The crystal structure of the α5β1 integrin reveals conformational changes and amino acids important for ligand binding.
Abstract
Integrin α5β1 is a major cellular receptor for the extracellular matrix protein fibronectin and plays a fundamental role during mammalian development. A crystal structure of the α5β1 integrin headpiece fragment bound by an allosteric inhibitory antibody was determined at a 2.9-Å resolution both in the absence and presence of a ligand peptide containing the Arg-Gly-Asp (RGD) sequence. The antibody-bound β1 chain accommodated the RGD ligand with very limited structural changes, which may represent the initial step of cell adhesion mediated by nonactivated integrins. Furthermore, a molecular dynamics simulation pointed to an important role for Ca2+ in the conformational coupling between the ligand-binding site and the rest of the molecule. The RGD-binding pocket is situated at the center of a trenchlike exposed surface on the top face of α5β1 devoid of glycosylation sites. The structure also enabled the precise prediction of the acceptor residue for the auxiliary synergy site of fibronectin on the α5 subunit, which was experimentally confirmed by mutagenesis and kinetic binding assays.
Introduction
Integrins are cell adhesion receptors that transmit bidirectional signals across the plasma membrane and link the extracellular environment to the actin cytoskeleton (Hynes, 2002). All integrins are noncovalently linked heterodimeric molecules consisting of one α and one β subunit, in which both subunits are required to create a functional binding site at the membrane-distal part of the cell surface receptor for specific extracellular ligands. Compared with other cell adhesion receptor classes, integrin’s ligand recognition mechanism is highly unique in three aspects. First, the ligand recognition specificity of each integrin heterodimer is determined combinatorially, in that both the α and the β subunits contribute to the selective ligand binding of the resultant heterodimeric receptor, and even the same β (or α) subunit will bind different ligands when paired with a different α (or β) subunit (Hynes, 2002). Second, in contrast to other divalent cation-dependent cell adhesion molecules such as cadherins in which metals do not directly bridge two molecules across the cell–cell junction (Patel et al., 2006), the core mechanism of integrin–ligand recognition involves a direct coordination bond between an Mg2+ bound on the integrin (called the metal ion–dependent adhesion site [MIDAS]) and a carboxylate oxygen from the ligand. Lastly, the ligand-binding affinity of integrins can be modulated allosterically via conformational changes that take place outside the binding pocket (Carman and Springer, 2003).
The determination of crystal structures of β3 integrin ectodomain fragments have contributed enormously to our understanding of the integrin–ligand interactions (Xiong et al., 2001, 2002, 2009; Xiao et al., 2004; Springer et al., 2008; Zhu et al., 2008, 2010). Specifically, the structures of αVβ3 and αIIbβ3 integrins in complex with their cognate peptide ligands represented by the Arg-Gly-Asp (RGD) sequence revealed how the small tripeptide portion is specifically recognized by integrins using a small binding cleft at the subunit interface and how the ligand binding is associated with the transition from the closed or low-affinity conformation to the open or high-affinity conformation of integrin. However, there remain important unanswered questions. For example, lack of an atomic resolution structure of integrin in complex with a protein ligand, which usually bears both a core binding motif such as RGD and a secondary synergy site, precludes the complete understanding of the basis for the physiological binding events. Also, two different conformations of β3 integrin were found in the ligand-bound state, leading to a controversy over the structural pathway that leads to the physiological activation/ligand binding for integrins. Another important issue is whether the same ligand recognition and affinity modulation mechanisms apply for integrins outside the β3 class. It is particularly important to obtain structural information about β1 integrins because they constitute the largest and probably the most ancient integrin subclass (Brower et al., 1997) and are fundamentally involved in mammalian development.
We report herein the crystal structure of a ligand-binding fragment of human α5β1 integrin, a prototypic integrin that functions as an RGD-dependent fibronectin receptor. The structure, solved as a complex with a Fab fragment of the anti–β1 inhibitory antibody SG/19, revealed high similarity to the ligand-unbound form of αVβ3 and αIIbβ3 integrins. Surprisingly, the RGD peptide can be introduced into the binding pocket by soaking, without causing any conformational change in integrin except for an ∼1-Å shift of one residue and the dissociation of Ca2+ from the adjacent to the MIDAS (ADMIDAS). Docking simulations and structure-based mutagenesis identified a single α5 residue responsible for the strong preference of α5β1 for fibronectin, establishing a basis for the combinatorial roles played by each subunit during the specific recognition of protein ligands.
Results
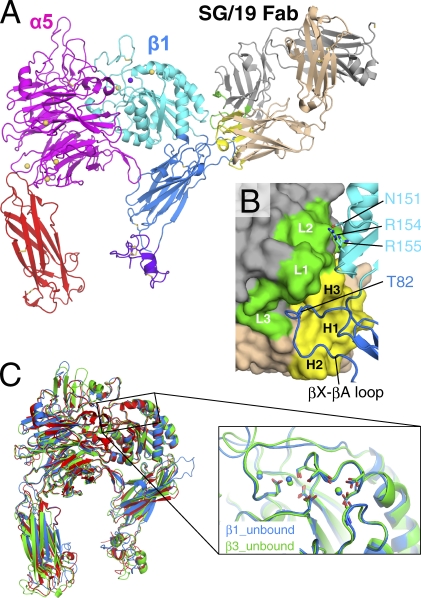
Despite extensive efforts, our initial attempts to crystallize the full-length α5β1 ectodomain fragment were unsuccessful. When we imaged various recombinant soluble integrins using negative-stain EM, we realized that the conformation of α5β1 was highly heterogeneous as a result of its flexible lower half (unpublished data), whereas full-length αVβ3 integrin showed uniformly compact bent conformation reminiscent of the crystal structure (Takagi et al., 2002). We reasoned that this conformational heterogeneity was to blame for the difficulty in crystallizing full-length α5β1 and thus focused on the truncated fragment devoid of the lower half (referred to hereafter as the headpiece fragment). A previous EM study showed that SG/19 binds at the junction between the βA and the hybrid domains of β1 subunit, thereby fixing the conformation of the hybrid domain, which is highly mobile in the context of the truncated fragment (Luo et al., 2004). We took advantage of this phenomenon to crystallize the α5β1 integrin headpiece fragment by making a complex with SG/19 Fab in the presence of physiological concentrations of Mg2+ and Ca2+. The structure, refined at a 2.9-Å resolution, contained all of the domains present in the constructs, including the β-propeller and thigh domains of α5 subunit, the plexin/semaphorin/integrin (PSI), hybrid and βA domains of β1, and the SG/19 Fab (Fig. 1 A; see also a rocking video in Video 1). Two α5β1–Fab complexes were contained in one crystallographic asymmetric unit and were essentially identical except for minor differences in the interdomain angles at the lower half of the molecule (Fig. S1). Therefore, we describe the structure of one complex (chains A, B, E, and F) in the following discussion.
Figure 1.
Structure of the α5β1 integrin headpiece in complex with SG/19 Fab. (A) Ribbon presentation of the overall structure with disulfide bonds in stick model. Individual domains are differently colored in magenta (β-propeller), red (thigh), cyan (βA), blue (hybrid), and purple (PSI), and bound metal ions are shown as spheres (yellow for Ca2+ and purple for Mg2+). SG/19 Fab is colored in gray (light chain) and wheat (heavy chain), with their CDR loop regions highlighted in green and yellow, respectively. (B) Close-up view of the SG/19-binding interface. SG/19 Fab and β1 chain are shown in surface and ribbon presentations, respectively, with the same color code used in A. CDR regions and important interface residues are labeled. (C) Superposition of three integrin headpiece structures in the ligand-unbound form. Blue, α5β1; red, αVβ3 (3IJE); green, αIIbβ3 (3FCS). On the right is a blowup of the region around the metal-binding sites in the β1 (blue) and β3 (green; 3FCS) βA domains.
