Abstract
The centrosome, which consists of two centrioles and the surrounding pericentriolar material, is the primary microtubule-organizing center (MTOC) in animal cells. Like chromosomes, centrosomes duplicate once per cell cycle and defects that lead to abnormalities in the number of centrosomes result in genomic instability, a hallmark of most cancer cells. Increasing evidence suggests that the separation of the two centrioles (disengagement) is required for centrosome duplication. After centriole disengagement, a proteinaceous linker is established that still connects the two centrioles. In G2, this linker is resolved (centrosome separation), thereby allowing the centrosomes to separate and form the poles of the bipolar spindle. Recent work has identified new players that regulate these two processes and revealed unexpected mechanisms controlling the centrosome cycle.
The centrosome duplication cycle
The centrosome of animal cells is comprised of centrioles and the surrounding pericentriolar material (PCM; Fig. 1; Paintrand et al., 1992; Bornens, 2002). The PCM is a meshwork of fibrous proteins that nucleates and anchors microtubules (MTs), whereas the centrioles reside at the core of the centrosomes and are important for centrosome integrity and centrosome duplication (Gould and Borisy, 1977; Piel et al., 2000). Despite certain variations on their structure, canonical centrioles consist of 9 MT triplets that form a cylinder with a length of ∼0.5 μm and a diameter of 0.2 μm (Bornens, 2002; Azimzadeh and Bornens, 2007).
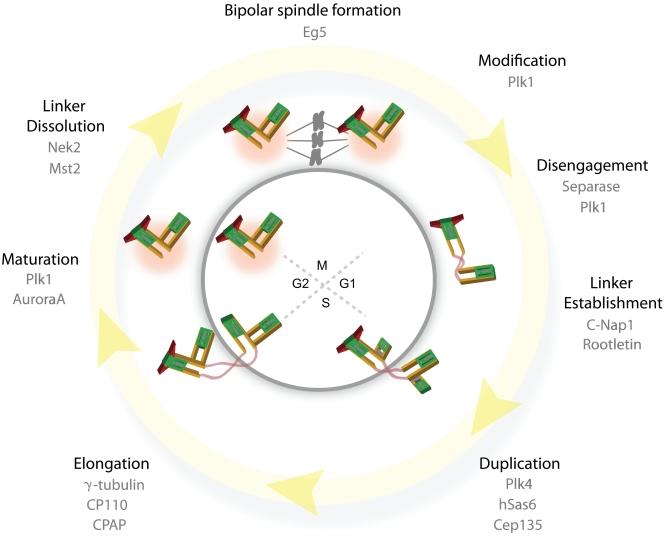
Figure 1.
The centrosome cycle of animal cells. Main events in the centrosome cycle are highlighted along with the key players that have been implicated in each process. Green-filled regions represent the centrin-positive distal regions of the centriole. As cells exit mitosis the daughter centriole disengages from the mother centriole, losing its orthogonal connection (centriole disengagement). Upon disengagement, the daughter centriole is connected to the mother by a flexible linker (pink strands). Centriole assembly factors accumulate in S phase and new centrioles are formed and gradually elongate throughout S and G2. At the G2/M transition the flexible linker that holds the centriole pairs together is lost (linker dissolution) and the centrioles accumulate more PCM (maturation) and constitute the poles of the mitotic spindle.
Centrosomes duplicate once per cell cycle (Bettencourt-Dias and Glover, 2007). During canonical centrosome duplication, one daughter centriole forms perpendicularly to each mother centriole in S phase (Fig. 1). The newly assembled centrioles remain tightly engaged with their mothers and gradually elongate throughout S and G2. At the G2/M transition, the centrioles accumulate more PCM and the two centrosomes (each carrying a mother and still tightly connected daughter centriole) start to separate by the dissolution of the linker that connects the two centrosomes (centrosome separation). The separated centrosomes then form the poles of the bipolar mitotic spindle (Fig. 1). As the cell exits mitosis each cell inherits one centrosome carrying a mother and a daughter centriole. The daughter centriole then separates from the mother centriole and the mother–daughter pair loses the orthogonal orientation (this process is termed centriole disengagement). Centriole disengagement is the prerequisite for another round of centrosome duplication in S phase.
Perturbations in the centrosome cycle can have catastrophic consequences, such as chromosome instability leading to tumorigenesis (Basto et al., 2008; Ganem et al., 2009). In addition, many genetic disorders are associated with defects in centrosome structure or number (Nigg, 2006; Nigg and Raff, 2009). To prevent such defects in centrosome propagation, the centrosome cycle is under strict control. Because several excellent reviews have provided an in-depth description of the centriole duplication cycle (Nigg, 2007; Strnad and Gönczy, 2008; Azimzadeh and Marshall, 2010), here we focus on recent advances in centriole disengagement and centrosome separation. We highlight newly identified players and outline the emerging models that arise from recent observations.
Centriole disengagement
A critical process intertwined with the duplication of the centrioles is the disengagement of the mother and daughter centrioles that breaks their orthogonal arrangement. This centriole configuration is established in S phase and persists until late mitosis/G1 phase. Centriole disengagement is crucial for the licensing of the two centrioles for duplication in G1/S and for limiting centriole duplication to one event per cell cycle (Tsou and Stearns, 2006). Centriole disengagement requires the proteolytic activity of separase (Tsou and Stearns, 2006; Tsou et al., 2009); however, the molecular details of this process have only recently begun to come to light (Fig. 2).
