specific G-quadruplex DNA ligand. 2001. p. 837. [DOI] [PMC free article] [PubMed]
- ROSSETTI L. FRANCESCHIN M. BIANCO A. ORTAGGI G. SAVINO M. Perylene diimides with different side chains are selective in inducing different G-quadruplex DNA structures and in inhibiting telomerase. Bioorg. Med. Chem. Lett. 2002;12:2527–2533. doi: 10.1016/s0960-894x(02)00504-8. [DOI] [PubMed] [Google Scholar]
- SCHMELLER T. LATZ-BRÜNING B. WINK M. Biochemical activities of berberine, palmatine and sanguinarine mediating chemical defence against microorganisms and herbivores. Phytochemistry. 1997;44:257–266. doi: 10.1016/s0031-9422(96)00545-6. [DOI] [PubMed] [Google Scholar]
- SCHMITT C.A. LOWE S.W. Apoptosis and therapy. J. Pathol. 1999;187:127–137. doi: 10.1002/(SICI)1096-9896(199901)187:1<127::AID-PATH251>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- SEENISAMY J. REZLER E.M. POWELL T.J. TYE D. GOKHALE V. JOSHI C.S. SIDDIQUI-JAIN A. HURLEY L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- SEHLSTEDT U. KIM S.K. CARTER P. GOODISMAN J. VOLLANO J.F. NORDEN B. DABROWIAK J.C. Interaction of cationic porphyrins with DNA. Biochemistry. 1994;33:417–426. doi: 10.1021/bi00168a005. [DOI] [PubMed] [Google Scholar]
- SIDDIQUI-JAIN A. GRAND C.L. BEARSS D.J. HURLEY L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMONSSON T. KUBISTA M. PECINKA P. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN D. GUO K. RUSCHE J.J. HURLEY L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN D. THOMPSON B. CATHERS B.E. SALAZAR M. KERWIN S.M. TRENT J.O. JENKINS T.C. NEIDLE S. HURLEY L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- SUN H. TANG Y. XIANG J. XU G. ZHANG Y. ZHANG H. XU L. Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg. Med. Chem. Lett. 2006;16:3586–3589. doi: 10.1016/j.bmcl.2006.03.087. [DOI] [PubMed] [Google Scholar]
- SUN H. XIANG J. TANG Y. XU G. Regulation and recognization of the extended G-quadruplex by rutin. Biochem. Biophys. Res. Commun. 2007;352:942–946. doi: 10.1016/j.bbrc.2006.11.125. [DOI] [PubMed] [Google Scholar]
- SUN H. XIANG J. ZHOU Q. YANG Q. XU G. TANG Y. Temperature-sensitive supramolecules self-assembled by G-quadruplex DNA. Int. J. Biol. Macromol. 2010;46:123–125. doi: 10.1016/j.ijbiomac.2009.10.005. [DOI] [PubMed] [Google Scholar]
- SUN H. ZHOU Q. XIANG J. TANG Y. Polyethylenimine effectively induces, stabilizes, and regulates intramolecular G-quadruplexes. Bioorg. Med. Chem. Lett. 2009;19:4669–4672. doi: 10.1016/j.bmcl.2009.06.082. [DOI] [PubMed] [Google Scholar]
- THOMPSON C.B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- TIAN M. ZHANG X. LI Y. JU Y. XIANG J. ZHAO C. TANG Y. Inducement of G-quadruplex DNA forming and down-regulation of oncogene C-MYC by bile acid-amino acid conjugate-BAA. Nucleosides Nucleotides Nucleic Acids. 2010;29:190–199. doi: 10.1080/15257771003704875. [DOI] [PubMed] [Google Scholar]
- WANG G. ZHANG J. Lü Q. XU R. DONG Q. Berbamine induces apoptosis in human hepatoma cell line SMMC7721 by loss in mitochondrial transmembrane potential and caspase activation. J. Zhejiang Univ. Sci. B. 2007;8:248–255. doi: 10.1631/jzus.2007.B0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG P. LEUNG C.H. MA D.L. YAN S.C. CHE C.M. Structure-based design of platinum (II) complexes as c-myc oncogene down-regulators and luminescent probes for G-quadruplex DNA. Chemistry. 2010;16:6900–6911. doi: 10.1002/chem.201000167. [DOI] [PubMed] [Google Scholar]
- WANG Z. GUO X. LIU Z. CUI M. SONG F. LIU S. Studies on alkaloids binding to GC-rich human survivin promoter DNA using positive and negative ion electrospray ionization mass spectrometry. J. Mass Spec. 2008;43:327–335. doi: 10.1002/jms.1320. [DOI] [PubMed] [Google Scholar]