The allosteric inhibitory antibody SG/19 makes extensive interactions with the long loop between β strands X and A (following the strand nomenclature by Xiong et al. [2001]) of β1 hybrid domain using its antigen-combining sites (Fig. 1 B). The species-specific Thr82 that had been identified as the epitope for SG/19 was in fact deeply inserted into the pocket formed by complementarity-determining region (CDR) loops H1, H2, H3, and L3. Furthermore, SG/19 also interacts with βA domain residues N151, R154, and R155 using its L2 loop outside the CDR core, anchoring the lower half of the α1 helix of βA domain. Because of this two-sided interaction, the H3 loop is wedged between βA and hybrid domains and prevents the outward swing of the hybrid domain. This resulted in the closed headpiece conformation with the tucked hybrid domain, similar to the ligand-unbound, low-affinity form of the β3 integrins. The similarity in the overall conformation can readily be appreciated upon structural superposition of the α5β1 headpiece onto the closed αvβ3 (3IJE, 1.92-Å root-mean-square deviation [RMSD] for 864 Cα atoms) and αIIbβ3 (3FCS, 1.52-Å RMSD for 820 Cα atoms; Fig. 1 C).
The three metal-binding sites found in β3 integrin were also present in β1, and we could see strong electron density at all sites (Fig. 2 A and Video 2), indicating that they were occupied. Based on the coordination geometries and similarity to the previously determined structures of β3 integrins, we assigned Mg2+ to the MIDAS and Ca2+ to the two flanking sites, the ligand-associated metal-binding site (LIMBS; also called SyMBS) and ADMIDAS. The correctness of the metal assignments was also supported by an experiment using a crystal treated with Mg2+ and EGTA, in which electron density for MIDAS remained unchanged, whereas that of ADMIDAS disappeared (unpublished data). Density for LIMBS remained after this treatment, but the completely buried nature of this ion may have prevented the access of EGTA. The coordination environment for MIDAS and ADMIDAS ions in the current complex was essentially identical to that of ligand-free β3 (Fig. 1 C). Therefore, binding of SG/19 not only induced the overall closed conformation of the headpiece (i.e., tucked hybrid domain) but also stabilized the low-affinity MIDAS configuration.
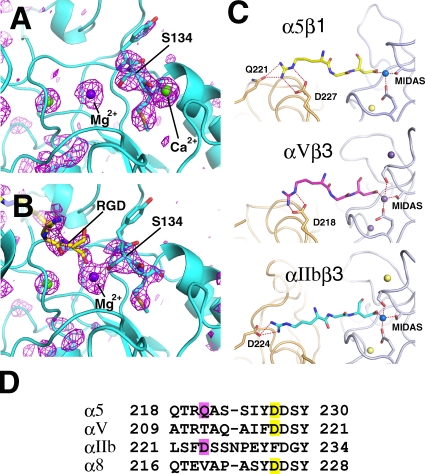
Figure 2.
Recognition of RGD peptide by α5β1. (A and B) Close-up views of the region surrounding MIDAS before (A) and after (B) ligand peptide soaking. Fo-Fc omit maps, in which RGD, metals, and β1 residues Tyr133–Met135 were removed from the models, are shown in magenta wireframes at a contour level of 3 σ. For reduction of model bias from the omitted atoms, the partial models were subjected to refinement before phase calculation. Note that the electron density corresponding to ADMIDAS Ca2+ disappeared in the RGD-bound structure. (C) Specific recognition of RGD peptide by three different classes of integrins: α5β1 (top), αVβ3 (1L5G; middle), and αIIbβ3 (2VDR; bottom). Hydrogen and coordination bonds are shown in dashed lines. Metal ions in the βA domain are shown in spheres. (D) Sequence alignment around the Arg acceptor residues of RGD-dependent α subunits. Head-on and side-on binding residues are highlighted in pink and yellow, respectively.
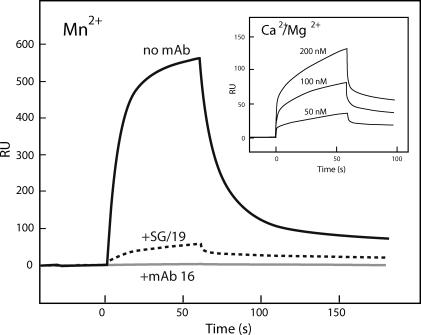
A previous kinetic study indicated that SG/19 attenuates, but not abolishes, the ligand-binding capability of Mn2+-activated full-length α5β1 integrin (Luo et al., 2004). We confirmed this finding in surface plasmon resonance assays using the truncated headpiece fragment of α5β1 (Fig. 3). Binding of the unclasped α5β1 headpiece fragment to the fibronectin ligand (Fig. 3, solid line) was greatly reduced but not completely abolished when bound by SG/19 (Fig. 3, dotted line), whereas complete inhibition was achieved by anti–α5 direct blocking antibody 16 (Fig. 3, gray line). This suggests that the SG/19-bound α5β1 integrin may become ligand occupied in the presence of a high-concentration ligand. We also confirmed that the headpiece fragment used in this study was capable of binding ligand in the divalent cation condition used in the crystallization (i.e., 1 mM Ca2+ and 1 mM Mg2+; Fig. 3, inset), although the binding was much weaker than that in the presence of Mn2+.
Figure 3.
Surface plasmon resonance binding analysis of the α5β1 headpiece fragment to Fn7–10. Purified and unclasped α5β1 headpiece fragment at a concentration of 100 nM was flowed for 60 s over the surface of immobilized Fn7–10 (∼1,400 resonance units [RU]) in a buffer containing 1 mM MnCl2. The integrin was preincubated with SG/19 Fab (10-fold molar excess; dotted line) or mAb 16 IgG (fourfold molar excess; gray line) or left untreated (solid line). (inset) Concentration-dependent binding sensorgrams of the same unclasped α5β1 headpiece fragment onto Fn7–10 in a buffer containing 1 mM CaCl2 and 1 mM MgCl2. This experiment was completed once.
In line with the aforementioned expectation, soaking of the SG/19–α5β1 complex crystal in a solution containing 1 mM hexapeptide GRGDNP resulted in an appearance of electron density above the MIDAS Mg2+, which we interpret as an RGD tripeptide segment (Fig. 2 B and Video 3). As the electron density of the ligand was weaker than the integrin part, we suspect that the occupancy of the peptide was not 100%, and the resultant structure, refined at 2.9 Å, may represent the mean of ligand-bound and -unbound forms. Nevertheless, overall structure of the RGD-bound α5β1 was very similar to that of ligand-free integrin (Fig. S2 and Video 4), which is consistent with the fact that the soaking did not change the diffraction quality of the crystal, nor did it affect the unit cell dimensions (Table 1). In the binding pocket, the Arg side chain makes hydrogen bonds with α5 residues Q221 and D227, and the Asp carboxylate directly coordinates the β1 MIDAS Mg2+, recapitulating the RGD-binding mode found in β3 integrins (Fig. 2 C). In the αVβ3–RGD complex structure (Xiong et al., 2002), the Arg of RGD is hydrogen bonded to αV D218 from the side, whereas in the αIIbβ3–RGD complex, it is hydrogen bonded to αIIb D224 head on (Fig. 2 C; Springer et al., 2008). The Arg recognition mode seen in α5β1 is a mixture of these, with both side-on (D227) and head-on (Q221) hydrogen bonds present. Sequence alignment around this region suggests that α8, another RGD-dependent integrin α subunit, has the side-on residue only (i.e., D225) and thus resembles αV (Fig. 2 D). As α5 is predicted to be the most ancient among the four (Hynes and Zhao, 2000), the head-on residue may have been lost during evolution to yield αV and α8 and then reappeared in αIIb with a concomitant loss of the side-on residue. In any case, the basic strategy for RGD recognition, in which the extended tripeptide portion is held by a two-point anchor situated at the well-shaped binding pocket at the subunit interface, seems to be shared among all integrins, as predicted earlier (Takagi, 2007).