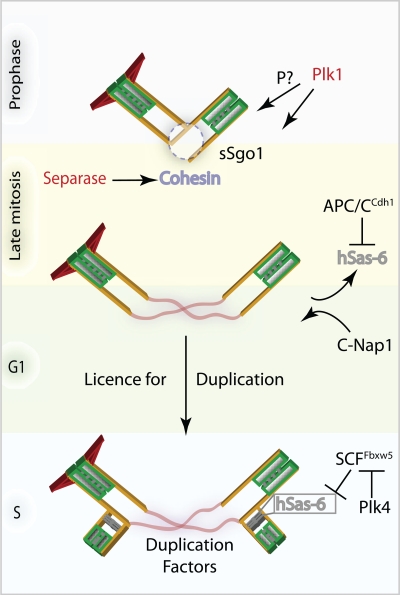
Figure 2.
Centriole disengagement and duplication. Main players of centriole disengagement are depicted. During mitosis, Plk1 and separase consecutively disjoin mother and daughter centrioles. In addition, Plk1-mediated modification of the centrioles defines their capability of becoming a fully functional centrosome. In late mitosis the main centriole assembly factor hSas-6 is degraded and its levels are kept low by APC/C- and SCF-mediated proteolysis until the duplication can be initiated. In addition, activation of Plk-4 inhibits this negative regulation to allow cartwheel formation.
The cysteine protease separase is well known for its function in chromosome segregation by its ability to cleave the cohesin complex subunit Scc1 (Nasmyth, 2002). Models in which separase relieves centriolar cohesion in the same way as it resolves chromosomes cohesion are attractive because they suggest a shared mechanism for regulating the cohesion of chromatids and centrioles, whose duplication are both restricted to one event per cell cycle. Results arising from the work of Tsou et al. (2009) initially suggested that separase dissolves the centriolar connection by targeting a substrate other than cohesin. This conclusion was based on the inability of a noncleavable cohesin subunit Scc1NC to prevent centriole disengagement while blocking sister chromatid separation. However, more recent data indicate that the cleavage of the cohesin complex is in fact required for centriole disengagement. Notably, artificial endoproteolysis of the cohesin complex subunits (Scc1 or Smc3) that carry engineered HRV/TEV cleavage sites by HRV/TEV proteases relieved centriolar cohesion in vitro and in vivo in the same way as it promoted sister chromatid separation under similar conditions (Schöckel et al., 2011). These results are consistent with earlier work by Nakamura et al. (2009) demonstrating that cohesin is involved in separase-dependent centriole disengagement through Aki1 kinase (Akt kinase interacting protein 1) that is required for the centrosomal recruitment of Scc1. Depletion of Aki1 results in loss of cohesin at centrioles and centriolar disengagement, which can be rescued by subsequent depletion of separase (Nakamura et al., 2009). Another protein that is intimately linked with centriole disengagement is Astrin. Similar to Aki1, Astrin depletion elevated the frequency of spindles containing prematurely separated centrioles, a phenotype that can be rescued by separase depletion. Unlike Aki1 depletion, however, down-regulation of Astrin activates separase directly and promotes premature centriole disengagement (Thein et al., 2007). In summary, although the molecular details of how centriolar cohesion is regulated by Aki and Astrin remains to be investigated, the emerging picture is that cohesin links the duplicated centrioles together. Subsequently, the proteolytic activity of separase cleaves the cohesin ring to induce centriole disjunction (Fig. 2).
Centromeric cohesin at centromeres is protected by an evolutionary conserved protein, Shugoshin (Sgo1) (McGuinness et al., 2005). Through direct interactions with the phosphatase PP2A, Sgo1 prevents phosphorylation of cohesin at centromeres during mitosis (Kitajima et al., 2006; Riedel et al., 2006). Interestingly, a shorter Sgo1 splice variant (sSgo1) localizes to centrosomes and was proposed to function as a protector of centriolar cohesion from separase during mitosis in a manner that is similar to its role in protecting cohesion from destruction during chromosome segregation. sSgo1 requires Plk1 for its function at centrioles, and in agreement with this notion, Plk1 associates with and phosphorylates sSgo1 (X. Wang et al., 2008). These data proposed a role of Plk1 in regulating centriole disengagement. Consistently, Tsou et al. (2009) demonstrated that separase activity is not the only factor controlling centriole disengagement because cells carrying a null allele of separase eventually disengaged their centrioles before the onset of mitosis. However, inhibition of Plk1 kinase completely abolishes centriole disengagement (Tsou et al., 2009). Recent findings suggest that Plk1 fosters centriole disengagement by promoting removal of cohesin from centrosomes in prophase and by stimulating separase to cleave cohesin at the centrioles during mitotic exit (Schöckel et al., 2011). Perplexingly, sSgo1 mutants in which the Plk1 target sites have been mutated to block sSgo1 phosphorylation increase the frequency of cells with split centrioles in mitosis because of a reduction in the recruitment of sSgo1 to centrosomes (X. Wang et al., 2008). This result suggests a dual function of Plk1: Plk1 may initially promote sSgo1 targeting and centriole engagement before contributing to cohesion cleavage and centriole disengagement during mitotic exit.
