A new method for visualizing translation in cells via standard immunofluorescence microscopy provides evidence for translation in the nucleoplasm and nucleolus.
Abstract
Whether protein translation occurs in the nucleus is contentious. To address this question, we developed the ribopuromycylation method (RPM), which visualizes translation in cells via standard immunofluorescence microscopy. The RPM is based on ribosome-catalyzed puromycylation of nascent chains immobilized on ribosomes by antibiotic chain elongation inhibitors followed by detection of puromycylated ribosome-bound nascent chains with a puromycin (PMY)-specific monoclonal antibody in fixed and permeabilized cells. The RPM correlates localized translation with myriad processes in cells and can be applied to any cell whose translation is sensitive to PMY. In this paper, we use the RPM to provide evidence for translation in the nucleoplasm and nucleolus, which is regulated by infectious and chemical stress.
Introduction
Protein translation is a central cellular function attracting increasing attention from cell biologists as they integrate gene product–specific information into a systems view of cellular function. Translation is heterogeneous, exhibiting global modifications tailored for differentiation states and environmental and infectious stress and specific adaptations to optimize translation of individual mRNAs (Mauro and Edelman, 2002; Komili et al., 2007). Recent findings support compartmentalized translation of specific mRNAs to facilitate cell migration (Rodriguez et al., 2006), embryogenesis, neuronal synapse formation and plasticity (Wang et al., 2010), viral infection (Katsafanas and Moss, 2007), and antigen processing (Dolan et al., 2010; Lev et al., 2010).
Traditionally, translation has been studied using amino acids with isotopic labels that enable detection via radioactivity or mass difference. Although it is possible to localize radioactivity in cells via light or electron microscopy to identify sites of translation, this method is cumbersome, insensitive, and poorly compatible with fluorescence detection. Amino acid analogues that can be identified after biotinylation or modification with haptens with secondary detection reagents can also be used to detect nascent proteins. Using these methods, however, a substantial fraction of the signal is associated with completed, released polypeptide chains and not nascent chains tethered to ribosomes.
Puromycin (PMY) is a Tyr-tRNA mimetic that enters the ribosome A site and terminates translation by ribosome-catalyzed covalent incorporation into the nascent chain C terminus (Pestka, 1971). Eggers et al. (1997) generated polyclonal antibodies to PMY and detected puromycylated nascent chains released from ribosomes by immunoblotting and immunoprecipitation. Fluorescent PMY can label nascent chains by microscopy (Starck et al., 2004), but as with other protein synthesis–based labeling methods, this approach does not distinguish attached from released nascent chains. Schmidt et al. (2009) extended this approach by using anti-PMY mAbs in flow cytometry to measure relative translation rates in living cells exposed to PMY to generate PMY-terminated cell surface proteins.
Here, we show that puromycylated nascent chains are immobilized on ribosomes by the chain elongation inhibitors cycloheximide (CHX) or emetine and describe the simple and generally applicable ribopuromycylation method (RPM). The RPM localizes actively translating ribosomes using anti-PMY mAbs via standard immunofluorescence of fixed and permeabilized cells or tissues. We use the RPM to address the critical question of whether compartmentalized translation occurs in the nucleus. First reported almost 60 yr ago (Allfrey, 1954; Allfrey et al., 1955) but falling into disfavor, nuclear translation was resurrected by Iborra et al. (2001) by demonstrating translation in isolated nuclei. Subsequently, these findings were attributed to contamination of nuclei with cytoplasmic ribosomes (Dahlberg et al., 2003; Nathanson et al., 2003). Using the RPM, we provide evidence for translation in the nucleus concentrated in the nucleolus.
Results
Biochemical basis of the RPM
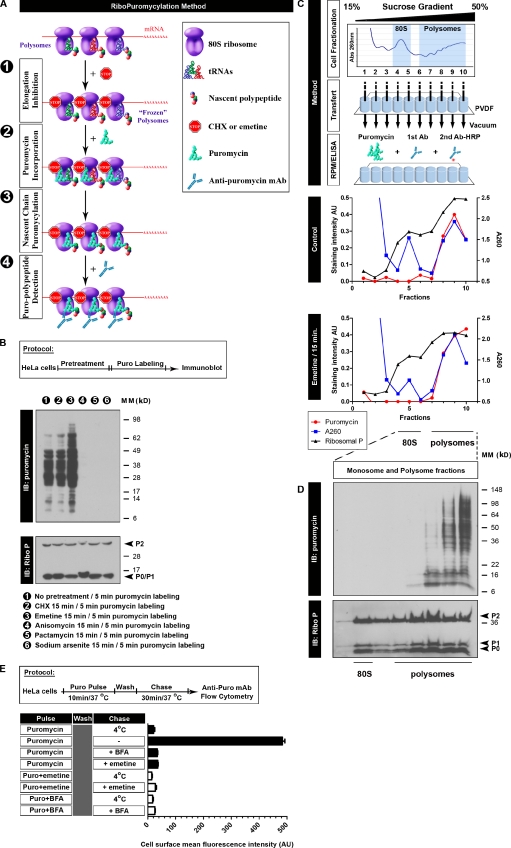
Although PMY has been extensively used for decades (Pestka, 1971; Prouty et al., 1975), the possible use of puromycylation as a method of identifying translating ribosomes in situ has not been previously explored. We reasoned that by briefly pretreating cells with translation elongation inhibitors like CHX or emetine (Pestka, 1971), we could freeze translation and then “puromycylate” immobilized nascent chains by incubating cells with PMY and detect actively translating ribosomes in permeabilized cells or cell extracts using the PMY-specific mAb 12D10 (Schmidt et al., 2009) to PMY tethered to ribosomes by a nascent chain (Fig. 1 A).
Figure 1.
Characterizing the RPM biochemically. (A) Schematic representation of the RPM. After freezing polysomes with an elongation inhibitor (step 1), PMY is added (step 2) to living cells or subcellular fractions, and nascent chains are puromycylated through ribosome catalysis (step 3). The anti-PMY mAb 12D10 (or other PMY mAbs that we have generated) detects puromycylated nascent chains via immunoblotting or indirect immunofluorescence (step 4). (B) Anti-PMY immunoblotting (IB) of total HeLa cell lysates from cells incubated with PMY for 5 min and other inhibitors as indicated. The smear of proteins represents C-terminally puromycylated nascent chains released from ribosomes. Chain elongation inhibitors do not effect (CHX) or enhance (emetine) nascent chain puromycylation, whereas protein synthesis inhibitors that deplete nascent chains by blocking initiation while allowing chain elongation and completion (pactamycin, direct initiation inhibitor, and arsenite, indirect initiation inhibitor) prevent puromycylation. Anisomycin blocks puromycylation by competing with PMY binding to ribosomes. On the bottom, blotting with anti–ribosomal P (Ribo P) human autoreactive antisera shows that the results cannot be attributed to lane loading discrepancies. (C) HeLa cells incubated or not incubated with emetine for 15 min were lysed and fractionated on 15–50% sucrose gradients. Fractions were bound to PVDF 96-well plates and incubated with PMY, which results in ribosome-catalyzed nascent chain puromycylation. Ribosomes were detected by A260 of fractions or by ELISA for the ribosomal P proteins (here resolved into the three known species) as detected by human autoimmune antibodies, which establishes that monosomes and 60S subunits bind well to PVDF. Puromycylation was detected by ELISA for PMY using 12D10 and clearly demonstrates that monosomes and free 60S subunits do not stably associate with PMY, which requires nascent chain puromycylation. (D) HeLa cells incubated with emetine were lysed and fractionated on 15–50% sucrose gradients. Monosome- and polysome-containing fractions were labeled with PMY on ice, and nascent chains were identified by immunoblotting with 12D10. Only polysomes demonstrate a significant anti-PMY signal, and from the pattern, it is clear that binding is based on nascent chain puromycylation. As expected, the mean size of puromycylated proteins increases with polysome size because, on average, faster sedimenting polysomes possess more ribosomes, translating longer mRNAs. (E) HeLa cells were pulsed with PMY with or without emetine or BFA. Cells were washed and chased with or without emetine/BFA. The control sample (pulse only, labeled 4°C) was kept cold during the chase. Expression of PMY (Puro) on the surface of live cells was determined by flow cytometry using 12D10. In the absence of inhibitors, some puromycylated nascent chains are sufficiently native to be delivered to and expressed on the cell surface with their C-terminal PMY exposed for detection (the basis for the SUnSET assay). Emetine blocks surface expression to the same extent as low temperature or BFA, which completely blocks egress of membrane proteins from the ER. This experiment functionally establishes that emetine prevents release of puromycylated nascent chains from ribosomes, as determined by the proxy population of cell surface proteins detected by the SUnSET method. Error bars represent the standard deviation of triplicate samples. Ab, antibody; AU, arbitrary units; MM, molecular mass.