Table 1.
Data collection and refinement statistics
| Crystal ID | Unliganded form | RGD peptide complex | |
| Data collection | Space group | P1 | P1 |
| Cell constant | a = 93.0, b = 102.8, c = 125.1 Å; α = 76.1°, β = 70.2°, γ = 71.3° | a = 92.8, b = 103.9, c = 125.3 Å; α = 75.7°, β = 70.2°, γ = 70.9° | |
| Resolution | 100–2.90 (3.00–2.90) | 100–2.90 (3.00–2.90) | |
| Unique reflection | 91,525 | 90,587 | |
| Completeness (%) | 98.8 (98.3) | 98.9 (98.4) | |
| Rsym (%) | 6.1 (46.2) | 7.6 (49.5) | |
| <I/σI> | 11.5 (2.1) | 9.4 (2.0) | |
| Multiplicity | 2.6 (2.6) | 3.8 (3.8) | |
| Refinement | Resolution | 100–2.90 (2.98–2.90) | 100–2.90 (2.98–2.90) |
| Rwork (%) | 21.1 (32.8) | 21.5 (33.0) | |
| Rfree (%) | 26.8 (38.4) | 27.4 (39.0) | |
| RMSDs from ideal values | |||
| Bond length (Å) | 0.007 | 0.007 | |
| Bond angle (degree) | 1.252 | 1.289 | |
| Mean B value (Å2) | 71.8 | 69.9 | |
| Ramachandran plot | |||
| Favored region (%) | 91.8 | 90.8 | |
| Outliers (%) | 0.9 | 1.1 |
Values for highest resolution shell are given in parentheses.
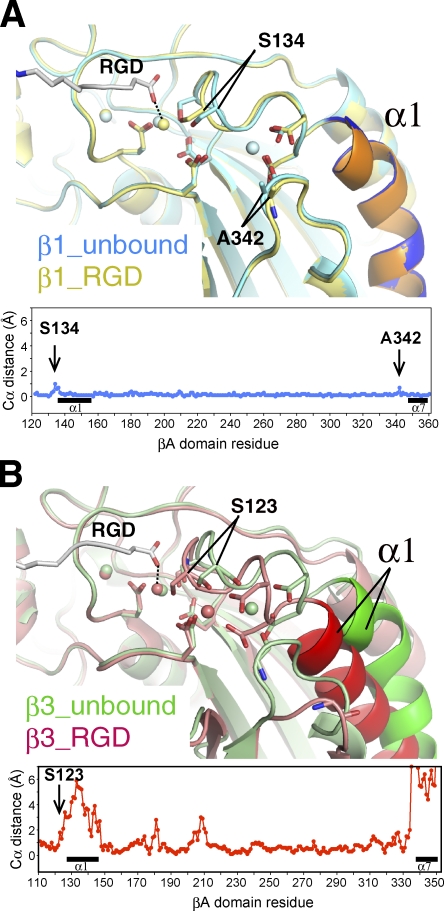
The structure of RGD-bound α5β1 was remarkably similar to that of the unliganded one, showing an RMSD of merely 0.27 Å for all Cα atoms (Fig. S2). In αVβ3 and αIIbβ3 integrins, binding of RGD or related ligands is accompanied by rearrangements of α1 and α7 helices and nearby segments in the βA domain, converting the MIDAS configuration to a high-affinity state that is characterized by a direct coordination of the last Ser residue in the conserved DXSXS motif to Mg2+ (Figs. 4 B and S3; Xiong et al., 2002; Springer et al., 2008). In α5β1, such structural rearrangements are largely prohibited by the bound SG/19, resulting in a surprisingly small RMSD of 0.19 Å between βA domains of unliganded and RGD-bound forms (Fig. 4 A). Nevertheless, Ser134 moves ∼1 Å toward MIDAS to directly coordinate Mg2+, resulting in the high MIDAS configuration (Figs. 4 A and S3). This indicates that the direct coordination of the ligand carboxylate and the Ser hydroxyl to MIDAS Mg2+ is internally coupled and can be accomplished even in the absence of the coordinated movements of the α1 and α7 helices. Similar rearrangement in the MIDAS coordination chemistry unaccompanied by α1 and α7 helix movements during activation has been recently reported for the αA domain of the α1 subunit (Lahti et al., 2011). In addition to the change in MIDAS configuration, there was another critical change induced by RGD binding; the electron density corresponding to the ADMIDAS Ca2+ was diminished in the RGD-soaked crystal (Figs. 2 B and S3 and Video 3), whereas that of LIMBS, MIDAS, and all other metals bound to the α5 subunit remained unchanged. This ADMIDAS-specific Ca2+ discharge seems inevitable because the two βA residues that shifted most upon RGD binding, S134 and A342 (Fig. 4 A), provided their backbone carbonyl to ADMIDAS coordination.
Figure 4.
Shape shifting of the βA domain upon ligand binding. (A) Structural superposition of the MIDAS region of β1 before (blue) and after (yellow) the binding of ligand. (B) Structural superposition of the MIDAS region of β3 before (3FCS; green) and after (2VDR; red) the binding of ligand. In both A and B, α1 helix is colored darker than the rest of the molecule, and the structural differences between the ligand-free and RGD-bound forms (plot of distances between Cα atoms in each βA domain) are shown at the bottom. Locations of the α1 and α7 helices are denoted by black horizontal bars.
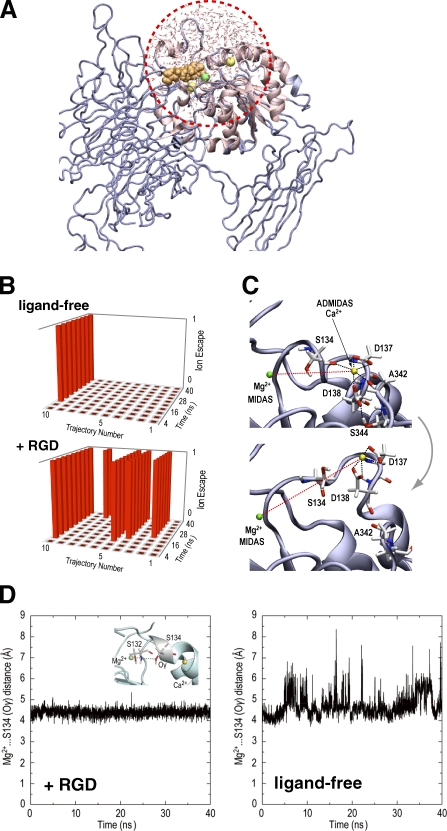
Very limited but characteristic changes in the β1 structure upon RGD binding prompted us to perform molecular dynamics (MD) simulations to see whether these changes can be reproduced in silico. To focus on the local environment surrounding the ligand-binding pocket and the metals, only the region within 20 Å from the Ser132 was subjected to the calculation (Fig. 5 A). In this condition, residues that are anchored by SG/19 were located outside the simulated region, in effect mimicking the conformational freezing by SG/19. First, 10 independent MD simulations (40 nanoseconds each) were performed on the ligand-unbound structure in which all the metal sites were occupied. The coordination environment of ADMIDAS remained largely undisturbed, and the Ca2+ moved from its original position in only 1 out of 10 simulations (Fig. 5 B, top). We then performed another set of simulations on the same structure after manually placing the RGD peptide at the binding pocket above the MIDAS Mg2+ to mimic the very first step of the ligand encounter. In this condition, Ca2+ at the ADMIDAS site became mobile and was expelled from the site in 5 out of 10 simulations (Fig. 5 [B and C] and Video 5). In contrast, both the MIDAS and LIMBS metals were highly stable and did not show major displacement from the original position. During the time frame of the simulation, the Ca2+ did not completely diffuse away from the site but remained attached to integrin through hydrogen bonds with D137 and D138, causing a large disturbance in the conformation of the β1-α1 loop containing these residues (and S134). To follow the complete dissociation process of Ca2+, much longer simulation (greater than a microsecond) and a more rigorous force field would be required. Therefore, the MD result is largely consistent with the ADMIDAS-specific metal discharge upon ligand binding, which was observed in the crystal of SG/19-bound low-affinity α5β1. Next, we focused on the movement of S134. Unlike in the crystal, significant movement of S134 toward Mg2+ was not observed in the RGD-bound structure, and the bond remained a water-mediated indirect coordination with a mean distance of 4.48 Å (Figs. 5 D [+RGD] and S4 and Video 6). In the absence of the RGD ligand, however, the loop harboring S134 exhibited a high degree of mobility, and S134 tended to move away from the Mg2+, and the distance between them frequently exceeded 7 Å (Figs. 5 D [ligand-free] and S4 and Video 7). These results indicate that ligand binding involving the direct coordination of the ligand carboxylate to Mg2+ acts favorably for the approach of S134 toward MIDAS, which is in agreement with the conformational shift that occurred during the RGD soaking experiment. Furthermore, the failure of MD to reproduce the switch from the indirect to direct coordination of S134 suggests that the high-affinity MIDAS configuration may not necessarily be ligand induced but could be ligand stabilized.