In summary, it becomes increasingly apparent that successive activities of Plk1 and separase are the major driving forces in dissolving centriole cohesion (Fig. 2). One attractive model is the coordination of centriole disengagement with chromosome segregation through the use of cohesin as the molecular “glue” in both cases. Although the evidence so far favors this idea, it is equally possible that separase merely initiates centriole disengagement. Complete spatial separation of the two disengaged centrioles may depend on MT-mediated forces to physically and mechanically push centrioles apart. Demonstrating localization of cohesin complex components and binding of separase to the centrosomes during the cell cycle and testing whether artificial cleavage of cohesin complex in S phase results in premature centriole disengagement will be important to help solve the unknowns of this process.
An additional interesting possibility is the involvement of PP2A phosphatase as Plk1 counteracting phosphatase that inhibits centriole disengagement. A Sgo1-interacting PP2A phosphatase complex was found at centromeres (Kitajima et al., 2006; Riedel et al., 2006). It is plausible that a similar PP2A–sSgo1 complex localizes to centrioles where it inhibits the disengagement of centrioles.
Finally, recent data revealed functions of Plk1 that go beyond the simple relief of centriolar cohesion, as Plk1 “modifies” proteins at the newly assembled daughter centrioles during mitosis to enable them to mature and organize PCM (“MTOC competent”). When centrioles lack this Plk1-dependent modification they lose the ability to duplicate and to develop into a fully functional MTOC (Wang et al., 2011). Taken together, these results suggest that modification of centriolar proteins by Plk1 is required for centriole duplication.
Resolving the centrosomal linker: Centrosome separation
In addition to the cohesin complex between the mother and daughter centrioles that is lost with mitotic exit (centriole disengagement), a second type of proteinaceous linker connects the proximal end of the two mother centrioles from G1 until onset of mitosis (Fig. 1, linker establishment; Bornens et al., 1987). This highly flexible linker is established with or slightly after centriole disengagement and persists until mitotic entry (Fig. 1, linker dissolution; O’Regan et al., 2007).
Two structural proteins have been identified as components of the centrosomal linker: C-Nap1 and rootletin. The NIMA-related Nek2A kinase phosphorylates these linker proteins in late G2, which initiates their displacement from the centrosomes. This then results in the separation of the two centrosomes. C-Nap1 mirrors Nek2A in localizing to the proximal ends of the mother centrioles (Fry et al., 1998). It serves as a docking site for rootletin, which is thought to physically connect the two mother centrioles (Bahe et al., 2005). Consistent with this idea, overexpression of rootletin leads to the formation of extensive fibers, which are able to recruit other interacting proteins such as Nek2A and C-Nap1. In addition, depletion of either C-Nap1 or rootletin results in premature centrosome separation independent of the cell cycle phase (Faragher and Fry, 2003). How C-Nap1 and rootletin are displaced from the centrosomes upon phosphorylation remains to be established. In any case, proteolytic degradation of C-Nap1 and rootletin is not required for their displacement from and subsequent separation of centrosomes (Mayor et al., 2000).
More recently, additional centrosomal proteins were identified that may function as centrosomal linker proteins. Cep68 and Cep215 (CDK5RAP2) were identified in an siRNA screen as putative linker components because removal of either molecule promoted premature centrosome separation. Cep68 localizes between the two centrioles, whereas Cep215 surrounds the centrioles (Graser et al., 2007). The loss of Cep68 from centrosomes upon mitotic entry of cells and its reliance upon the presence of other linker proteins for recruitment to centrosomes is certainly consistent with Cep68 being a target of Nek2A. The differing behavior and localization of Cep215 favors an alternative mode of regulation, perhaps directly through Plk1.
β-Catenin is best known for its role as an effector of the Wnt signaling pathway (Dierick and Bejsovec, 1999). It has also been linked to the maintenance of centrosomal integrity. It localizes to proximal and distal regions of the centrioles and the region between the centrosomes. In addition, β-catenin is an in vitro and in vivo substrate of Nek2A. However, its depletion phenotype does not display the characteristics of premature centrosome separation exhibited upon depletion of other linker proteins. Nonetheless, β-catenin localization does seem to require rootletin and C-Nap1, although unlike these established linker molecules it fails to dissociate from the centrosomes during mitosis (Bahmanyar et al., 2008).
The function of Nek2A and its upstream regulation
As for other mitotic kinases, it is hardly surprising that protein levels and activity of Nek2A are subject to cell cycle–dependent regulation. Nek2A protein levels peak in late S and G2 phases followed by an APC/C-mediated degradation in prometaphase that is completed by the time of nuclear envelope breakdown (NEBD; Hayes et al., 2006). It was therefore proposed that the ensuing changes in Nek2A levels and activity reach a critical threshold in prophase that is sufficiently high to dissolve the centrosomal linker (Hayes et al., 2006). Subsequently, the lack of any impact of siRNA depletion of Nek2A upon cell cycle progression challenged this idea (Fletcher et al., 2004). However, recent studies revealed that Nek2A activity is subject to both positive and negative control and that a parallel pathway can substitute for the loss of Nek2A in centrosomal linker dissolution (Pugacheva and Golemis, 2005; Mardin et al., 2010, 2011; Matsuo et al., 2010).