Despite CHX pretreatment nearly completely blocking translation as measured by incorporation of [35S]methionine into acid-insoluble proteins (Fig. S1 A), its continued presence has no significant effect on nascent chain puromycylation after 5-min exposure to PMY, as detected by anti-PMY immunoblotting of total cell lysates (Fig. 1 B). Emetine, an irreversible and highly effective translation elongation inhibitor, actually enhances puromycylation (Fig. 1 B), likely because of some combination of greater effectiveness (it is irreversible) and lack of interference with PMY association with ribosomes (Pestka et al., 1972). Consequently, we preferentially used emetine for the RPM. In contrast, blocking translation initiation with pactamycin (an initiation inhibitor) or arsenite (an oxidizing stressor) inhibits puromycylation, which we attribute to the release of nascent chains as their translation is completed, generating nontranslating monosomes (Table 1). Importantly, anisomycin, a competitive inhibitor of PMY binding to ribosomes (Pestka et al., 1972; Sugita et al., 1995; Hansen et al., 2003), inhibits puromycylation (Fig. 1 B).
Table 1.
Properties of protein synthesis inhibitors
| Translation inhibitor | Reversibility | Mechanism of action | Effect on ribosome | Reference |
| CHX | Reversible | Binds E site, 60S ribosome subunit | Blocks translocation during elongation; stabilizes polysomes | Schneider-Poetsch et al., 2010 |
| Emetine | Irreversible | Binds 40S ribosome subunit | Freezes translation during elongation; stabilizes polysomes | Jiménez et al., 1977 |
| Anisomycin | Reversible | Binds A site, 60S ribosome subunit; competes with PMY | Blocks peptidyl transferase activity; stabilizes polysomes | Hansen et al., 2003 |
| Pactamycin | Reversible | Binds 40S ribosome subunit in initiation complex | Blocks initiation step of translation; inhibits polysome formation | Kappen et al., 1973 |
| Sodium arsenite | Reversible | Oxidative stressor induces eIF2a phosphorylation | Induces stress granule; inhibits polysome formation | Kedersha et al., 2002 |
| Harringtonine | Partially reversible | Prevents peptide bond formation at the initiation complex | Blocks initiation step of translation; inhibits polysome formation | Fresno et al., 1977 |
To further establish the biochemical mechanism of puromycylation in the presence of CHX or emetine, we developed an ELISA that measures puromycylation on nascent chains present on ribosomes extracted from cells. We isolated ribosomes by fractionating HeLa cell lysates on a 15–50% sucrose gradient (Stephens et al., 2008), bound gradient fractions to polyvinylidene fluoride (PVDF) in a 96-well format, briefly incubated with PMY, and detected bound PMY by ELISA. This revealed that anti-PMY mAb binding is proportional to the amounts of polysomes (as opposed to monosomes) in the gradient fractions (Fig. 1 C). Concurrent ELISA for the large ribosome subunit acidic P proteins (consisting of P0, P1, and P2, for which highly specific antibodies in the form of human autoimmune anti–ribosomal P sera are commercially available) demonstrates that the failure to detect PMY in monosome (fraction 5) and free large subunit (fraction 4) fractions cannot be attributed to the lack of binding of ribosomes to PVDF. Treating cells with emetine before lysis increased the ratio of polysomes to monosomes and recapitulated the relationship between PMY binding and polysome abundance.
Anti-PMY immunoblotting of monosome- and polysome-containing fractions exposed to PMY in solution confirmed the lack of puromycylated proteins in the 80S (monosome) fraction and revealed parallel increases in puromycylated nascent chains with polysome sedimentation (Fig. 1 D). Blotting with anti–ribosomal P antibodies confirmed the presence of ribosomal proteins on the PVDF immunoblotting membrane for monosome fractions (Fig. 1 D).
These findings demonstrate that PMY binding to ribosomes correlates with the presence of puromycylated nascent chains and is not affected by emetine modification of ribosomes. Although PMY is capable of binding to ribosomes without puromycylation, we note that this interaction is of relatively low affinity (high micromolar) and is rapidly reversible (accounting for the reversibility of PMY on protein synthesis), which will not be detected when PMY is removed and ribosomes are subjected to multiple high volume washes. The low affinity binding of PMY with ribosomes is why previous investigators turned to N-bromoacetyl PMY to permanently label ribosomes for structural experiments by covalently linking PMY to ribosomes (Lührmann et al., 1981).
Functional evidence for stability of puromycylated nascent chain–ribosome association
Having established that PMY will only stably bind ribosomes via nascent chain puromycylation, we next used the surface sensing of translation (SUnSET) method (Schmidt et al., 2009) to test whether puromycylated nascent chains are released in the presence of emetine. We pulsed cells for 10 min with PMY, washed to remove free PMY, incubated cells for 30 min at 37°C in the presence of various inhibitors, and measured the amount of puromycylated proteins transported to the cell surface via flow cytometry using 12D10 (Figs. 1 E and S1 B; Schmidt et al., 2009). Because puromycylated proteins reach the surface via the Golgi complex, blocking export from the ER with brefeldin A (BFA) sets the baseline level for complete blockade of nascent protein cell surface expression. As expected, 12D10 staining with cells incubated with BFA is equivalent to cells incubated at 4°C, which also prevents exocytosis.
Importantly, PMY labeling cells in the presence of emetine reduced PMY surface staining to background levels obtained with labeling with BFA, functionally demonstrating that emetine completely blocks release of puromycylated nascent chains by this measure. Adding emetine only during the chase provides a blockade indistinguishable from adding BFA only during the chase, showing that nascent chains remain associated with ribosomes for at least several minutes after puromycylation.
These findings demonstrate that emetine prevents the exocytosis of PMY-labeled nascent chains, consistent with the interpretation that they remain associated with ribosomes. Although SUnSET exclusively detects plasma membrane proteins with surface-exposed C termini, it is likely that other proteins are similarly retained on ribosomes, supporting the use of the RPM to detect translating ribosomes.
RPM visualizes active translation
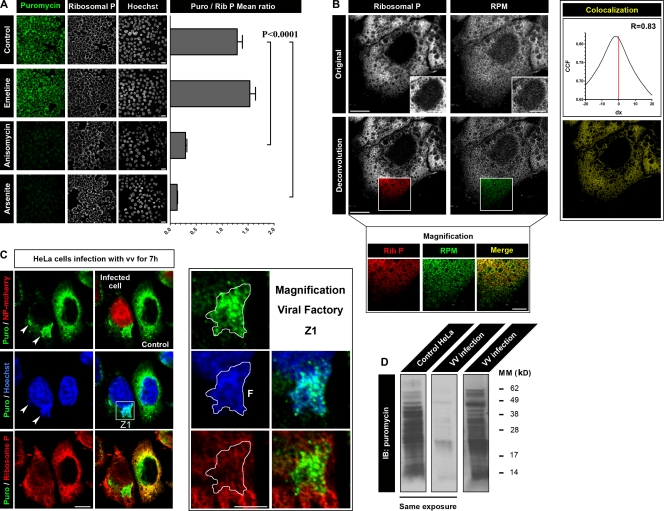
We next visualized ribosome-associated nascent chains in cells via laser-scanning confocal microscopy by incubating HeLa cells with PMY and CHX for 5 min at 37°C (all the images that follow were generated by laser-scanning confocal microscopy). We then treated cells with digitonin to release PMY not associated with nascent chains, fixed cells with PFA, and performed standard indirect immunofluorescence with the anti-PMY mAb and anti–ribosomal P antibodies. Arsenite-induced translation inhibition reduced anti-PMY staining to background levels (Fig. 2 A). Emetine pretreatment increased staining, as opposed to its inhibition by anisomycin, which competes with PMY for binding to ribosomes.
Figure 2.