Figure 5.
Effect of ligand binding on the conformation of the MIDAS/ADMIDAS region studied by MD simulations. (A) Illustration of the simulating system with GSBP. The region inside the sphere with a radius of 20 Å centered on Ser132 (red dotted circle) was allowed to undergo explicit dynamics, whereas atoms in the rest of the system are taken into account implicitly. (B) Occurrence of the ADMIDAS dissociation in 10 independent 40-nanosecond simulations with and without RGD. The ADMIDAS Ca2+ was considered to have escaped when the three interactions holding ADMIDAS (Ca2+…Ser134, Ca2+…Ala342, and Asp138…Ser344) were all broken and when the distance between MIDAS Mg2+ and ADMIDAS Ca2+ became >11 Å. Each trajectory is divided into 10 bins (4 nanoseconds for each), and the dissociation state for each bin is indicated on the vertical axis (state = 1 means that the event occurrence exceeded 50%). (C) Example of the ADMIDAS escape. Structural snapshots before (0 nanoseconds; top) and after (40 nanoseconds; bottom) the MD simulation in trajectory no. 10 of +RGD simulation reveals loss of two coordination bonds (black dotted lines) and an increase in the distance between MIDAS and ADMIDAS (red dotted lines). See also Video 5. (D) Stabilization of the water-mediated coordination of S134 to Mg2+ by RGD binding. Distances between the Oγ of S134 and the MIDAS Mg2+ (inset) were traced over time. Representative tracings for simulations in the presence (+RGD, trajectory no. 8; see also Video 6) or the absence (ligand-free, trajectory no. 9; see also Video 7) are shown. See Fig. S4 for tracings of all trajectories.
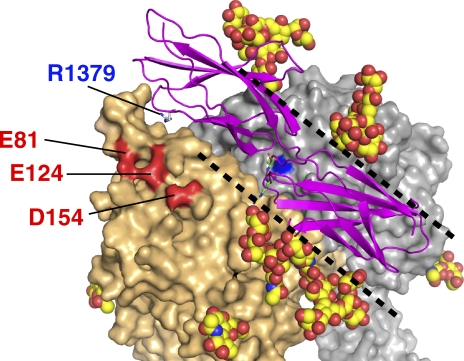
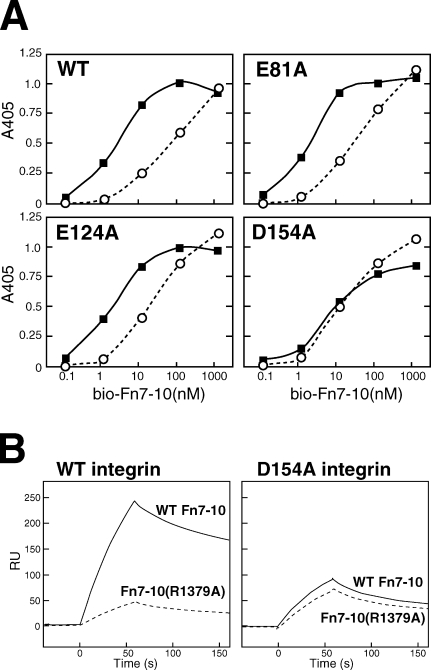
The structure of the aforementioned RGD–α5β1 complex can be used to simulate how the protein ligand (i.e., fibronectin) binds to integrin. As shown in Fig. 6, the RGD-binding pocket is surrounded by several N-glycan chains, leaving a trenchlike exposed surface along the subunit interface. This results in a limited choice of docking orientation when the elongated fibronectin molecule tries to make close contact. Simple docking of 30 nuclear magnetic resonance structures of core integrin-binding fibronectin fragments (Fn9-10) onto α5β1 using the RGD segment as a guide resulted in only a handful of complex models compatible with both structures. Fig. 6 shows one of the most successful models, in which the Fn9 module approaches the α5 subunit. In this model, the residue known to be most important for the synergy activity of the fibronectin (Arg1379) is pointing toward the negatively charged surface created by Glu81, Glu124, and Asp154 of α5, which is likely to constitute the basis for the synergy effects. We individually mutated these residues to Ala and prepared recombinant soluble α5β1 ectodomain fragments carrying these mutations and tested their ability to bind fibronectin. In the solid-phase binding assay, wild-type (WT) integrin α5β1 showed ∼10–50-fold lower affinity toward the fibronectin fragment lacking the three synergy residues (R1374, P1376, and R1379) than the WT fibronectin fragment (Fig. 7 A, top left graph; Takagi et al., 2003). This synergy site dependency was also observed with mutant α5β1 carrying either the E81A or the E124A mutation (Fig. 7 A), indicating that these residues are not required for the synergy site recognition. In contrast, the D154A integrin mutant could no longer distinguish between WT and mutant fibronectin (Fig. 7 A, bottom right graph), suggesting the critical importance of this residue for the synergy effect. Similar results were obtained in the surface plasmon resonance analysis using immobilized Fn7–10 and solution-phase integrin (Fig. 7 B). As a single residue mutant of Fn7–10 (i.e., R1379A) was used in this experiment instead of the triple mutant used in the solid-phase assay, we conclude that Asp154 of α5 and Arg1379 of fibronectin constitute the major interacting pair, as predicted from the docking simulation.
Figure 6.
A model of the Fn9-10–α5β1 complex. The no. 10 molecule from the mouse Fn9-10 fragment nuclear magnetic resonance structures (Protein Data Bank accession no. 2MFN) docked onto the α5β1 using the bound RGD peptide (green stick model) as a guide is shown as a magenta ribbon model, with the Arg1379 side chain highlighted. Carbohydrate moieties are shown by modeling GlcNAc2Man5 residues (space-filling model) at the tip of Asn residues. α5 and β1 chains are surface rendered in wheat and gray, respectively, except for the three acidic residues (red) and MIDAS Mg2+ (blue). The trenchlike exposed surface is marked by black dotted lines.
Figure 7.
