Abstract
Although the physical separation of transcription in the nucleus and translation in the cytoplasm has presided as a fundamental tenet of cell biology for decades, it has not done so without recurring challenges and contentious debate. In this issue, David et al. (2012. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201112145) rekindle the controversy by providing convincing experimental evidence for nuclear translation.
Translation in the nucleus was first described more than half a century ago by Allfrey (1954), who reported a rapid incorporation of radioactive amino acids into nuclear proteins. These experiments were followed in the 1970s by a prominent study by Goidl et al. (1975), demonstrating the isolation of polyribosomes from nuclei. This study came to the already controversial conclusion that the nucleus can be an active site of protein synthesis. These data notwithstanding, the concept of nuclear translation has engaged many but convinced few. Perhaps the most compelling argument against nuclear translation is the problem of delivery of the translation machinery (ribosomes, initiation and elongation factors, and charged tRNAs) to the nucleus. Optical imaging experiments of the subcellular distributions of several GFP-tagged translation factors have indicated, for example, very low levels of translation factors in the nucleus, presenting a difficulty for canonical translation (Bohnsack et al., 2002). It is also established that small and large ribosomal subunits undergo maturation in the cytosol, and thus, it is possible that intranuclear ribosomal subunits are functionally compromised (Udem and Warner, 1973; Ford et al., 1999) And although there is evidence for aminoacylated tRNAs in the nucleus (Lund and Dahlberg, 1998), the combined results are generally agreed to strongly disfavor nuclear translation as a biological reality.
The question of nuclear translation was revived in a landmark study by the Cook laboratory, which used biotinylated lysyl-tRNA to visualize translation in isolated mammalian nuclei (Iborra et al., 2001). These probes were readily detected in the nucleus, colocalized with sites of active transcription, and their incorporation was suppressed by inhibition of RNA polymerase II, raising the possibility that translation and transcription may be, to some degree, coupled as they are in bacteria. A subsequent study found that, in polytene chromosomes, ribosomes colocalize with transcriptional components (Brogna et al., 2002), further substantiating the coordination of translation and transcription.
Despite multiple approaches pointing toward the conclusion that some translation occurs in the nucleus, doubt remained because of concerns of contamination from cytosolic or endoplasmic reticulum–bound ribosomes and antibody specificity (Nathanson et al., 2003; Dahlberg and Lund, 2004). In using both a novel method for identifying translation sites, termed ribopuromycylation, and by imaging translation in intact cells, David et al. (in this issue) circumvent many of these concerns and provide significant new evidence for nuclear translation. In the ribopuromycylation technique, cells are first treated with an inhibitor of translation elongation to yield stalled polyribosomes with associated nascent peptide chains. Subsequent treatment with puromycin results in its covalent incorporation into nascent chains. Importantly, the puromycylated nascent chains retain their association with the ribosome, thereby enabling imaging of translation with antipuromycin antibodies. Chemical and viral inhibitors of translation serve as negative controls to show that this technique does indeed capture active translational elongation. In this carefully performed series of experiments, David et al. (2012) find a substantial signal corresponding to translation in the nucleus and that is particularly enriched in the nucleolus. That the antipuromycin signal was substantially colocalized with an antiribosomal protein antibody lends additional credence to the observation.
Combined with the aforementioned studies, a parsimonious conclusion would be that an appreciable amount of ribosomal translation activity occurs within the nucleus. Although the noted absence of some initiation factors is of interest, it is important to recognize that the ribosome is, at its core, an enzyme with some degree of promiscuity. Indeed, the initial characterization of the ribosome relied on its ability to translate polyuridylic acid to yield polyphenylalanine (Crick et al., 1961). Nuclear ribosomes could, therefore, plausibly rely on a subset of factors outside of the core initiation complexes, as we understand them.