Live-cell RPM detects translating ribosomes. (A) HeLa cells pretreated for 15 min with the inhibitor indicated were incubated with PMY (Puro) for 5 min at 37°C before digitonin extraction and PFA fixation. Samples were then incubated with 12D10 to detect PMY. For each condition, multiple fields were acquired, and the mean fluorescence ratio of PMY/ribosome staining for each field was quantitated using ImageJ. Values are plotted on the right (means ± SEM). Statistical analysis, two-tailed unpaired t test. Bars, 20 µm (B) Higher magnification images of a HeLa cell pretreated with emetine and labeled live with PMY at 37°C as in A. (bottom) The same image after deconvolution to minimize unfocused light from other focal planes. Nuclear magnification in top insets show weak but well above background RPM and ribosomal P (Rib P) labeling. Although overall colocalization is extensive (right column), magnification shows that there are imbalances in intensity of the staining. Bars: (main images and top insets) 10 µm; (bottom magnification) 5 µm. (C) Active translation was localized in HeLa cells 7 h after infection with a recombinant VV (rVV) expressing NP-mCherry by live RPM staining. In the images shown, only one of the two cells is infected, as demonstrated by the presence of NP-mCherry in the nucleus. Translation is nearly completely localized to viral factories in infected cells, identified as juxtanuclear DNA-positive structures (arrowheads). Even within a factory, ribosomes display different RPM intensities, indicative of differential translation rates and seen in the zoomed image, Z1. Bars: (main images) 10 µm; (Z1 images) 2 µm. F, factory. (D) Uninfected or VV-infected HeLa cells were labeled with PMY after pretreatment with emetine and were extracted directly in culture flasks with 1% NP-40. Puromycylated nascent chains were analyzed by immunoblotting with 12D10. VV infection diminishes the total PMY signal consistent with its effect on overall translation, and the translation profile differs from uninfected cells, consistent with detection of viral versus host proteins, as clearly shown from a longer exposure (right) to normalize levels of puromycylation. CCF, cross-correlation function; IB, immunoblot; MM, molecular mass.
We correlated the effects of detergent extraction of ribosomes and puromycylated nascent chains on immunofluorescence versus immunoblotting (Fig. S2). Digitonin treatment results in minimal loss of ribosomes and no detectable release of puromycylated nascent chains. In contrast, treating cells with NP-40 releases both ribosomes and puromycylated nascent chains and greatly reduces cytoplasmic immunofluorescence (see next section). This provides biochemical evidence supporting the conclusion that puromycylated nascent chains remain tethered to ribosomes.
High resolution confocal imaging (Fig. 2 B) shows considerable but incomplete colocalization of PMY and ribosomal P, expected because of the absence of ribosomal P from some ribosomes (Gonzalo and Reboud, 2003) as well as the detection of nontranslating monosomes, free ribosome heavy subunits, and free ribosomal P by anti–ribosomal P antibodies. The ability of the RPM to discriminate translating from nontranslating ribosomes is demonstrated in vaccinia virus (VV)–infected cells (Fig. 2 C). VV, like many viruses, shuts down host protein synthesis and shifts translation to viral mRNAs. VV assembles in cytoplasmic “factories,” identified by cytoplasmic DNA (stained by Hoechst 3358; Fig. 2 C, arrowheads) that recruit ribosomes and translation factors (Katsafanas and Moss, 2007), implicating factories as sites of viral protein synthesis. 7 h after infection with VV expressing the karyophilic fusion protein influenza A virus (IAV) nucleoprotein (NP)-mCherry, translation principally localizes to a large DNA containing a viral factory (Fig. 2 C, magnification Z1). Notably, many ribosomes remain outside of viral factories and now do not stain with anti-PMY antibodies, indicating the specificity of the RPM for translating ribosomes. Anti-PMY immunoblotting of PMY-exposed VV-infected versus uninfected HeLa cells confirms that VV infection results in a diminished overall translation rate caused by host shutdown and also a distinct pattern of protein puromycylation, consistent with translation of viral proteins (Fig. 2 D).
The RPM detects robust nuclear translation in HeLa cells
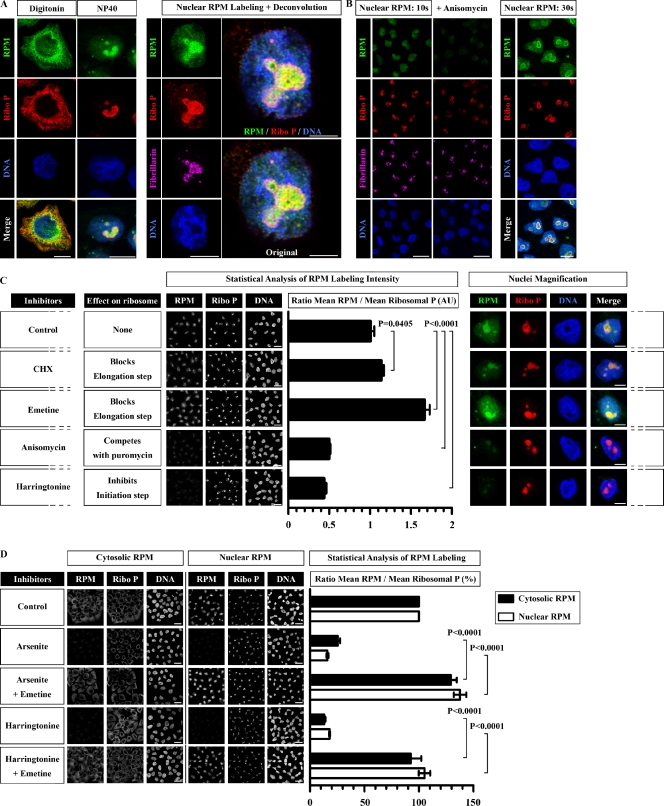
We frequently noted a clear above background but less intense PMY and ribosomal P staining of the HeLa cell nucleoplasm using the digitonin RPM protocol. Although nuclei harbor large numbers of ribosomes, particularly in nucleoli in which ribosomes are assembled, digitonin poorly permeabilizes the nuclear membrane and nucleoplasm (Griffiths, 1993), limiting antibody access to the nucleus and particularly the nucleolus, which is not stained by antibodies to ribosomal P proteins (Fig. 3 A). NP-40 permeabilization of nuclear membranes before fixation greatly increased nuclear RPM staining, particularly in nucleoli identified by diminished DNA staining and binding of antibodies specific for the nucleolar protein fibrillarin (Fig. 3 A). A potential artifact of nuclear translation is that the puromycylated nuclear proteins detected are synthesized by cytoplasmic ribosomes and transported to the nucleus during the labeling period. This artifact is unlikely to be a problem with the RPM because, as we show functionally (Fig. 1 E) and biochemically (Fig. S2), puromycylated nascent chains remain tethered to ribosomes by translation elongation inhibitors and cannot traffic to nuclei. To be more certain, however, we determined that incubating cells with PMY for as short as 10 s suffices to clearly label the nuclear body and nucleoli, with intense staining occurring after only 30 s of PMY exposure (Fig. 3 B).
Figure 3.
RPM detects nuclear/nucleolar translation. (A) HeLa cells were labeled live with emetine and PMY and processed for RPM staining using the regular digitonin extraction method or NP-40 extraction before fixation. (right) Higher magnification series of the nucleus stained after NP-40 extraction demonstrates intense RPM staining of a fibrillarin-positive nucleolus. Merged image is shown with and without deconvolution to minimize unfocused light. Bars: (main images) 10 µm; (large images) 5 µm. (B) HeLa cells were labeled with PMY for 10 or 30 s before NP-40 extraction. Pretreating cells with anisomycin, a competitive inhibitor of PMY, reduces RPM to background values, demonstrating the ribosome dependence of nuclear RPM staining. Bars: (left) 20 µm; (right) 10 µm. (C) HeLa cells pretreated for 20 min with the inhibitor indicated were incubated for 5 min with PMY before RPM staining using NP-40 extraction. Five fields were acquired for each condition, and the mean fluorescence ratio of PMY/ribosome staining for each field was quantitated using ImageJ software. Values are plotted on the right (means ± SEM). Statistical analysis, two-tailed unpaired t test. Bars: (left) 20 µm; (right) 5 µm. (D) HeLa cells pretreated for 20 min with the inhibitors indicated were incubated for 5 min with PMY before RPM staining using either digitonin or NP-40 extraction. Emetine blocks the inhibitory effects of arsenite or harringtonine on cytoplasmic and nuclear RPM staining (digitonin vs. NP-40 extraction), demonstrating that RPM staining is based on the presence of ribosome-associated nascent chains. As for C, five fields were acquired for each condition, and the mean fluorescence ratio of PMY/ribosome staining for each field was quantitated using ImageJ. Values are plotted on the right (means ± SEM), considering the control ratio as 100%. Statistical analysis, two-tailed unpaired t test. Bars, 20 µm. AU, arbitrary unit; Ribo P, ribosomal P.