Determination of the synergy site acceptor residue on α5 subunit. (A) Solid-phase equilibrium binding using immobilized full-length α5β1 ectodomain and solution-phase biotinylated Fn7–10. Binding experiments with WT (closed squares) or R1374A/P1376A/R1379A mutant (open circles) Fn7–10 fragments to four different α5β1 preparations were performed, and data from one representative of three independent experiments are shown. (B) Effect of synergy residue mutations on the binding kinetics studied by surface plasmon resonance analysis. WT (solid lines) and R1379A mutant (dotted lines) biotinylated Fn7–10 were immobilized separately onto a streptavidin-coated chip (∼1,000 resonance units [RU]). The surface was infused for 60 s with either WT (left) or D154A-purified (right) full-length recombinant α5β1 integrin at 20 nM in a running buffer containing 1 mM Mn2+, and the dissociation phase was followed for 100 s. The data are from a single representative experiment out of three repeats.
Discussion
β1 integrins constitute the largest integrin subfamily and recognize a wide range of ligands depending on the partner α subunit. α5β1 was one of the first integrin heterodimers to be identified, and its function as a fibronectin receptor has been studied extensively (Tamkun et al., 1986). Especially, the discovery of the site on fibronectin important for binding α5β1 integrin (synergy site) has set the basis for our general understanding of the integrin–ligand recognition mechanism, in which the combination of a core interaction (e.g., RGD site) with a secondary interaction (e.g., synergy site) ensures both high affinity and specificity (Obara et al., 1988; Aota et al., 1994). It is known that R1379 makes the greatest contribution to the synergy effect on the fibronectin side (Redick et al., 2000). Through structure-guided mutation experiments, we have now successfully identified the specific residue (Asp154) that distinguishes α5 from other α subunits and results in its strong preference for fibronectin over other RGD ligands.
Binding between α5β1 and fibronectin has been extensively studied through biochemical, immunochemical, and cell biological approaches (Humphries, 2000), and most of those results can be rationalized by the current structure. For example, the epitope for the anti–α5 antibody mAb 16 has been mapped to E116/L118 that sits next to the RGD-binding pocket, which is in line with the strong and direct function-blocking ability of this antibody (Burrows et al., 1999). Leu212, the epitope residue for another function-blocking antibody P1D6, is located adjacent to D154, explaining why this antibody preferentially blocks synergy site interaction (Mould et al., 1997). Ala mutation of I210 was reported to severely compromise the interaction with the synergy region (Mould et al., 2003b). As the side chain of I210 makes van der Waals contact with D154 to orient its side chain outward, the I210A mutation may indirectly affect the synergy site binding by partially burying the D154 carboxylate. Also, it has been reported that W157 is responsible for the α5 subunit’s preference for the RGDGW peptide (Humphries et al., 2000). The side chain of W157 rises on one side of the RGD-binding pocket, and it is likely that the bound RGDGW peptide makes a reverse turn at the second Gly to allow the Trp-Trp ring packing. In addition to providing these fine residue-wise structural insights into the ligand recognition mechanism, the structure offers a unique view of how the ligand-binding specificity might be regulated by sugar chains. In contrast to the β3 chain, which is devoid of N-glycosylation sites on the top face of the βA domain, β1 possesses two conserved N-glycosylation sites near the ligand-binding site (Fig. 6). These glycans, together with two more glycans attached to α5, would occupy considerable space because of their conformational flexibility. Such a canopy may function to selectively capture only the ligands that have the RGD sequence at an apex of a protruding loop (Leahy et al., 1996; Copié et al., 1998) or to restrict the preferential orientation of transient docking of fibrillar ligands so that the RGD site along the fiber can be scanned efficiently. As differences in the glycosylation state of α5β1 on the cell surface have been reported to affect the biological function of the receptor (Seales et al., 2005; Isaji et al., 2006, 2009), it would be valuable to test these possibilities in a cell-based system.
A previous EM study predicted that SG/19 exerts its inhibitory effect allosterically by preventing the hybrid domain swing-out in β1 (Luo et al., 2004), and the current study confirmed the lack of direct occlusion of the fibronectin-binding surface by this antibody. The crystal structure also revealed that the binding of SG/19 not only restricted the global domain orientation but also fixed the local conformation of the βA domain by directly binding to the lower half of the α1 helix. In β3, this helix moves inward to push the α7 helix during the transition to the high-affinity state, resulting in the approach of ADMIDAS toward MIDAS (Fig. 4 B; Xiong et al., 2002; Luo and Springer, 2006). We predict that a similar conformational change can take place in β1 during the ligand binding, as suggested earlier by Mould et al. (2002), if the α1 helix is not frustrated by SG/19. As this inward movement of the α1 helix was observed in all ligand-bound β3 structures reported so far regardless of the conformation of the hybrid domain, it is likely that the SG/19 exerts its inhibitory action primarily through the α1 helix rather than the hybrid domain. In fact, epitopes for a series of anti–chicken β1 antibodies with function-blocking activity have been mapped exactly to this region (Shih et al., 1997), suggesting an intimate relationship between the ligand binding and the conformational freedom of the α1 helix. Although there is no anti–human β1 antibody mapped to this region as a result of the sequence conservation between human and mouse β1, there are many function-modulating (i.e., inhibitory and activating) antibodies mapped to the neighboring α2 helix (Takada and Puzon, 1993; Luo et al., 2004). These antibodies are likely to recognize residues in α1 helix as well, just like in the case of SG/19. Therefore, the segregation of epitopes for many function-modulating antibodies in this region suggests that this region exists in at least two alternative conformations (i.e., high- and low-affinity forms) and that each antibody affects the ligand-binding ability depending on the conformer it stabilizes.
High-resolution structures of the αIIbβ3 headpiece revealed that the β3 MIDAS always assumes octahedral geometry, with the top axial coordination site reserved for a water or ligand carboxylate. The configuration of the MIDAS metal can be grouped into two types that differ by only one coordination site, one with a direct coordination from the side chain of the last Ser residue of the DXSXS motif (MIDAShigh configuration) and the other with the same bond replaced with a water-mediated indirect coordination (MIDASlow configuration; Fig. S3). In β3 integrins, binding of a ligand or a ligand-mimetic analog is tightly coupled with the MIDAShigh configuration (Zhu et al., 2010). Here, we showed that the same coupling exists in β1 integrin. Our MD simulation data also hinted an energetic linkage between the two coordination bonds. Further quantum mechanical calculations would reveal the precise mechanism of the coupling.
As we used the allosteric inhibitory antibody SG/19 to minimize the interdomain flexibility intrinsic to α5β1 integrin to facilitate crystallization, the RGD-bound α5β1 structure we obtained must be regarded as somewhat artificial. Nevertheless, this procedure may have served to visualize a structural state corresponding to the very first step of physiological ligand binding by integrins on the resting cell surface. Thus, a ligand can approach the resting integrin with MIDASlow to produce transient ligand docking. The ligand-bound MIDASlow would quickly be converted to MIDAShigh by pulling the α1-β1 loop (and the Ser134 therein) closer, together with the Ca2+-loaded ADMIDAS. In our crystal structure, however, only the S134 (and the following two residues) are forced to move toward Mg2+ to fulfill the requirement for MIDAShigh configuration because full-loop movement is prohibited by SG/19 anchorage of the C terminus of the α1 helix. At the same time, this movement destabilized the coordination environment of the ADMIDAS, resulting in the release of Ca2+.