The localized activity of Nek2A kinase toward its substrates C-Nap1 and rootletin is regulated by two Hippo pathway components: the Mst2 kinase and the scaffold protein hSav1 (Mardin et al., 2010). Conventionally, Hippo pathway proteins are well known to function in growth control and apoptosis by regulating the localization of the transcriptional coactivators YAP and TAZ (Edgar, 2006; Pan, 2010). However, increasing evidence supports the notion that Hippo pathway components can function independently of their role in the conventional pathway (Yang et al., 2004; Yabuta et al., 2007; Chiba et al., 2009; Hergovich et al., 2009; Oh and Irvine, 2010), the control of centrosome duplication and separation being prime examples.
Independently of the rest of the Hippo pathway, the Mst1/Mob1/NDR signaling cassette contributes to the control of centrosome duplication in human cells (Hergovich et al., 2007, 2009). In addition, to control centrosome separation, Mst2 kinase assisted by the scaffold protein hSav1 phosphorylates Nek2A kinase. This phosphorylation is crucial for the formation of the hSav1–Nek2A–Mst2 complexes and the targeting of phosphorylated Nek2A to centrosomes. The accumulation of Nek2A at the centrosomes is critical for centrosome separation because defects in this step lead to decreased C-Nap1 phosphorylation (Mardin et al., 2010).
Interestingly, the interaction between hSav1, Mst2, and Nek2A is mediated by a type of coiled-coil domain, known as the SARAH (for Sav/RASSF/Hpo) domain (Mardin et al., 2010). This domain is present at the extreme C termini of the Hippo pathway components hSav1, Mst1/2, and Rassf1 and mediates mutual interactions between SARAH domain proteins (Scheel and Hofmann, 2003) through homo- and heterodimerization via head-to-tail anti-parallel arrangements of the coiled-coil structure (Hwang et al., 2007). A SARAH-like domain at the extreme C terminus of Nek2A promotes binding of Nek2A to Mst2 and hSav1. These SARAH domain–mediated interactions are essential for centrosome separation (Mardin et al., 2010).
The timely activation of Nek2A in centrosome separation is under direct control of the cell cycle machinery. Upon mitotic entry, Aurora A and Plk1 kinases, which are both involved in centrosome maturation, promote centrosome separation by stimulating the phosphorylation and displacement of linker proteins at the centrosome (Fig. 3, after NEBD; Mardin et al., 2011). The main function of Aurora A in centrosome separation is probably the activation of the kinase activity of Plk1 (Mardin et al., 2011). This is consistent with previous observations that established that Aurora A elevates Plk1 kinase activity via phosphorylation of T210 in the T loop (Macůrek et al., 2008; Seki et al., 2008). In turn, Plk1 binds to and phosphorylates Mst2 kinase. This phosphorylation is important to mediate the interactions between Nek2A and the phosphatase PP1γ (Helps et al., 2000; Mi et al., 2007). In vivo PP1γ antagonizes Nek2A by dephosphorylating C-Nap1 rather than dephosphorylating Nek2A. Therefore, the level of PP1γ associated with Nek2A is a critical aspect of the control of the phosphorylation status of C-Nap1. Mst2 binding to PP1γ-Nek2A modulates this well-balanced level of phosphorylation. Of note, it is regulation of binding affinity via Plk1 phosphorylation of Mst2 that determines the dissociation of Mst2–Nek2A–PP1γ complexes. The kinase activity of Mst2 does probably not participate in the regulation of PP1γ binding to Nek2A (Mardin et al., 2011). Thus, phosphorylation of Mst2 by Plk1 leads to a reduction in the levels of PP1γ in the Mst2–Nek2A–PP1γ complex. Nek2A kinase at centrosomes therefore is able to phosphorylate C-Nap1 beyond the critical threshold that is required to promote the dissolution of the centrosomal linker (Mardin et al., 2010).
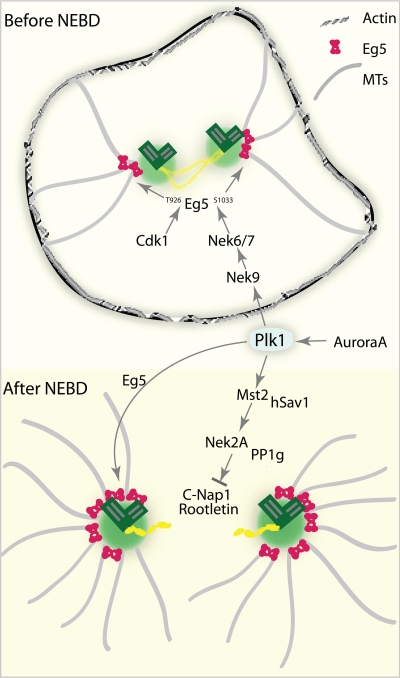
Figure 3.
Centrosome separation before and after NEBD. Plk1 has a central role in regulating the centrosome separation via different pathways. In G2, Plk1 activates Nek9/6/7 cascade to regulate the accumulation of Eg5. Moreover, Cdk1 is important for Eg5 binding to MTs. After NEBD, Plk1 regulates the phospho-balance on the centrosomal linker by controlling the association of Nek2 with PP1γ through phosphorylation of Mst2. In addition, Plk1 is involved in the targeting of Eg5 to the spindle poles.