Nuclear RPM is caused by nuclear protein synthesis and not import or trapping of cytoplasmic puromycylated nascent chains
Four lines of evidence support the conclusion that the RPM detects puromycylation of nascent chains in nuclei/nucleoli and not PMY bound to other cellular structures or puromycylated proteins that originate in the cytosol. First, CHX and emetine enhance RPM staining of the nucleoplasm and nucleolus (Fig. 3 C). If RPM nuclear staining is caused by transport of cytoplasmic puromycylated proteins, these inhibitors would reduce and not increase nuclear staining because they inhibit release of puromycylated proteins from cytoplasmic ribosomes. Second, anisomycin, which competes with PMY for A-site binding, blocks nuclear RPM staining, linking staining to ribosomal-based catalysis and essentially ruling out PMY association with other nuclear targets (Fig. 3, B and C) because PMY and anisomycin are structurally dissimilar and unlikely to competitively bind the same off-target nuclear structure.
Third, harringtonine, which blocks initial peptide bond formation but allows completion of nascent chains and induces ribosome dissociation (Table 1), inhibits nuclear RPM staining, demonstrating that PMY staining does not reflect PMY binding to nuclear ribosomes (already extremely unlikely because of the low binding affinity of PMY for ribosomes in the absence of the nascent chain to puromycylate). Moreover, harringtonine inhibition of nuclear RPM staining (or cytoplasmic staining) is overridden if chain elongation is blocked by simultaneous addition of emetine (Fig. 3 D). Thus, harringtonine inhibition of nuclear RPM staining is caused by the specific absence of nascent chains on nuclear and cytoplasmic ribosomes and not other effects on cells. We made identical findings using emetine with arsenite as an alternative treatment to convert protein-synthesizing polysomes to translationally inactive monosomes (Fig. 3 D).
Fourth, blocking translation initiation via VV shutdown of host protein synthesis inhibits nuclear RPM staining in parallel with inhibiting synthesis outside of viral factories, when cells are examined 2, 4, and 5 h after infection (Fig. 4, A and B). Furthermore, nuclear RPM staining is blocked in parallel with the shutoff of host protein synthesis mediated by three RNA-different viruses. Vesicular stomatitis virus (VSV; a rhabdovirus; Fig. 4 C) and Semliki forest virus (SFV; an alphavirus; Fig. 4 D) nearly completely abrogate RPM staining of the nucleoplasm and nucleolus. IAV (an orthomyxovirus) selectively inhibits nucleolar RPM, sparing RPM staining in the nucleoplasm (Fig. 4 E).
Figure 4.
Stress-regulated nuclear translation detected by RPM. (A) Uninfected HeLa cells or HeLa cells infected for 2, 4, or 5 h with rVV expressing NP-mCherry at a high MOI to infect all cells in fields before digitonin (cytoplasmic)- or NP-40 (nuclear)–based RPM staining. It is clear that nuclear RPM staining decreases in parallel with shunting of protein synthesis to viral factories and increased NP-mCherry expression. (B) HeLa cells were infected with rVV expressing NP-mCherry at a lower MOI to infect ∼50% of cells before nuclear RPM staining (NP-40 extraction). (left) At 6 h after infection, the three infected cells of the six in the field demonstrate greatly reduced RPM staining in the nucleoplasm and, particularly, nucleoli. At 7 h after infection, one infected cell (identified by its DNA labeled viral factory) of two in the field demonstrates no detectable nuclear RPM staining. Arrowheads show the nucleus of VV-infected cells as identified by DNA staining of factory. (C) Uninfected HeLa cells or HeLa cells infected for 7 h with recombinant VSV expressing NP-EGFP before RPM staining and extraction with either digitonin (cytoplasmic RPM) or NP-40 (nuclear RPM). Five fields were acquired for each condition, and the mean fluorescence ratio of PMY/ribosome staining for each field was quantitated using ImageJ. Values are plotted on the right (means ± SEM). Statistical analysis, two-tailed unpaired t test. VSV infection clearly results in diminished nuclear translation despite robust cytoplasmic synthesis of viral proteins, further ruling out the possibility that the nuclear/nucleolar RPM is caused by trafficking/trapping of cytoplasmic puromycylated proteins. (D) Uninfected HeLa cells or HeLa cells infected for 7 h with recombinant SFV expressing IAV NP before RPM staining and extraction with either digitonin (cytoplasmic RPM) or NP-40 (nuclear RPM). As previously reported (Berglund et al., 2007), when expressed by SFV, NP principally localizes to mitochondria because of downstream initiation (cytoplasmic staining), with a minor full-length cohort localizing to the nucleus (nuclear staining). In SFV-infected cells (arrowheads), it is clear that both cytoplasmic and nuclear RPM staining are greatly diminished compared with surrounding uninfected cells. High magnification of cytoplasmic RPM staining reveals multiple translation foci in which viral proteins are likely translated. (E) HeLa cells either pretreated for 15 min with the inhibitor indicated or infected with IAV (Flu) were incubated with PMY for 5 min at 37°C in the presence of emetine. In this experiment, cells were simultaneously fixed and permeabilized by incubation with PBS containing NP-40 and PFA. This allows antibody access to nucleoli and fixes cytoplasmic ribosomes to enable direct comparison between nuclear and cytoplasmic RPMs. Cells were stained for PMY (green), large ribosomal subunit (red), DNA, and fibrillarin. This experiment shows that the nucleolar RPM staining is robust compared with the cytoplasmic RPM and is inhibited in parallel with inhibitors that block protein synthesis. Diminished nucleolar RPM in IAV-infected cells provides additional evidence against trapping of cytoplasmic puromycylated proteins in the nucleoli during fixation. Bars, 10 µm. AU, arbitrary units; Ribo P, ribosomal P.
The four viruses examined inhibit host protein synthesis by distinct mechanisms (Bushell and Sarnow, 2002), minimizing the possibility that their nuclear RPM staining inhibition is caused by other effects that the viruses have on host cell metabolism or structure. For each virus infection, nucleoli are intact, as clearly shown by either fibrillarin or ribosomal P staining, so the lack of nucleolar staining cannot be attributed to a lack of nucleoli themselves. For VV and SFV, which greatly reduce cytoplasmic RPM staining, surrounding uninfected cells provide an abundant source of puromycylated proteins yet do not result in nuclear staining of infected cells, providing further evidence against trapping of puromycylated cytoplasmic proteins in the nucleoplasm/nucleolus. The same is true for IAV and VSV, in which robust cytoplasmic RPM staining provides a more proximal source of puromycylated nascent chains.
To approximate the ratio of nuclear to cytoplasmic RPM staining, we developed a cofixation/NP-40 permeabilization modified protocol (simultaneously extracting cells with NP-40 while fixing with 3% PFA) that enables simultaneous visualization of staining in the nucleus and cytoplasm and recapitulates the enhancing effect of emetine and inhibiting effects of harringtonine and arsenite on a nuclear/cytoplasmic RPM (Fig. 4 E). Notably, RPM nuclear and cytoplasmic signals were similar to the values obtained via sequential procedures using NP-40 and digitonin, respectively. The cofixation/permeabilization method confirmed that nuclear RPM staining represents a significant fraction of a total cellular RPM.
Using this method, we also found a selective effect of IAV in blocking nucleolar but not nucleoplasmic or cytoplasmic RPM staining. This provides strong evidence against nucleolar trapping of puromycylated proteins translated elsewhere, particularly because IAV encodes seven proteins targeted to the nucleus (NS1, NP, M1, PB1-F2, and the three polymerases), three of which are translated at high levels (NS1, NP, and M1).