Although the physiological importance of ADMIDAS in integrin function is well appreciated, the exact role played by this metal remains somewhat controversial. Mould et al. (2003a) reported that mutation to ADMIDAS in α5β1 resulted in a diminished ligand-binding ability. They also observed that the mutant became resistant to the artificial activation by an activating antibody, TS2/16, and that activation by Mn2+ no longer exposed the activation epitope. From these observations, the authors concluded that ADMIDAS was critical in maintaining the active conformation of the receptor. On the other hand, Chen et al. (2003) found that a similar mutation in the ADMIDAS of α4β7 integrin expressed on the cell surface canceled the default low-affinity state of the integrin, resulting in the overall up-regulation of cell adhesion. This led to a proposal that the ADMIDAS represents a negative regulatory site for integrin function. These seemingly contradicting results can be reconciled if we take a view that the function of ADMIDAS is to physically link the change in the ligand–MIDAS bond to the change elsewhere in the molecule, particularly the hybrid domain via the α1 helix. Thus, the mutation of ADMIDAS residues would decouple the conformational relay in both directions, resulting in either activation or deactivation of a particular integrin depending on its default affinity state.
There has been a debate over how the integrin conformation and the ligand binding are mutually related. Particularly, the hypothesis that a global conformational change (i.e., a switchblade-like integrin extension) is linked to the local conformational up-regulation of the ligand-binding MIDAS has been challenged by Adair et al. (2005), who observed a compact (bent) conformer of αVβ3 upon fibronectin binding. This discrepancy can be reconciled with the current structure because it unequivocally showed that even an integrin permanently maintained in a low-affinity conformation by an inhibitory antibody is capable of ligand binding via the high-affinity form of MIDAS. In addition, we have recently found, using EM, that β1 integrins do not assume acutely bent conformation under the nonactivating condition (unpublished data). It is possible that the ligand binding and the overall integrin conformation are less tightly coupled than originally thought, especially in β1 integrins. Nevertheless, the existence of local conformational coupling between the ligand-binding pocket and the α1 helix–hybrid domain region in β subunit is supported by an overwhelming number of studies (Mould et al., 2002; Xiong et al., 2002; Xiao et al., 2004; Zhu et al., 2010). Exactly how this conformational relay system, exerted mainly by ADMIDAS, contributes to the inside-out and outside-in integrin signal transduction needs to be explored in more detail using structural as well as cell biological analysis.
Materials and methods
Preparation of the α5β1 integrin headpiece and SG/19 Fab fragment
The expression construct for the α5 subunit contained residues 1–623 followed by a 30-residue ACID-Cys peptide, and the construct for β1 contained residues 1–445 followed by a Tobacco etch virus (TEV) protease recognition sequence, a 30-residue BASE-Cys peptide, and a hexahistidine tag. When combined, the C-terminal ACID-Cys and BASE-Cys segments form an intersubunit disulfide-bridged α-helical coiled-coil (called clasp) that can be released by a treatment with TEV protease (Takagi et al., 2002). These constructs were cotransfected into CHO Lec 3.2.8.1 cells to establish stable cell lines. Recombinant integrins were purified from the culture supernatants by an immunoaffinity chromatography using anti–coiled-coil antibody 2H11 (Chang et al., 1994) followed by a gel filtration on a Superdex 200 HR column (1.6 × 60 cm; GE Healthcare) equilibrated with 20 mM Tris and 150 mM NaCl, pH 7.5 (TBS), containing 1 mM CaCl2 and 1 mM MgCl2. The peak fraction was concentrated to ∼1 mg/ml and stored at −80°C until used. Removal of the C-terminal coiled-coil clasp and the hexahistidine tag was achieved by treatment with TEV protease at room temperature for ∼16 h. SG/19 IgG from mouse hybridoma cell culture supernatant was purified using a Protein A column (GE Healthcare). SG/19 Fab fragment was prepared by papain digestion using immobilized papain (Thermo Fisher Scientific). The DNA sequences for the variable region of the SG/19 heavy and light chains were determined as follows: in brief, total RNA was isolated from the hybridoma cells using the SV Total RNA isolation kit (Promega), and the cDNA was amplified by RT-PCR reaction using the OneStep RT-PCR kit (QIAGEN) with Ig-Primer Sets (EMD). The PCR products were cloned into pDrive vector (QIAGEN) and then sequenced. Sequences have been deposited in GenBank/EMBL/DDBJ under accession no. HE578877 (for heavy chain) and accession no. HE578878 (for light chain).
Crystallization and data collection
Unclasped α5β1 fragment was incubated with a saturating concentration of SG/19 Fab fragment, and the resultant integrin–Fab complex was purified on a Superdex 200 column equilibrated with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, and 1 mM MgCl2. Purified protein was concentrated to 6 mg/ml for crystallization using an Ultrafree-0.5 centrifugal concentrator (10-kD molecular mass cutoff; Millipore). Initial screening for crystallization conditions was performed using Index (Hampton Research). For this screen, a mosquito crystallization robot (TTP LabTech) was used to dispense 200 nanoliters of protein solution mixed in a 1:1 ratio with the reservoir solution. Drops were equilibrated over 100 µl of reservoir solution using the sitting drop vapor diffusion method at 293 K. The initial crystallization condition (0.1 M bis-tris, pH 6.5, and 20% polyethylene glycol monomethyl ether 5000) was further optimized using a 24-well crystallization plate with the hanging drop vapor diffusion method. Each well contained 500 µl of reservoir solution, and the drop volume was a mixture of 0.3 µl of protein solution and 0.3 µl of reservoir solution. The reproducibility of the crystals was greatly improved by the addition of microseed crystals in the crystallization drop. Diffraction-quality crystals were obtained under a condition of 0.1 M bis-tris, pH 6.5, and 20% polyethylene glycol 8000 at 293 K and grew to dimensions of ∼100 × 100 × 20 µm within 2 wk. The RGD peptide complex crystals were prepared by soaking the crystals of ligand-free form in 1 mM RGD peptide (NH2-GlyArgGlyAspAsnPro-COOH) for 30 min at 293 K.
Before x-ray diffraction experiments, crystals were soaked in reservoir solution containing an additional 20% ethylene glycol and flash cooled in liquid nitrogen. X-ray diffraction datasets for the crystals were collected at 95 K on beamline BL17A at Photon Factory using a wavelength of 1.000 Å and a charge-coupled device detector (Quantum 270; Area Detector Systems Corporation). All datasets were processed and scaled using HKL2000 program suite (Otwinowski and Minor, 1997).
Structure determination and refinement
Initial phase determination was performed by molecular replacement using the program MOLREP (Vagin and Teplyakov, 1997) in the CCP4 program suite. The atomic coordinates of the αVβ3 headpiece (Protein Data Bank accession no. 1L5G) and a murine Fab fragment (Protein Data Bank accession no. 1FGN) were used as search models. The orientation and position of β-propeller and thigh domain of α5 subunit and βA domain of β1 subunit were initially determined. Subsequently, the positions of variable and constant regions of SG/19 Fab fragment were determined. After several cycles of rigid body refinement by the program REFMAC5 (Murshudov et al., 1997) of the CCP4 program suite, the hybrid domain of β1 subunit could be introduced. Model reconstruction was conducted manually with Coot (Emsley and Cowtan, 2004). Crystallographic refinement was performed using the program REFMAC5. As a final check on the model, the stereochemical quality was assessed using the program Molprobity (Lovell et al., 2003). The final models included the following residues: chain A, 1–600; chain B, 6–29 and 43–445; chain C, 1–512, 518–553, and 560–600; chain D, 5–33 and 43–445; chains E and L, 1–219; and chains F and H, 1–218. Data collection and refinement statistics are summarized in Table 1. All figures were prepared using PyMOL (DeLano Scientific). Atomic coordinates and structural factors have been deposited in the Protein Data Bank under accession no. 3VI3 (unliganded form) and accession no. 3VI4 (RGD peptide complex).