Conversely, in cells lacking Plk1 activity or expressing nonphosphorylatable Mst2 mutants, the levels of PP1γ in the Mst2–Nek2A–PP1γ complex increase, leading to a reversal of C-Nap1 phosphorylation by Nek2A and to a block in centrosome separation (Mardin et al., 2011). Plk1 activity is well known to peak in G2 and in mitosis (Golsteyn et al., 1995). Thus, it is plausible that Plk1 controls the balance of phosphorylation/dephosphorylation of the centrosome linker and thereby determines the timing of centrosome separation.
This positive regulation of Nek2A by Hippo pathway components might be counteracted through negative control by pericentrin (kendrin) and the focal adhesion scaffolding protein HEF1. HEF1 has been shown to inhibit Nek2A accumulation and activity at the centrosomes (Pugacheva and Golemis, 2005), whereas pericentrin acts as an inhibitor of Nek2A kinase activity. Consistently, siRNA-mediated depletion of pericentrin causes premature centrosome separation in interphase. How pericentrin down-regulates Nek2A kinase activity is unknown; however, the authors proposed a mechanism wherein pericentrin binding changes the overall structure of the protein, flipping the structure of the catalytic domain of Nek2A into an inhibitory conformation (Matsuo et al., 2010). Interestingly, pericentrin and Cep215 localization also depends on Plk1, suggesting an additional level of regulation, either through Nek2A or by direct phosphorylation of some proteins involved in centrosomal cohesion (Graser et al., 2007; Haren et al., 2009). When viewed together, these findings clearly show that the precise control of localization and activity of the Nek2A kinase is important for the timely separation of the two centrosomes in G2.
Surprisingly, Nek2A depletion does not cause significant defects in cell cycle progression (Fletcher et al., 2004; unpublished data). However, under conditions when Eg5 motor activity is reduced but not completely blocked, the hSav1–Nek2A–Mst2 pathway becomes essential for the formation of the bipolar spindle (Mardin et al., 2010). This suggests a two-step process for centrosome separation: phosphorylation-dependent dissolution of the centrosome linker via Mst2–Nek2A kinases and the force-dependent separation of the centrosomes by Eg5 activity (discussed further in the next section).
Regulation of mammalian kinesin-5, Eg5
After the initial separation of the centrosomes by the displacement of linker proteins, motor proteins act via their anti-parallel sliding activity on MTs to physically separate the two centrosomes. The kinesin Eg5 is the principal force generator that drives centrosome separation. Eg5 belongs to the kinesin-5 subfamily of motor proteins, which are homotetrameric, plus end–directed motors (Sawin et al., 1992; Cole et al., 1994). Inhibition of Eg5 by small molecule inhibitors such as monastrol or depletion of the protein by siRNA results in prometaphase-arrested cells with monopolar spindles (Whitehead and Rattner, 1998; Kapoor et al., 2000). It is well established that while dispersed in the cytoplasm in interphase, the Eg5 motor first accumulates at spindle poles during prophase, after which it is found along the metaphase spindle (Sawin and Mitchison, 1995). How Eg5 is targeted to the spindle poles is a longstanding question. In Xenopus egg extracts transport of Eg5 to the poles is dependent upon a direct interaction between Eg5 and components of the minus end–directed dynein/dynactin motor complex (Uteng et al., 2008). However, in human cells this seems not to be the case, as depletion of dynein heavy chain does not affect centrosomal localization of Eg5 (Tanenbaum et al., 2008).
Previous data suggest that Cdk1 promotes centrosome separation by phosphorylating and activating Eg5 at the spindle poles (Blangy et al., 1995). This phosphorylation (Thr 926) is important for the binding of Eg5 to MTs. However, at least in chicken DT40 cells Cdk1 by itself is not required for the enrichment of Eg5 at centrosomes (Smith et al., 2011). Instead, growing evidence argues that in human cells the targeting of Eg5 to spindle poles requires Plk1 activity (Fig. 3, after NEBD; Bertran et al., 2011; Mardin et al., 2011; Smith et al., 2011). Inhibition of Plk1 prevents accumulation of Eg5 at centrosomes without altering the cellular levels of the protein (Mardin et al., 2011). How Plk1 regulates Eg5 localization is presently unclear. A direct phosphorylation-dependent regulation is unlikely because Plk1 is unable to phosphorylate Eg5 in vitro (Smith et al., 2011). In addition, phospho-analysis of Eg5 in human mitotic cells failed to identify additional sites other than T926 (Cdk1 phosphorylation), as reported before (Blangy et al., 1995), and S1033 (Nek6 kinase phosphorylation; Rapley et al., 2008).
Importantly, the Plk1-regulated targeting of Eg5 to the centrosome requires an intact MT cytoskeleton. In cells incubated with low concentrations of the MT-depolymerizing drug nocodazole, Eg5 localized to remnants of MTs; however, in the complete absence of MTs, Eg5 no longer bound to centrosomes (Mardin et al., 2011). Lack of Plk1 might trap Eg5 on MTs to prevent its subsequent targeting to the poles. Alternatively, because Plk1 inhibition decreases the MT nucleation capacity of spindle poles (Lane and Nigg, 1996), the lower density of MTs at centrosomes might indirectly affect Eg5 localization.