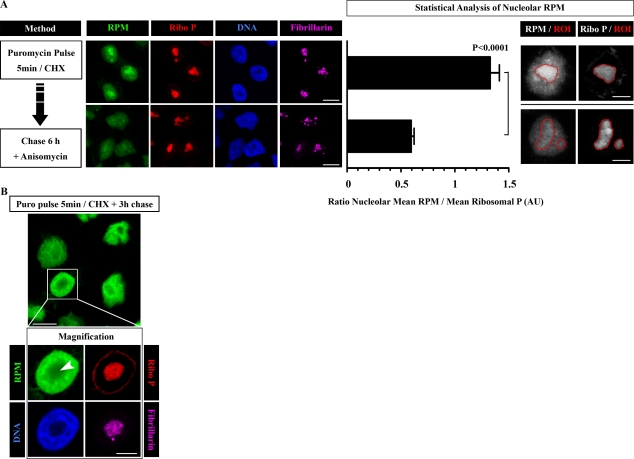
To “chase” nucleolar RPM staining, we incubated cells with anisomycin after PMY pulse. This was necessary to block further puromycylation, whose occurrence we hypothesize reflects the creation of an intracellular PMY depot during pulsing. After 6-h incubation, nucleolar PMY staining was significantly diminished, whereas staining in the nucleoplasm was constant (Fig. 5 A). Even in the absence of anisomycin during the chase, the balance of RPM staining moves from the nucleoli to the nuclear body (Fig. 5 B), providing additional evidence that the nucleolar RPM staining is not caused by import or trapping of puromycylated proteins from the nucleoplasm.
Figure 5.
Nucleolar RPM labeling can be chased. (A) HeLa cells were pulse labeled for 5 min with PMY in the presence of CHX and then chased for 6 h in the presence of anisomycin to prevent further PMY incorporation. For time 0 and time 6 h, cells were RPM stained using NP-40 extraction. Nucleolar RPM staining was measured by defining regions of interest (ROIs) around 15 different nucleoli and calculating the ratio between RPM and ribosomal P (Ribo P) nucleolar mean intensities (mean ± SEM). Statistical analyses, two-tailed unpaired t test. (B) HeLa cells were labeled with PMY CHX for 5 min and then chased for 3 h in the absence of inhibitors. Cells were RPM stained using NP-40 extraction. The arrowhead shows the nucleolus. Bars: (A [left] and B [main image]) 10 µm; (A [right] and B [magnification]) 5 µm. AU, arbitrary unit.
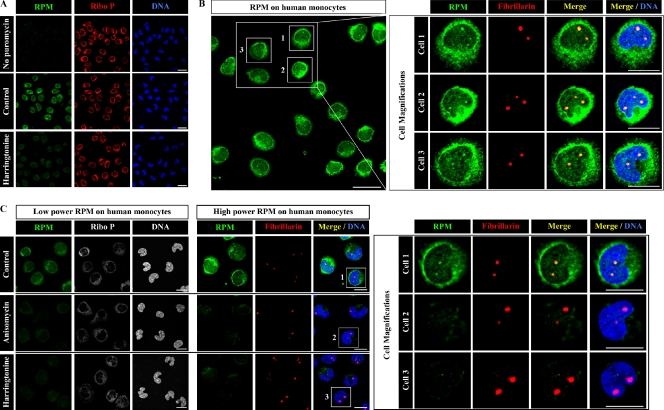
RPM staining detects nuclear translation in human monocytes
HeLa cells have been cultured for dozens of generations, are highly aneuploid, and are likely to demonstrate aberrant translation in some aspects relative to normal vertebrate cells. To show that nuclear translation is a normal process in human cells, we performed the RPM using purified blood monocytes from healthy donors (Fig. 6 A). As with HeLa cells, monocytes demonstrate clear staining of the nucleoplasm and nucleoli after 5-min exposure to PMY and emetine (Fig. 6 B). Moreover, anisomycin and harringtonine each inhibit both cytoplasmic and nuclear RPM staining (Fig. 6 C), recapitulating our previous observations in HeLa cells. Thus, nuclear translation also occurs in normal human cells ex vivo.
Figure 6.
Nuclear RPM staining in monocytes. (A) Elutriated human peripheral blood monocytes pretreated or not pretreated with harringtonine 15 min before RPM staining (5-min PMY pulse in the presence of emetine) were fixed and extracted simultaneously with polysome buffer containing PFA and digitonin. Note that in the absence of PMY no RPM staining is visible. (B) Same PMY staining protocol as in A. Intense RPM staining clearly colocalizes with nucleoli (red), which are smaller in monocytes than HeLa cells as shown by fibrillarin staining. (C) Elutriated monocytes pretreated or not pretreated with harringtonine or anisomycin for 15 min before RPM staining were fixed and extracted simultaneously with polysome buffer containing PFA and digitonin. RPM staining in cytoplasm and nucleoli is blocked by either inhibitor as seen in increasing magnifications from left to right, demonstrating the dependence of ribosome-catalyzed puromycylation of the nascent chain. Bars: (A and B [main image]) 20 µm; (B [magnification] and C) 10 µm. Ribo P, ribosomal P.
Discussion
The RPM is a unique tool for visualizing translation sites that will facilitate understanding of translation compartmentalization, as illustrated by selective translation activity at VV factories and clear evidence for translation in the nucleoplasm and nucleolus. The principal advantage of the RPM over traditional methods using radiolabeled or modified amino acids is that the RPM detects nascent chains attached to translating ribosomes without detecting released polypeptides. Even during a brief exposure to tagged amino acids, some chains will terminate (∼1% per second, given a mean protein of 500 residues and a five-residue translation rate per second), meaning that some of the nascent chains detected were released from ribosomes. We provide clear functional evidence that puromycylated nascent chains remain associated with ribosomes when cells are preincubated with chain elongation inhibitors (Fig. 1 E).
The RPM provides direct histological evidence for the existence of nuclear translation. We find that nuclear RPM staining mostly occurs in the nucleolus, echoing initial autoradiography experiments demonstrating intense nucleolar labeling after a brief exposure of cells to tritiated amino acids (Birnstiel, 1967). Despite skepticism regarding nuclear translation (Dahlberg et al., 2003; Nathanson et al., 2003; Dahlberg and Lund, 2004), an ever increasing list of translation components has been detected in the nucleus and nucleolus, including amino acid–acylated tRNA, aminoacyl-tRNA synthetases, and translation initiation factors (Lejbkowicz et al., 1992; Lund and Dahlberg, 1998; Dostie et al., 2000; Nathanson and Deutscher, 2000; Iborra et al., 2001; McKendrick et al., 2001; Ferraiuolo et al., 2004; Gunasekera et al., 2004).
An obvious future extension of our findings entails identifying the proteins translated in the nucleus. Initial studies describing nucleolar protein synthesis suggest that histones are synthesized in nucleoli (Birnstiel and Flamm, 1964; Zimmerman et al., 1969). To maximize efficiency, it is logical that some nuclear/nucleolar proteins, including ribosome subunits themselves, would be synthesized at the site of ribosome assembly in the nucleolus. An intriguing possibility is the suggestion that newly assembled ribosomes are quality control tested for protein synthetic capacity before their export from the nucleolus (Pederson and Politz, 2000).
Part of the nuclear translation controversy is the extent to which nonsense-mediated decay (NMD) occurs in the nucleus (Bhalla et al., 2009) or nucleolus (Kim et al., 2009), so this is an obvious area for future exploration. Nuclear translation of nascent messages is a parsimonious explanation for NMD regulation of mRNA in the nucleus itself and for the related phenomenon of nonsense-associated altered splicing of mRNAs with premature termination codons (Wang et al., 2002). Nuclear synthesis of defective ribosomal products via NMD pioneer round translation (Apcher et al., 2011) may generate peptides presented by major histocompatibility complex class I molecules for immunosurveillance (Yewdell and Nicchitta, 2006). Peptides might also be generated in the nucleus by the “translasome,” a supercomplex of ribosomes, initiation factors, aminoacyl-tRNA synthetases, and proteasomes (Sha et al., 2009). We recently provided evidence that antigenic peptides might be synthesized in the nucleus of IAV-infected cells (Dolan et al., 2010), which would be consistent with the continued protein synthesis in the nucleoplasm of the IAV-infected cells we observe here.