MD simulation
All atom MD simulations of the βA domain of α5β1 were performed using the CHARMM (c35b2) software package (Brooks et al., 1983). The CHARMM27 force field parameter (MacKerell et al., 1998) with CMAP correction (MacKerell et al., 2004) was used for the protein and the RGD ligand. The parameter developed by Babu and Lim (1999) was used for Ca2+ and Mg2+. The TIP3P model (Jorgensen et al., 1983) was used for water molecules. The crystal structure of the ligand-free form was used as the starting structure without modification, except for the manual placement of the RGD peptide at location defined by the RGD complex structure when calculating the effect of ligand binding. We used a generalized solvent boundary potential (GSBP; Im et al., 2001) and focused dynamics of protein around the metal-binding sites. To set up the system for use with GSBP, the region surrounding three metal-binding sites (LIMBS, MIDAS, and ADMIDAS) was solvated using a preequilibrated solvent sphere with a radius of 20 Å centered on Ser132 of β1. Solvent molecules within 2.6 Å of any nonhydrogen atom of the protein were removed. The 10-picosecond equilibration and the aforementioned solvation process were iteratively repeated until the number of newly added solvent molecules became less than five. The final equilibrated structure was used for 10 independent MD runs. We assigned different initial velocity for each run. MD simulations were performed using Langevin dynamics at 300 K with a friction constant corresponding to a relaxation time of 5 picoseconds−1 applied to the nonhydrogen atoms. The geometry of the water was kept fixed using SHAKE (Ryckaert et al., 1977). A time step of 2 femtoseconds was used. After 500 picoseconds of equilibration (100 picoseconds of solvent relaxation, 200 picoseconds of protein relaxation with the fixed MIDAS coordination structure, and another 200 picoseconds of full relaxation), we performed 40 nanoseconds of MD simulation for each run. The trajectories without showing the Ca2+ dissociation were used for all analysis, except for analyzing the dissociation itself.
Fibronectin-binding assays
All recombinant fibronectin fragments encompassing the 7th to 10th Fn3 repeats (Fn7–10) were produced using a bacterial expression system as previously described (Takagi et al., 2003). In brief, a segment corresponding to residues 1,142–1,509 of human fibronectin with one Cys residue added after residue 1,509 was cloned into pET11c vector (EMD) and expressed in Escherichia coli. Protein was purified from bacterial lysates by anion-exchange chromatography on a HiTrap Q column (GE Healthcare) and biotinylated via the sulfhydryl group of the Cys with polyethylene oxide–maleimide-activated biotin (Thermo Fisher Scientific) according to the manufacturer’s recommendation. Synergy site mutants with triple (R1374A/P1376A/R1379A) or single (R1379A) mutations were prepared by QuikChange mutagenesis. To produce full-length α5β1 ectodomain fragments, 293T cells were transiently transfected with vectors encoding α5 (residues 1–954) and β1 (residues 1–708) with the C-terminal coiled-coil, and the covalently linked heterodimeric proteins carrying α5 mutations (E81A, E124A, or D154A) or the WT sequence were purified from the culture supernatants. Solid-phase binding assay was performed as follows: solutions of purified full-length α5β1 ectodomains (5 µg/ml in TBS containing 1 mM CaCl2 and 1 mM MgCl2) were used to coat 96-well microtiter plates (MaxiSorp; Thermo Fisher Scientific) by an overnight incubation at 4°C. Coating with BSA was used to determine the background values of unspecific binding. After a 1-h blocking step (1% BSA in TBS), varying concentrations of biotinylated Fn7–10 were incubated in the presence of 1 mM Mn2+ for 3 h at room temperature. After washing, bound Fn7–10 was chromogenically detected by peroxidase-streptavidin conjugate and substrate. For the surface plasmon resonance binding experiments, biotinylated Fn7–10 was directly captured on streptavidin-conjugated Sensor Chip SA (GE Healthcare). Various α5β1 integrin heterodimers were used as analytes at a flow rate of 20 µl/min. Between each run, bound integrin was completely stripped off from the ligand by regeneration of the surface with 50 mM NaOH containing 20 mM EDTA.
Online supplemental material
Figs. S1 and S2 show the structural superpositions between two α5β1–SG/19 complexes contained in the asymmetric unit (Fig. S1) and between the ligand-free and RGD-bound complexes (Fig. S2). Fig. S3 shows the detailed depictions of metal coordination geometries in the MIDAS and ADMIDAS of β1 and β3 chains. Fig. S4 shows the S134(Oγ)-Mg2+ distance tracings for all trajectories during the MD simulations. Videos 1 and 4 present rocking videos showing the overall structure of the α5β1 headpiece–SG/19 Fab complex in the absence (Video 1) and presence (Video 4) of RGD ligand. Videos 2 and 3 present rocking videos of the 2Fo-Fc electron density map of the ligand-free (Video 2) and RGD-bound (Video 3) integrin near the MIDAS region. Videos 5–7 show the close-up view videos of metal-binding sites during the MD simulation from the trajectory obtained in the +RGD simulation (Videos 5 and 6) or the ligand-free simulation (Video 7). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111077/DC1.
Acknowledgments
We thank Keiko Tamura-Kawakami and Maiko Nampo for their excellent technical support, Mayumi Nakano for preparation of the manuscript, and Samuel Thompson for critical editing of the manuscript. We are also grateful to the staff of the beamlines at Photon Factory, SPring-8, and National Synchrotron Radiation Research Center for providing data collection facilities and support.
This work was partly supported by the Grants-in-Aid for Scientific Research (A) and by the Grants-in-Aid for Scientific Research on Innovative Areas (transient macromolecular complexes), both from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Abbreviations used in this paper:
- CDR
- complementarity-determining region
- GSBP
- generalized solvent boundary potential
- LIMBS
- ligand-associated metal-binding site
- MD
- molecular dynamics
- MIDAS
- metal ion–dependent adhesion site
- PSI
- plexin/semaphorin/integrin
- RMSD
- root-mean-square deviation
- TEV
- Tobacco etch virus
- WT
- wild type
References
- Adair B.D., Xiong J.P., Maddock C., Goodman S.L., Arnaout M.A., Yeager M. 2005. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J. Cell Biol. 168:1109–1118 10.1083/jcb.200410068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S., Nomizu M., Yamada K.M. 1994. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 269:24756–24761 [PubMed] [Google Scholar]
- Babu C.S., Lim C. 1999. Theory of ionic hydration: Insights from molecular dynamics simulations and experiment. J. Phys. Chem. B. 103:7958–7968 10.1021/jp9921912 [DOI] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217 10.1002/jcc.540040211 [DOI] [Google Scholar]
- Brower D.L., Brower S.M., Hayward D.C., Ball E.E. 1997. Molecular evolution of integrins: Genes encoding integrin beta subunits from a coral and a sponge. Proc. Natl. Acad. Sci. USA. 94:9182–9187 10.1073/pnas.94.17.9182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows L., Clark K., Mould A.P., Humphries M.J. 1999. Fine mapping of inhibitory anti-alpha5 monoclonal antibody epitopes that differentially affect integrin-ligand binding. Biochem. J. 344:527–533 10.1042/0264-6021:3440527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman C.V., Springer T.A. 2003. Integrin avidity regulation: Are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 15:547–556 10.1016/j.ceb.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Chang H.C., Bao Z., Yao Y., Tse A.G., Goyarts E.C., Madsen M., Kawasaki E., Brauer P.P., Sacchettini J.C., Nathenson S.G., et al. 1994. A general method for facilitating heterodimeric pairing between two proteins: Application to expression of alpha and beta T-cell receptor extracellular segments. Proc. Natl. Acad. Sci. USA. 91:11408–11412 10.1073/pnas.91.24.11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Salas A., Springer T.A. 2003. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat. Struct. Biol. 10:995–1001 10.1038/nsb1011 [DOI] [PubMed] [Google Scholar]