The regulation of Eg5 is even more complex, as the NIMA kinase family members Nek9, Nek6, and Nek7 also contribute to the targeting of Eg5 to centrosomes. Active Nek9 accumulates at the centrosomes in mitosis and directly phosphorylates Nek6/Nek7 (Roig et al., 2002, 2005; Belham et al., 2003). By releasing the auto-inhibitory conformation of Nek6/7, Nek9 activates both kinases (Richards et al., 2009). Activated Nek6 and Nek7 kinases then control mitotic progression and contribute to bipolar spindle formation (O’Regan and Fry, 2009).
Bertran et al. (2011) have now demonstrated that the Nek9/6/7 signaling cascade functions downstream of Plk1 but upstream of Eg5 in centrosome separation. When phosphorylated and activated by Plk1, Nek9, in conjunction with Nek6/Nek7, is able to target Eg5 to the centrosome. The authors propose that the ability of Nek6 to phosphorylate Eg5 on Ser1033 is responsible for Plk1-mediated targeting of Eg5 to the spindle poles. Consistently, Eg5 mutations that block this phosphorylation fail to concentrate at the centrosomes, although the underlying mechanism behind this observation remains to be established (Bertran et al., 2011).
Interestingly, overexpression of either Nek9 or Nek6 promotes premature centrosome separation in interphase cells. Remarkably, this premature centrosome separation is distinct from Nek2A-induced centrosome separation because it is completely dependent on forces provided by Eg5. Additional experiments will reveal if Plk1 targets Eg5 to the centrosomes via two distinct pathways before and after NEBD, as outlined in Fig. 3, or if Plk1–Nek9–Nek6/7 regulation persists in mitosis. Development of specific inhibitors toward Nek kinases may help to solve this issue.
Additional forces that contribute to centrosome separation
After NEBD, Eg5 activity is counteracted by the minus end–directed motor dynein in human cells. The dynein/dynactin complex generates an inward, minus end–directed force that antagonizes Eg5 action during centrosome separation, such that simultaneous impairment of both Eg5 and dynein activities is conducive to bipolar spindle formation (Tanenbaum et al., 2008). In contrast, before NEBD dynein cooperates with Eg5 in centrosome separation, as inhibition of both molecules at this stage of mitosis produces more severe defects in centrosome separation than inhibition of either molecule alone (Tanenbaum et al., 2008). Therefore, centrosome separation before and after NEBD is driven by distinct processes (Tanenbaum and Medema, 2010).
Recently, kinetochores (KTs) were implicated in centrosome separation after NEBD (Toso et al., 2009). KTs contribute to centrosome separation by stabilizing MTs to form K-fibers and the generation of an outward pushing force on these K-fibers. This process becomes essential when Aurora A is inactivated. However, KT-based forces are not sufficient to overcome centrosome separation defects arising from Eg5 inhibition. The authors suggest that KTs contribute to the overall force that separates the centrosomes via the generation of a MT poleward flux.
The actin cytoskeleton has also been suggested to contribute to centrosome separation, particularly before NEBD (Whitehead et al., 1996; Uzbekov et al., 2002; W. Wang et al., 2008). Centrosomes fail to separate when F-actin is depolymerized by specific drugs, although the underlying mechanism is poorly understood. Recently, actin depolymerization has been shown to reverse the failure of centrosome separation arising from Plk1 inhibition during G2 phase (Smith et al., 2011). How actin contributes to the force generation to separate the centrosomes before NEBD is presently unclear, but it is possible that it could act as a stable matrix upon which MT-dependent motor proteins could exert their function.
Future prospects
Despite this recent rapid advance in our understanding of the molecular mechanisms of centriole/centrosome separation, much remains to be learned. Considering the central importance of separase and the cohesin complex in centriole disengagement, it will be crucial to investigate what is embraced by cohesin to maintain the centriole cohesion. The findings of Schöckel et al. (2011) raise the possibility of cohesin encircling DNA; however, so far evidence for the presence of DNA in centrosomes is lacking, whereas specific RNAs were found at this organelle (Alliegro et al., 2006). Another important question concerns the possible link between the centriole disengagement and the assembly of the centriolar linker. What is the basis of Plk1-mediated modification of centrioles and what are the relevant substrates for Plk1 kinase? Careful analysis of the centriole markers demonstrated that Plk1-modified centrioles accumulate C-Nap1, whereas non-modified centrioles are associated with hSas-6 (Wang et al., 2011). Thus, it will be important to elucidate how the assembly of the centrosomal linker is connected with centriole disengagement.
Finally, recent pioneering studies for the in vitro assembly of the centrioles and centriole disengagement (Kitagawa et al., 2011; Schöckel et al., 2011; van Breugel et al., 2011) have shown that it is possible to reconstitute aspects of the centrosome cycle in vitro. Certainly, newly developed cell biological tools, combined with such in vitro analyses, will continue to shed light on the unknowns of centrosome biology.
Acknowledgments
We apologize to those of our colleagues whose important contributions could not be acknowledged due to space constraints. We would like to thank Iain Hagan, Ingrid Hoffmann, and Joan Roig for comments on the manuscript.