Although we have limited our experiments to mammalian cells and focused on nuclear translation, the RPM can potentially be applied to all cell types with ribosomes that catalyze puromycylation, including invertebrate multicellular organisms and single-cell eukaryotic and prokaryotic organisms. We believe that the RPM will catalyze discoveries that increase understanding of the role of translation compartmentalization in cellular function and translational control in multicellular organisms.
Materials and methods
Cells and treatments
HeLa cells (American Type Culture Collection) were propagated in DME and 7.5% FBS and passaged 1 d before each experiment. Drugs were used at the indicated concentrations: 355 µM CHX (Sigma-Aldrich), 208 µM emetine (EMD), 9.4 µM anisomycin (EMD), 0.5 µM pactamycin, 500 µM sodium arsenite (Sigma-Aldrich), and 17.8 µM BFA (Sigma-Aldrich). Elutriated monocytes were obtained from healthy anonymous donors at the National Institutes of Health blood bank. After collection, cells were frozen in FBS supplemented with 10% DMSO. For culture, 107 cells were thawed and plated in 10 ml serum-free RPMI 1640 for 90 min at 37°C, and nonattached cells were removed by aspiration. 10 ml of fresh RPMI 1640 was added to attached monocytes, and cells were cultured for 3 h at 37°C before the RPM.
VV infection.
HeLa cells were infected with VV at a multiplicity of 1 plaque-forming unit (pfu)/cell in saline supplemented with 0.1% BSA. After adsorption at 37°C for 1 h, infected monolayers were overlaid with DME containing 7.5% FBS and incubated for an additional 6 h.
VSV infection.
HeLa cells were infected with VSV at a multiplicity of 10 pfu/cell in DME. After adsorption at 37°C for 1 h, infected monolayers were overlaid with DME containing 7.5% FBS and incubated for an additional 6 h.
SFV infection.
SFV was activated with 0.5 mg/ml chymotrypsin (Sigma-Aldrich) for 40 min on ice followed by 10-min incubation with 0.4 mg/ml aprotinin (Sigma-Aldrich). Then, HeLa cells were infected with SFV in DME. After adsorption at 37°C for 90 min, infected monolayers were overlaid with DME containing 7.5% FBS and incubated for an additional 5.5 h.
IAV infection.
HeLa cells were infected with the Influenza A/Puerto Rico/8/34 strain at a multiplicity of 10 pfu/cell in Autopow infection medium, pH 6.8. After adsorption at 37°C for 1 h, infected monolayers were overlaid with DME containing 7.5% FBS and incubated for an additional 5 h.
Ribosome ELISA
2 × 106 HeLa cells were used per condition. Cells were treated for 15 min with various inhibitors, trypsinized, and washed twice with cold PBS. Cells were resuspended in 1 ml of ice-cold homogenization buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 0.2 M sucrose [MP Biomedicals], 0.5% NP-40 [Thermo Fisher Scientific], 100 µg/ml CHX, EDTA-free protease inhibitors [Roche], 10 U/ml RNaseOut [Invitrogen], and diethylpyrocarbonate water). Cells were stroked 10 times with a stainless steel pestle-type homogenizer on ice, and cell lysates were centrifuged (centrifuge 5417 R; rotor F45-30-11; Eppendorf) at 13,000 rpm for 10 min at 4°C. Lysates were layered over a 15–50% sucrose gradient in polysome buffer (homogenization buffer without NP-40). After centrifugation at 35,000 rpm (rotor SW 41 Ti; Beckman Coulter) for 2.5 h at 4°C, each gradient was fractionated. 10 fractions were collected manually from the top of the gradient, and 100 µl of each fraction was added to wells of a 96-well PVDF plate (MultiScreenHTS; Millipore). Two identical plates were loaded, and one of them was used as a control. Each plate was incubated for 10 min on ice, vacuumed, and washed once with 200 µl/well polysome buffer. Then, 100 µl PMY (91 µM) was added in polysome buffer to the experimental plate and the polysome buffer in the control plate. Plates were incubated for 10 min on ice and washed three times with 200 µl polysome buffer, and 100 µl of 3% PFA was added before incubating plates for 10 min. Wells were washed with PBS, incubated with 100 µl blocking buffer (StartingBlock; Thermo Fisher Scientific) for 30 min at RT, vacuumed, and incubated for 1 h with 2.5 µg/ml anti-PMY mAb at RT. Next, wells were washed three times with PBS and incubated with secondary rabbit anti–mouse HRP conjugate (Cappel; MP Biomedicals) for 1 h at RT. 100 µl 3,3,5,5-tetramethylbenzidine substrate (KPL) was added to each well, and plates were incubated for 10 min at RT before stopping the reaction using 50 µl of 1.6% HCl. The liquid in each well was transferred to a plastic plate with a transparent bottom, and HRP activity was measured by absorbance at 450 nm. Levels of background staining were determined using the mean of unlabeled wells from the control plate and subtracted from test values. Data were graphed using Prism software (GraphPad Software).
RPM
HeLa cells were transferred to coverslips the day before the experiment to reach 60–80% confluence at the time of the experiment. Coverslips were transferred to 24-well plates. Then, depending on the experiment, cells were pretreated with different inhibitors. HeLa cells on coverslips were incubated with DME with 7.5% FBS supplemented with 91 µM PMY and 208 µM emetine for 5 min at 37°C. All extraction procedures were performed on ice using reagents prechilled to ice temperature. Cells were incubated for 2 min with 500 µl/well permeabilization buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 355 µM CHX, EDTA-free protease inhibitors, and 10 U/ml RNaseOut containing 0.015% digitonin [Wako Chemicals USA]). After this extraction step, cells were washed once with 500 µl polysome buffer and fixed with 500 µl of 3% PFA (Electron Microscopy Sciences) for 15 min at RT. PFA was aspirated, PBS was added, and cells were maintained at 4°C before immunofluorescence staining. In place of the regular digitonin extraction, a coextraction/fixation procedure can be performed (Fig. 4 D) using polysome buffer supplemented with 1% NP-40 and 3% PFA for 20 min on ice.
RPM of monocytes.
Human peripheral blood monocytes were incubated on coverslips pretreated with Alcian blue (Sigma-Aldrich) in warm RPMI 1640 supplemented with Hepes for 5 min. Cells were pretreated or not pretreated with harringtonine or anisomycin for 15 min. Then, warm labeling medium (RPMI 1640, 208 µM emetine, and 91 µM PMY) was added for 5 min. Cells were washed with cold PBS and fixed/permeabilized in polysome buffer supplemented with 0.015% digitonin and 3% PFA for 20 min on ice.
Nuclear RPM.
HeLa cells on coverslips were incubated with DME and 7.5% FBS supplemented with 91 µM PMY and 208 µM emetine for 10 s to 5 min at 37°C. Cells were then incubated on ice for 5 min with 1% (vol/vol) NP-40, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and EDTA-free protease inhibitors, gently washed once with the same buffer lacking NP-40, and fixed with 3% PFA for 15 min at RT before staining.
For chase experiments, HeLa cells on coverslips were labeled with PMY and CHX for 5 min at 37°C, washed twice with cold DME, and chased with warm DME and 7.5% FBS supplemented or not supplemented with 9.4 µM anisomycin. After three or six chases, cells were then NP-40 extracted and PFA fixed as described in the preceding paragraph.
Antibodies
The mouse PMY-specific mAb (clone 12D10) has been previously described (Schmidt et al., 2009). Since then, our laboratory developed two other anti-PMY mAbs (2A4 and 5B12) with indistinguishable properties in immunoblots and immunofluorescence (Fig. S3). These mAbs will be available to the scientific community after signing a standard National Institutes of Health material transfer agreement. The anti–ribosomal P antibody is a human polyclonal autoimmune antisera (Immunovision). As secondary antibodies, we used goat anti–mouse A488 (Invitrogen), donkey anti–rabbit Texas red, and donkey anti–human Cy5 (Jackson ImmunoResearch Laboratories, Inc.). DNA was labeled with Hoechst 3358 (Invitrogen). Polyclonal rabbit anti–mouse Ig–HRP (Dako) and goat anti–human Ig–HRP (Jackson ImmunoResearch Laboratories, Inc.) were used for immunoblotting.