- Copié V., Tomita Y., Akiyama S.K., Aota S., Yamada K.M., Venable R.M., Pastor R.W., Krueger S., Torchia D.A. 1998. Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: Comparison with the human fibronectin crystal structure. J. Mol. Biol. 277:663–682 10.1006/jmbi.1998.1616 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Humphries J.D., Askari J.A., Zhang X.P., Takada Y., Humphries M.J., Mould A.P. 2000. Molecular basis of ligand recognition by integrin alpha5beta 1. II. Specificity of arg-gly-Asp binding is determined by Trp157 OF THE alpha subunit. J. Biol. Chem. 275:20337–20345 10.1074/jbc.M000568200 [DOI] [PubMed] [Google Scholar]
- Humphries M.J. 2000. Integrin structure. Biochem. Soc. Trans. 28:311–339 10.1042/0300-5127:0280311 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Hynes R.O., Zhao Q. 2000. The evolution of cell adhesion. J. Cell Biol. 150:F89–F96 10.1083/jcb.150.2.F89 [DOI] [PubMed] [Google Scholar]
- Im W., Berneche S., Roux B. 2001. Generalized solvent boundary potential for computer simulations. J. Chem. Phys. 114:2924–2937 10.1063/1.1336570 [DOI] [Google Scholar]
- Isaji T., Sato Y., Zhao Y., Miyoshi E., Wada Y., Taniguchi N., Gu J. 2006. N-glycosylation of the beta-propeller domain of the integrin alpha5 subunit is essential for alpha5beta1 heterodimerization, expression on the cell surface, and its biological function. J. Biol. Chem. 281:33258–33267 10.1074/jbc.M607771200 [DOI] [PubMed] [Google Scholar]
- Isaji T., Sato Y., Fukuda T., Gu J. 2009. N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: Identification of the minimal N-glycosylation requirement for alpha5beta1. J. Biol. Chem. 284:12207–12216 10.1074/jbc.M807920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935 10.1063/1.445869 [DOI] [Google Scholar]
- Lahti M., Bligt E., Niskanen H., Parkash V., Brandt A.M., Jokinen J., Patrikainen P., Käpylä J., Heino J., Salminen T.A. 2011. Structure of collagen receptor integrin α(1)I domain carrying the activating mutation E317A. J. Biol. Chem. 286:43343–43351 10.1074/jbc.M111.261909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy D.J., Aukhil I., Erickson H.P. 1996. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 84:155–164 10.1016/S0092-8674(00)81002-8 [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Davis I.W., Arendall W.B., III, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. 2003. Structure validation by Cα geometry: Φ,ψ and Cβ deviation. Proteins. 50:437–450 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Luo B.H., Springer T.A. 2006. Integrin structures and conformational signaling. Curr. Opin. Cell Biol. 18:579–586 10.1016/j.ceb.2006.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.H., Strokovich K., Walz T., Springer T.A., Takagi J. 2004. Allosteric beta1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J. Biol. Chem. 279:27466–27471 10.1074/jbc.M404354200 [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Bashford D., Bellott M., Dunbrack R.L., Jr, Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Feig M., Brooks C.L., III 2004. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 126:698–699 10.1021/ja036959e [DOI] [PubMed] [Google Scholar]
- Mould A.P., Askari J.A., Aota S., Yamada K.M., Irie A., Takada Y., Mardon H.J., Humphries M.J. 1997. Defining the topology of integrin alpha5beta1-fibronectin interactions using inhibitory anti-alpha5 and anti-beta1 monoclonal antibodies. Evidence that the synergy sequence of fibronectin is recognized by the amino-terminal repeats of the alpha5 subunit. J. Biol. Chem. 272:17283–17292 10.1074/jbc.272.28.17283 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Askari J.A., Barton S., Kline A.D., McEwan P.A., Craig S.E., Humphries M.J. 2002. Integrin activation involves a conformational change in the alpha 1 helix of the beta subunit A-domain. J. Biol. Chem. 277:19800–19805 10.1074/jbc.M201571200 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Barton S.J., Askari J.A., Craig S.E., Humphries M.J. 2003a. Role of ADMIDAS cation-binding site in ligand recognition by integrin alpha 5 beta 1. J. Biol. Chem. 278:51622–51629 10.1074/jbc.M306655200 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Symonds E.J., Buckley P.A., Grossmann J.G., McEwan P.A., Barton S.J., Askari J.A., Craig S.E., Bella J., Humphries M.J. 2003b. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J. Biol. Chem. 278:39993–39999 10.1074/jbc.M304627200 [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Obara M., Kang M.S., Yamada K.M. 1988. Site-directed mutagenesis of the cell-binding domain of human fibronectin: Separable, synergistic sites mediate adhesive function. Cell. 53:649–657 10.1016/0092-8674(88)90580-6 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Patel S.D., Ciatto C., Chen C.P., Bahna F., Rajebhosale M., Arkus N., Schieren I., Jessell T.M., Honig B., Price S.R., Shapiro L. 2006. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 124:1255–1268 10.1016/j.cell.2005.12.046 [DOI] [PubMed] [Google Scholar]
- Redick S.D., Settles D.L., Briscoe G., Erickson H.P. 2000. Defining fibronectin’s cell adhesion synergy site by site-directed mutagenesis. J. Cell Biol. 149:521–527 10.1083/jcb.149.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Seales E.C., Shaikh F.M., Woodard-Grice A.V., Aggarwal P., McBrayer A.C., Hennessy K.M., Bellis S.L. 2005. A protein kinase C/Ras/ERK signaling pathway activates myeloid fibronectin receptors by altering beta1 integrin sialylation. J. Biol. Chem. 280:37610–37615 10.1074/jbc.M508476200 [DOI] [PubMed] [Google Scholar]
- Shih D.T., Boettiger D., Buck C.A. 1997. Epitopes of adhesion-perturbing monoclonal antibodies map within a predicted alpha-helical domain of the integrin beta 1 subunit. J. Cell Sci. 110:2619–2628 [DOI] [PubMed] [Google Scholar]
- Springer T.A., Zhu J., Xiao T. 2008. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 182:791–800 10.1083/jcb.200801146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Puzon W. 1993. Identification of a regulatory region of integrin beta 1 subunit using activating and inhibiting antibodies. J. Biol. Chem. 268:17597–17601 [PubMed] [Google Scholar]
- Takagi J. 2007. Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 19:557–564 10.1016/j.ceb.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., Springer T.A. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–11 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., Walz T. 2003. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22:4607–4615 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W., DeSimone D.W., Fonda D., Patel R.S., Buck C., Horwitz A.F., Hynes R.O. 1986. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell. 46:271–282 10.1016/0092-8674(86)90744-0 [DOI] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. 1997. MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 30:1022–1025 10.1107/S0021889897006766 [DOI] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., Springer T.A. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D.L., Joachimiak A., Goodman S.L., Arnaout M.A. 2001. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 294:339–345 10.1126/science.1064535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 296:151–155 10.1126/science.1069040 [DOI] [PubMed] [Google Scholar]
- Xiong J.P., Mahalingham B., Alonso J.L., Borrelli L.A., Rui X., Anand S., Hyman B.T., Rysiok T., Müller-Pompalla D., Goodman S.L., Arnaout M.A. 2009. Crystal structure of the complete integrin αVβ3 ectodomain plus an α/β transmembrane fragment. J. Cell Biol. 186:589–600 10.1083/jcb.200905085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., Springer T.A. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., Negri A., Provasi D., Filizola M., Coller B.S., Springer T.A. 2010. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 116:5050–5059 10.1182/blood-2010-04-281154 [DOI] [PMC free article] [PubMed] [Google Scholar]