The work of E. Schiebel is supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) Schi 295/3.
Footnotes
Abbreviations used in this paper:
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
- PCM
- pericentriolar material
- Sgo1
- Shugoshin
- sSgo1
- short Sgo1
References
- Alliegro M.C., Alliegro M.A., Palazzo R.E. 2006. Centrosome-associated RNA in surf clam oocytes. Proc. Natl. Acad. Sci. USA. 103:9034–9038 10.1073/pnas.0602859103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. 2007. Structure and duplication of the centrosome. J. Cell Sci. 120:2139–2142 10.1242/jcs.005231 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Marshall W.F. 2010. Building the centriole. Curr. Biol. 20:R816–R825 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahe S., Stierhof Y.D., Wilkinson C.J., Leiss F., Nigg E.A. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171:27–33 10.1083/jcb.200504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S., Kaplan D.D., Deluca J.G., Giddings T.H., Jr, O’Toole E.T., Winey M., Salmon E.D., Casey P.J., Nelson W.J., Barth A.I. 2008. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22:91–105 10.1101/gad.1596308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belham C., Roig J., Caldwell J.A., Aoyama Y., Kemp B.E., Comb M., Avruch J. 2003. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278:34897–34909 10.1074/jbc.M303663200 [DOI] [PubMed] [Google Scholar]
- Bertran M.T., Sdelci S., Regué L., Avruch J., Caelles C., Roig J. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30:2634–2647 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D.M. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Blangy A., Lane H.A., d’Hérin P., Harper M., Kress M., Nigg E.A. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 83:1159–1169 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25–34 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M.C., Karsenti E. 1987. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 8:238–249 10.1002/cm.970080305 [DOI] [PubMed] [Google Scholar]
- Chiba S., Ikeda M., Katsunuma K., Ohashi K., Mizuno K. 2009. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr. Biol. 19:675–681 10.1016/j.cub.2009.02.054 [DOI] [PubMed] [Google Scholar]
- Cole D.G., Saxton W.M., Sheehan K.B., Scholey J.M. 1994. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 269:22913–22916 [PMC free article] [PubMed] [Google Scholar]
- Dierick H., Bejsovec A. 1999. Cellular mechanisms of wingless/Wnt signal transduction. Curr. Top. Dev. Biol. 43:153–190 10.1016/S0070-2153(08)60381-6 [DOI] [PubMed] [Google Scholar]
- Edgar B.A. 2006. From cell structure to transcription: Hippo forges a new path. Cell. 124:267–273 10.1016/j.cell.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Faragher A.J., Fry A.M. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 14:2876–2889 10.1091/mbc.E03-02-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher L., Cerniglia G.J., Nigg E.A., Yend T.J., Muschel R.J. 2004. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat. Res. 162:128–135 10.1667/RR3211 [DOI] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn R.M., Mundt K.E., Fry A.M., Nigg E.A. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 129:1617–1628 10.1083/jcb.129.6.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould R.R., Borisy G.G. 1977. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. Cell Biol. 73:601–615 10.1083/jcb.73.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Nigg E.A. 2007. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120:4321–4331 10.1242/jcs.020248 [DOI] [PubMed] [Google Scholar]
- Haren L., Stearns T., Lüders J. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE. 4:e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M.J., Kimata Y., Wattam S.L., Lindon C., Mao G., Yamano H., Fry A.M. 2006. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat. Cell Biol. 8:607–614 10.1038/ncb1410 [DOI] [PubMed] [Google Scholar]
- Helps N.R., Luo X., Barker H.M., Cohen P.T. 2000. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 349:509–518 10.1042/0264-6021:3490509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Lamla S., Nigg E.A., Hemmings B.A. 2007. Centrosome-associated NDR kinase regulates centrosome duplication. Mol. Cell. 25:625–634 10.1016/j.molcel.2007.01.020 [DOI] [PubMed] [Google Scholar]
- Hergovich A., Kohler R.S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B.A. 2009. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 19:1692–1702 10.1016/j.cub.2009.09.020 [DOI] [PubMed] [Google Scholar]
- Hwang E., Ryu K.S., Pääkkönen K., Güntert P., Cheong H.K., Lim D.S., Lee J.O., Jeon Y.H., Cheong C. 2007. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc. Natl. Acad. Sci. USA. 104:9236–9241 10.1073/pnas.0610716104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P., Steinmetz M.O. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell. 144:364–375 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 441:46–52 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- Lane H.A., Nigg E.A. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713 10.1083/jcb.135.6.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., Medema R.H. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 455:119–123 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Mardin B.R., Lange C., Baxter J.E., Hardy T., Scholz S.R., Fry A.M., Schiebel E. 2010. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12:1166–1176 10.1038/ncb2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin B.R., Agircan F.G., Lange C., Schiebel E. 2011. Plk1 controls the Nek2A-PP1g antagonism in centrosome disjunction. Curr. Biol. 21:1–7 10.1016/j.cub.2011.05.047 [DOI] [PubMed] [Google Scholar]
- Matsuo K., Nishimura T., Hayakawa A., Ono Y., Takahashi M. 2010. Involvement of a centrosomal protein kendrin in the maintenance of centrosome cohesion by modulating Nek2A kinase activity. Biochem. Biophys. Res. Commun. 398:217–223 10.1016/j.bbrc.2010.06.063 [DOI] [PubMed] [Google Scholar]