SUnSET
HeLa cells in suspension were incubated for 10 min at 37°C with or without 4.5 µM PMY supplemented or not supplemented with emetine or 17.8 µM BFA. Cells were washed twice and incubated for 30 min at 37°C in DME/7.5% FBS supplemented or not supplemented with emetine or BFA. Control samples (pulse only) were immediately shifted to ice. Cells were stained with anti-PMY mAb 2A4 and secondary antibody anti–mouse Alexa Fluor 647 (Invitrogen). Each condition was performed independently. The mean fluorescence intensity from PMY-unexposed HeLa cells was subtracted from each value.
Immunofluorescence and microscopy
All staining was performed using staining buffer (SB; 0.05% saponin, 10 mM glycine, 5% FBS, and PBS) as previously described (Lelouard et al., 2004). In brief, coverslips were incubated in SB for 15 min and then incubated with antibodies diluted in SB for 1 h at RT. After washing three times with PBS, coverslips were incubated with secondary antibodies diluted in SB for 1 h at RT. Coverslips were washed twice and incubated with 1 µg/ml Hoechst 3358 diluted in SB for 5 min at RT. Coverslips were washed twice with distilled water and mounted on slides with Fluoromount-G (SouthernBiotech). Images were acquired at 37°C using a laser-scanning confocal microscope (TCS SP5; Leica) with an HCX Plan Apochromat λ blue 63.0×, 1.40 NA oil UV objective, type FF immersion liquid (Cargille), and LAS AF software (V2.3.1; Leica). Images were processed using Imaris (Bitplane), Huygens Essentials software for image deconvolution using the classic maximum likelihood estimation algorithm (version 3.6; Scientific Volume Imaging), and Photoshop (CS2; Adobe) to change contrast and levels without manipulating the γ function. Each set of images for a given experiment was processed identically to maintain the image intensity ratio. ImageJ (National Institutes of Health) and Prism software were used for quantitation and statistical analysis. Groups were analyzed for statistical significance with a two-tailed unpaired t test. Error bars represent the SEM.
Biochemistry
HeLa cells were extracted with either permeabilization puffer (polysome buffer complemented with 0.015% digitonin) or NP-40 buffer (containing 1% NP-40, 150 mM NaCl, PBS, and protease inhibitors). For analyzing total cell lysates, HeLa cell pellets were directly resuspended in 95°C sample buffer (SDS protein gel loading solution 2×; Quality Biological, Inc.) supplemented with 10 mM DTT.
Extraction protocol for cells in suspension.
HeLa cells (106 cells per condition) were trypsinized and washed twice with PBS. Cells were treated with different inhibitors at 37°C at 106 cells/ml, centrifuged, washed once with warm PBS, and resuspended in 1 ml DME and 7.5% FBS containing 91 µM PMY and 355 µM CHX (except for the control in Fig. 2 A), either on ice (with ice-cooled PMY labeling buffer) or at 37°C (with warm PMY labeling buffer). In both cases, cells were labeled for 5 min (or more for cold labeling), washed twice with cold PBS, and pelleted. Then, cells were resuspended in sample buffer and incubated for 20 min at 95°C before immunoblotting.
Extraction protocol in flasks.
Cells were cultured and incubated with different inhibitors at 37°C, washed once with warm DME and 7.5% FCS, and incubated for 5 min at 37°C with DME and 7.5% FCS containing 91 µM PMY and 355 µM CHX for 5 min. After washing twice with cold PBS supplemented with 355 µM CHX, cells were extracted with 500 µl NP-40 buffer for 5 min on ice. Extracts were collected and centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was removed for immunoblotting.
Two-step extraction protocol in flasks.
HeLa cells were cultured and incubated with different inhibitors at 37°C, washed once with warm DME with 7.5% FCS, and incubated for 5 min at 37°C with DME and 7.5% FCS containing 91 µM PMY and 355 µM CHX for 5 min. After washing twice with cold PBS supplemented with 355 µM CHX, cells were extracted with 500 µl permeabilization buffer for 2 min on ice. Extracts were collected for immunoblotting. Then, cells were extracted again with NP-40 buffer for 5 min on ice. These extracts were also collected for immunoblotting.
Cell fractionation.
HeLa cells were cultured, lysed, and fractionated on 15–50% sucrose gradient as previously described (see Ribosome ELISA). Each fraction was incubated with 91 µM PMY for 20 min on ice. Then, sample buffer was added to each fraction before 10-min incubation at 95°C and immunoblotting.
Immunoblotting.
Samples were electrophoresed in 4–12% NuPAGE polyacrylamide gels (Invitrogen) or 10–20% Tris/glycine polyacrylamide gels (Bio-Rad Laboratories). Proteins were transferred to PVDF membranes (Bio-Rad Laboratories), and membranes were stained for 10 min with Coomassie blue (50% methanol, 10% acetic acid, and 0.05% Coomassie blue R-250 [MP Biomedicals]) and washed with 50% methanol to confirm transfer uniformity. Membranes were incubated with blocking buffer (StartingBlock), and then, 1.66 µg/ml anti-PMY mAb (12D10) and human anti–ribosomal P antibody (1:3,000) were added in buffer (StartingBlock) and incubated overnight at 4°C. After washing three times with wash buffer (PBS and 0.1% Tween 20), secondary antibodies were added in buffer (StartingBlock) and incubated for 1 h. Membranes were washed three times, and ECL substrate (SuperSignal; Thermo Fisher Scientific) was used to detect staining by exposure to x-ray film.
Metabolic labeling.
HeLa cells were trypsinized and washed twice with DME without methionine (Invitrogen), and 106 cells were incubated for 5 min at 37°C in methionine-free DME supplemented with 0.1 mCi/ml [35S]methionine alone, with 91 µM PMY, or PMY with 355 µM CHX. After washing three times with ice-cold PBS, pelleted cells were lysed in 500 µl of 1.5% SDS, 150 mM NaCl, and PBS and incubated for 20 min at 95°C. 2 µl of each sample was applied per spot of a 96-well filter mat (six replicates per condition). After drying at 60°C, the mat was incubated in 10% TCA for 30 min at RT, washed twice in 70% ethanol solution (10 min/wash), dried at 60°C, and placed in a scintillation bag with 5 ml scintillation liquid (PerkinElmer). Radioactivity was quantitated using a β counter liquid scintillation counter (1450 MicroBeta TriLux; PerkinElmer).
Online supplemental material
Fig. S1 shows the effectiveness of PMY. Fig. S2 shows immunoblotting of detergent fractions of PMY-labeled cells. Fig. S3 shows the characterization of anti-PMY mAbs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201112145/DC1.
Acknowledgments
We thank Tom Dever (National Institute of Child Health and Human Development, Rockville, MD) and Jon Dinman (University of Maryland, College Park, MD) for invaluable advice and discussions.
This work was generously supported by the Division of Intramural Research and the National Institute of Allergy and Infectious Diseases and by grants to P. Pierre from Ligue National Contre le Cancer, the Human Frontier Science Program, and the European Network of Excellence DC-THERA.