- WONG H.M. PAYET L. HUPPERT J.L. Function and targeting of G-quadruplexes. Curr. Opin. Mol. Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- WU H.L. HSU C.Y. LIU W.H. YUNG B.Y.M. Berberine-induced apoptosis of human leukemia HL-60 cells is associated with down-regulation of nucleophosmin/B23 and telomerase activity. Int. J. Cancer. 1999;81:923–929. doi: 10.1002/(sici)1097-0215(19990611)81:6<923::aid-ijc14>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- YANG D. OKAMOTO K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Q. XIANG J. YANG S. LI Q. ZHOU Q. GUAN A. ZHANG X. ZHANG H. TANG Y. XU G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: II. The binding characterization with specific intramolecular G-quadruplex and the recognizing mechanism. Nucleic Acids Res. 2010a;38:1022–1033. doi: 10.1093/nar/gkp1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Q. XIANG J. YANG S. ZHOU Q. LI Q. GUAN A. ZHANG X. ZHANG H. TANG Y. XU G. Recognizing hybrid/mixed G‐quadruplex in human telomeres by using a cyanine dye supramolecule with confocal laser scanning microscopy. Chin. J. Chem. 2010b;28:1126–1132. [Google Scholar]
- YANG Q. XIANG J. YANG S. ZHOU Q. LI Q. TANG Y. XU G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. Recognizing mixed G-quadruplex in human telomeres. Chem. Commun. 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- YANG S. XIANG J. YANG Q. LI Q. ZHOU Q. ZHANG X. TANG Y. XU G. Formation of human telomeric G-quadruplex structures induced by the quaternary benzophenanthridine alkaloids: sanguinarine, nitidine, and chelerythrine. Chin. J. Chem. 2010c;28:771–780. [Google Scholar]
- YANG S. XIANG J.F. YANG Q.F. ZHOU Q.J. ZHANG X.F. LI Q. TANG Y.L. XU G.Z. An important functional group, benzo [1, 3] dioxole, of alkaloids induces the formation of the human telomeric DNA G-quadruplex. Chin. Sci. Bull. 2011;56:613–617. [Google Scholar]
- YIN F. LIU J. PENG X. Effects of triethylene tetraamine on telomerase activity and proliferation in HeLa cells. Cell Biol. Int. 2004;28:287–291. doi: 10.1016/j.cellbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- ZAHLER A.M. WILLIAMSON J.R. CECH T.R. PRESCOTT D.M. Inhibition of telomerase by G-quartet DMA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- ZHANG X. ZHANG H. XIANG J. LI Q. YANG Q. SHANG Q. ZHANG Y. TANG Y. The binding modes of carbazole derivatives with telomere G-quadruplex. J. Mol. Struct. 2010;982:133–138. [Google Scholar]
- ZHAO T. WANG X. RIMANDO A.M. CHE C. Folkloric medicinal plants: Tinospora sagittata var. cravaniana and Mahonia bealei. Planta Med. 1991;57:505. doi: 10.1055/s-2006-960188. [DOI] [PubMed] [Google Scholar]
- ZHOU Q. LI L. XIANG J. SUN H. TANG Y. Fast screening and structural elucidation of G-quadruplex ligands from a mixture via G-quadruplex recognition and NMR methods. Biochimie. 2009;91:304–308. doi: 10.1016/j.biochi.2008.10.011. [DOI] [PubMed] [Google Scholar]
- ZHOU Q. LI L. XIANG J. TANG Y. ZHANG H. YANG S. LI Q. YANG Q. XU G. Screening potential antitumor agents from natural plant extracts by G-quadruplex recognition and NMR methods. Angew. Chem. 2008a;120:5672–5674. doi: 10.1002/anie.200800913. [DOI] [PubMed] [Google Scholar]
- ZHOU Q. LI L. XIANG J. TANG Y. ZHANG H. YANG S. LI Q. YANG Q. XU G. Screening Potential Antitumor Agents from Natural Plant Extracts by G‐Quadruplex Recognition and NMR Methods. Angew. Chem. 2008b;120:5672–5674. doi: 10.1002/anie.200800913. [DOI] [PubMed] [Google Scholar]
- ZHU X. KUMAR R. MANDAL M. SHARMA N. SHARMA H.W. DHINGRA U. SOKOLOSKI J.A. HSIAO R. NARAYANAN R. Cell cycle-dependent modulation of telomerase activity in tumor cells. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6091–6095. doi: 10.1073/pnas.93.12.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.