- Mayor T., Stierhof Y.D., Tanaka K., Fry A.M., Nigg E.A. 2000. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151:837–846 10.1083/jcb.151.4.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness B.E., Hirota T., Kudo N.R., Peters J.M., Nasmyth K. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3:e86 10.1371/journal.pbio.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J., Guo C., Brautigan D.L., Larner J.M. 2007. Protein phosphatase-1alpha regulates centrosome splitting through Nek2. Cancer Res. 67:1082–1089 10.1158/0008-5472.CAN-06-3071 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Arai H., Fujita N. 2009. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J. Cell Biol. 187:607–614 10.1083/jcb.200906019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science. 297:559–565 10.1126/science.1074757 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 2006. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 119:2717–2723 10.1002/ijc.22245 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 2007. Centrosome duplication: of rules and licenses. Trends Cell Biol. 17:215–221 10.1016/j.tcb.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- O’Regan L., Fry A.M. 2009. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29:3975–3990 10.1128/MCB.01867-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L., Blot J., Fry A.M. 2007. Mitotic regulation by NIMA-related kinases. Cell Div. 2:25 10.1186/1747-1028-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Irvine K.D. 2010. Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 20:410–417 10.1016/j.tcb.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. 1992. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108:107–128 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Pan D. 2010. The hippo signaling pathway in development and cancer. Dev. Cell. 19:491–505 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E.N., Golemis E.A. 2005. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat. Cell Biol. 7:937–946 10.1038/ncb1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J., Nicolàs M., Groen A., Regué L., Bertran M.T., Caelles C., Avruch J., Roig J. 2008. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 121:3912–3921 10.1242/jcs.035360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M.W., O’Regan L., Mas-Droux C., Blot J.M., Cheung J., Hoelder S., Fry A.M., Bayliss R. 2009. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell. 36:560–570 10.1016/j.molcel.2009.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Gálová M., Petronczki M., Gregan J., Cetin B., et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441:53–61 10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- Roig J., Mikhailov A., Belham C., Avruch J. 2002. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 16:1640–1658 10.1101/gad.972202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J., Groen A., Caldwell J., Avruch J. 2005. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell. 16:4827–4840 10.1091/mbc.E05-04-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Mitchison T.J. 1995. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA. 92:4289–4293 10.1073/pnas.92.10.4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., Mitchison T.J. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Scheel H., Hofmann K. 2003. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. 13:R899–R900 10.1016/j.cub.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Schöckel L., Möckel M., Mayer B., Boos D., Stemmann O. 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 13:966–972 10.1038/ncb2280 [DOI] [PubMed] [Google Scholar]
- Seki A., Coppinger J.A., Jang C.Y., Yates J.R., Fang G. 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E., Hégarat N., Vesely C., Roseboom I., Larch C., Streicher H., Straatman K., Flynn H., Skehel M., Hirota T., et al. 2011. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 30:2233–2245 10.1038/emboj.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. 2008. Mechanisms of procentriole formation. Trends Cell Biol. 18:389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Medema R.H. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Galjart N., Medema R.H. 2008. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27:3235–3245 10.1038/emboj.2008.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein K.H., Kleylein-Sohn J., Nigg E.A., Gruneberg U. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178:345–354 10.1083/jcb.200701163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., McAinsh A.D. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., Jallepalli P.V. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng M., Hentrich C., Miura K., Bieling P., Surrey T. 2008. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 182:715–726 10.1083/jcb.200801125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R., Kireyev I., Prigent C. 2002. Centrosome separation: respective role of microtubules and actin filaments. Biol. Cell. 94:275–288 10.1016/S0248-4900(02)01202-9 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.O., Robinson C.V., et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science. 331:1196–1199 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Wang W., Chen L., Ding Y., Jin J., Liao K. 2008. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J. Cell Sci. 121:1334–1343 10.1242/jcs.018176 [DOI] [PubMed] [Google Scholar]
- Wang W.J., Soni R.K., Uryu K., Tsou M.F. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Y., Duan Q., Jiang N., Huang Y., Darzynkiewicz Z., Dai W. 2008. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell. 14:331–341 10.1016/j.devcel.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead C.M., Rattner J.B. 1998. Expanding the role of HsEg5 within the mitotic and post-mitotic phases of the cell cycle. J. Cell Sci. 111:2551–2561 [DOI] [PubMed] [Google Scholar]
- Whitehead C.M., Winkfein R.J., Rattner J.B. 1996. The relationship of HsEg5 and the actin cytoskeleton to centrosome separation. Cell Motil. Cytoskeleton. 35:298–308 [DOI] [PubMed] [Google Scholar]
- Yabuta N., Okada N., Ito A., Hosomi T., Nishihara S., Sasayama Y., Fujimori A., Okuzaki D., Zhao H., Ikawa M., et al. 2007. Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 282:19259–19271 10.1074/jbc.M608562200 [DOI] [PubMed] [Google Scholar]
- Yang X., Yu K., Hao Y., Li D.M., Stewart R.A., Insogna K.L., Xu T. 2004. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat. Cell Biol. 6:609–617 10.1038/ncb1140 [DOI] [PubMed] [Google Scholar]