Footnotes
Abbreviations used in this paper:
- BFA
- brefeldin A
- CHX
- cycloheximide
- IAV
- influenza A virus
- NMD
- nonsense-mediated decay
- NP
- nucleoprotein
- pfu
- plaque-forming unit
- PMY
- puromycin
- PVDF
- polyvinylidene fluoride
- RPM
- ribopuromycylation method
- rVV
- recombinant VV
- SFV
- Semliki forest virus
- SUnSET
- surface sensing of translation
- VSV
- vesicular stomatitis virus
- VV
- vaccinia virus
References
- Allfrey V.G. 1954. Amino acid incorporation by isolated thymus nuclei. I. The role of desoxyribonucleic acid in protein synthesis. Proc. Natl. Acad. Sci. USA. 40:881–885 10.1073/pnas.40.10.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey V.G., Mirsky A.E., Osawa S. 1955. Protein synthesis in isolated cell nuclei. Nature. 176:1042–1049 10.1038/1761042a0 [DOI] [PubMed] [Google Scholar]
- Apcher S., Daskalogianni C., Lejeune F., Manoury B., Imhoos G., Heslop L., Fåhraeus R. 2011. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc. Natl. Acad. Sci. USA. 108:11572–11577 10.1073/pnas.1104104108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund P., Finzi D., Bennink J.R., Yewdell J.W. 2007. Viral alteration of cellular translational machinery increases defective ribosomal products. J. Virol. 81:7220–7229 10.1128/JVI.00137-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla A.D., Gudikote J.P., Wang J., Chan W.K., Chang Y.F., Olivas O.R., Wilkinson M.F. 2009. Nonsense codons trigger an RNA partitioning shift. J. Biol. Chem. 284:4062–4072 10.1074/jbc.M805193200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. 1967. The nucleolus in cell metabolism. Annu. Rev. Plant Physiol. 18:25–58 10.1146/annurev.pp.18.060167.000325 [DOI] [Google Scholar]
- Birnstiel M.L., Flamm W.G. 1964. Intranuclear site of histone synthesis. Science. 145:1435–1437 10.1126/science.145.3639.1435 [DOI] [PubMed] [Google Scholar]
- Bushell M., Sarnow P. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395–399 10.1083/jcb.200205044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J.E., Lund E. 2004. Does protein synthesis occur in the nucleus? Curr. Opin. Cell Biol. 16:335–338 10.1016/j.ceb.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Dahlberg J.E., Lund E., Goodwin E.B. 2003. Nuclear translation: what is the evidence? RNA. 9:1–8 10.1261/rna.2121703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan B.P., Knowlton J.J., David A., Bennink J.R., Yewdell J.W. 2010. RNA polymerase II inhibitors dissociate antigenic peptide generation from normal viral protein synthesis: a role for nuclear translation in defective ribosomal product synthesis? J. Immunol. 185:6728–6733 10.4049/jimmunol.1002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Lejbkowicz F., Sonenberg N. 2000. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 148:239–247 10.1083/jcb.148.2.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers D.K., Welch W.J., Hansen W.J. 1997. Complexes between nascent polypeptides and their molecular chaperones in the cytosol of mammalian cells. Mol. Biol. Cell. 8:1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Lee C.S., Ler L.W., Hsu J.L., Costa-Mattioli M., Luo M.J., Reed R., Sonenberg N. 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. USA. 101:4118–4123 10.1073/pnas.0400933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M., Jiménez A., Vázquez D. 1977. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72:323–330 10.1111/j.1432-1033.1977.tb11256.x [DOI] [PubMed] [Google Scholar]
- Gonzalo P., Reboud J.P. 2003. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 95:179–193 10.1016/S0248-4900(03)00034-0 [DOI] [PubMed] [Google Scholar]
- Griffiths G. 1993. Fine Structure Immunocytochemistry. Springer-Verlag, Berlin: 459 pp [Google Scholar]
- Gunasekera N., Lee S.W., Kim S., Musier-Forsyth K., Arriaga E. 2004. Nuclear localization of aminoacyl-tRNA synthetases using single-cell capillary electrophoresis laser-induced fluorescence analysis. Anal. Chem. 76:4741–4746 10.1021/ac049567e [DOI] [PubMed] [Google Scholar]
- Hansen J.L., Moore P.B., Steitz T.A. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061–1075 10.1016/S0022-2836(03)00668-5 [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson D.A., Cook P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science. 293:1139–1142 10.1126/science.1061216 [DOI] [PubMed] [Google Scholar]
- Jiménez A., Carrasco L., Vázquez D. 1977. Enzymic and nonenzymic translocation by yeast polysomes. Site of action of a number of inhibitors. Biochemistry. 16:4727–4730 10.1021/bi00640a030 [DOI] [PubMed] [Google Scholar]
- Kappen L.S., Suzuki H., Goldberg I.H. 1973. Inhibition of reticulocyte peptide-chain initiation by pactamycin: accumulation of inactive ribosomal initiation complexes. Proc. Natl. Acad. Sci. USA. 70:22–26 10.1073/pnas.70.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas G.C., Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2:221–228 10.1016/j.chom.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Koroleva O.A., Lewandowska D., Pendle A.F., Clark G.P., Simpson C.G., Shaw P.J., Brown J.W. 2009. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 21:2045–2057 10.1105/tpc.109.067736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S., Farny N.G., Roth F.P., Silver P.A. 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell. 131:557–571 10.1016/j.cell.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz F., Goyer C., Darveau A., Neron S., Lemieux R., Sonenberg N. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA. 89:9612–9616 10.1073/pnas.89.20.9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H., Ferrand V., Marguet D., Bania J., Camosseto V., David A., Gatti E., Pierre P. 2004. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 164:667–675 10.1083/jcb.200312073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev A., Princiotta M.F., Zanker D., Takeda K., Gibbs J.S., Kumagai C., Waffarn E., Dolan B.P., Burgevin A., Van Endert P., et al. 2010. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc. Natl. Acad. Sci. USA. 107:6964–6969 10.1073/pnas.0910997107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann R., Bald R., Stöffler-Meilicke M., Stöffler G. 1981. Localization of the puromycin binding site on the large ribosomal subunit of Escherichia coli by immunoelectron microscopy. Proc. Natl. Acad. Sci. USA. 78:7276–7280 10.1073/pnas.78.12.7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 282:2082–2085 10.1126/science.282.5396.2082 [DOI] [PubMed] [Google Scholar]
- Mauro V.P., Edelman G.M. 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA. 99:12031–12036 10.1073/pnas.192442499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKendrick L., Thompson E., Ferreira J., Morley S.J., Lewis J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21:3632–3641 10.1128/MCB.21.11.3632-3641.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson L., Deutscher M.P. 2000. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 275:31559–31562 10.1074/jbc.C000385200 [DOI] [PubMed] [Google Scholar]
- Nathanson L., Xia T., Deutscher M.P. 2003. Nuclear protein synthesis: a re-evaluation. RNA. 9:9–13 10.1261/rna.2990203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T., Politz J.C. 2000. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148:1091–1095 10.1083/jcb.148.6.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. 1971. Inhibitors of ribosome functions. Annu. Rev. Microbiol. 25:487–562 10.1146/annurev.mi.25.100171.002415 [DOI] [PubMed] [Google Scholar]
- Pestka S., Rosenfeld H., Harris R., Hintikka H. 1972. Studies on transfer ribonucleic acid-ribosome complexes. XXI. Effect of antibiotics on peptidyl-puromycin synthesis by mammalian polyribosomes. J. Biol. Chem. 247:6895–6900 [PubMed] [Google Scholar]
- Prouty W.F., Karnovsky M.J., Goldberg A.L. 1975. Degradation of abnormal proteins in Escherichia coli. Formation of protein inclusions in cells exposed to amino acid analogs. J. Biol. Chem. 250:1112–1122 [PubMed] [Google Scholar]
- Rodriguez A.J., Shenoy S.M., Singer R.H., Condeelis J. 2006. Visualization of mRNA translation in living cells. J. Cell Biol. 175:67–76 10.1083/jcb.200512137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 6:275–277 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z., Brill L.M., Cabrera R., Kleifeld O., Scheliga J.S., Glickman M.H., Chang E.C., Wolf D.A. 2009. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell. 36:141–152 10.1016/j.molcel.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck S.R., Green H.M., Alberola-Ila J., Roberts R.W. 2004. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem. Biol. 11:999–1008 10.1016/j.chembiol.2004.05.011 [DOI] [PubMed] [Google Scholar]
- Stephens S.B., Dodd R.D., Lerner R.S., Pyhtila B.M., Nicchitta C.V. 2008. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol. Biol. 419:197–214 10.1007/978-1-59745-033-1_14 [DOI] [PubMed] [Google Scholar]
- Sugita M., Morita T., Yonesaki T. 1995. Puromycin induces apoptosis of developing chick sympathetic neurons in a similar manner to NGF-deprivation. Zoolog. Sci. 12:419–425 10.2108/zsj.12.419 [DOI] [PubMed] [Google Scholar]
- Wang D.O., Martin K.C., Zukin R.S. 2010. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 33:173–182 10.1016/j.tins.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chang Y.-F., Hamilton J.I., Wilkinson M.F. 2002. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell. 10:951–957 10.1016/S1097-2765(02)00635-4 [DOI] [PubMed] [Google Scholar]
- Yewdell J.W., Nicchitta C.V. 2006. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 27:368–373 10.1016/j.it.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Zimmerman E.F., Hackney J., Nelson P., Arias I.M. 1969. Protein synthesis in isolated nuclei and nucleoli of HeLa cells. Biochemistry. 8:2636–2644 10.1021/bi00834a058 [DOI] [PubMed] [Google Scholar]