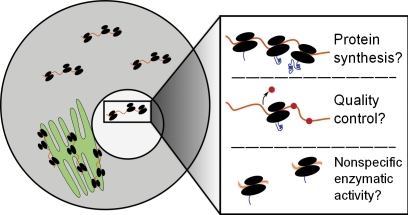
With the phenomenon of nuclear translation on more stable footing, several important questions regarding its mechanism and biological function can be asked. Most important is a fundamental distinction: is the primary function of ribosomes in the nucleus to synthesize nuclear proteins or to serve in a quality control role, such as nonsense-mediated decay (NMD)? There is some evidence for each possibility. NMD, in which ribosomes scan for mRNAs with premature stop codons and mark them for degradation (Belgrader et al., 1993), is appealing to imagine as a nuclear process. Live-cell imaging of mRNAs with a premature stop codon found that nonsense mutations are identified cotranscriptionally (de Turris et al., 2011), and components of the NMD machinery can physically associate with the transcriptional complex (Iborra et al., 2004). On the other hand, there have been studies of substantial buildup of newly synthesized proteins in the nucleus (Birnstiel and Flamm, 1964), and as we have discussed, intact polyribosomes have been detected in the nucleus (Goidl et al., 1975). Some combination of these two functional roles may be at play, each of which stands to contribute to our understanding of posttranscriptional gene regulation (Fig. 1). A critical missing piece of the puzzle is insight into the identities of the (presumably) mRNAs that serve as substrates for nuclear translation. Here, genome-scale ribosome footprinting (Ingolia et al., 2009) could be quite informative, assuming that sufficient biochemical evidence could be marshaled to convincingly demonstrate that the nuclear polyribosome fraction in question originates from intranuclear sites rather than the outer nuclear envelope, which is itself richly arrayed with polyribosomes.
Figure 1.
Potential roles of nuclear ribosomal activity. We suggest three general classes of potential activities for nuclear ribosomes. First is bona fide protein synthesis, in which mature proteins (likely those that are functional in the nucleus) are synthesized on polyribosomes. A second possibility is that nuclear ribosomes participate in a quality control process, either of mRNAs or of the ribosomes themselves. Depicted is the removal of the exon junction complex in NMD, a process that targets mRNAs with premature stop codons for degradation. A final possibility is that ribosomes react with RNA substrates in a nonspecific manner while in the nucleus, producing proteins with no particular coherence.
Some tantalizing clues about the biological function of nuclear translation are provided by the experiments by David et al. (2012) using viral infection to inhibit translation. Instead of a uniform reduction in translation, influenza infection specifically inhibits translation in the nucleolus. This may point toward an inhibition of NMD, which is used against viruses (LeBlanc and Beemon, 2004), or some particularly unique mode of inhibition. Here, too, insights into the RNAs undergoing translation would be very informative. Again, and given that the preponderance of experimental evidence convincingly demonstrates that intranuclear ribosomes are enzymatically active, the challenge is to distinguish between enzymatic “noise,” which may be an intrinsic behavior of intranuclear ribosomes, or biologically relevant mRNA translation. If the latter, such results would fit into our increasingly complex understanding of the cellular architecture of protein synthesis, which includes not only cytosolic protein synthesis but nuclear, endoplasmic reticular (Reid and Nicchitta, 2012), cytoskeletal (Kislauskis et al., 1997; Sharp et al., 2011), and mitochondrial-directed (Corral-Debrinski et al., 2000) translation.
Although considered skepticism is crucial to the progression of science, it is a valuable exercise to pose the questions what if and why. The history of science offers myriad examples of new ideas that overturned widely accepted models, many of which encountered fierce resistance in spite of fundamentally sound evidence. The question of nuclear translation may prove to be one such case study.
References
- Allfrey V.G. 1954. Amino acid incorporation by isolated thymus nuclei. I. The role of desoxyribonucleic acid in protein synthesis. Proc. Natl. Acad. Sci. USA. 40:881–885 10.1073/pnas.40.10.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L.E. 1993. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. USA. 90:482–486 10.1073/pnas.90.2.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M.L., Flamm W.G. 1964. Intranuclear site of histone synthesis. Science. 145:1435–1437 10.1126/science.145.3639.1435 [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Regener K., Schwappach B., Saffrich R., Paraskeva E., Hartmann E., Görlich D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21:6205–6215 10.1093/emboj/cdf613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S., Sato T.A., Rosbash M. 2002. Ribosome components are associated with sites of transcription. Mol. Cell. 10:93–104 10.1016/S1097-2765(02)00565-8 [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon C., Jacq C. 2000. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20:7881–7892 10.1128/MCB.20.21.7881-7892.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H., Barnett L., Brenner S., Watts-Tobin R.J. 1961. General nature of the genetic code for proteins. Nature. 192:1227–1232 10.1038/1921227a0 [DOI] [PubMed] [Google Scholar]
- Dahlberg J.E., Lund E. 2004. Does protein synthesis occur in the nucleus? Curr. Opin. Cell Biol. 16:335–338 10.1016/j.ceb.2004.03.006 [DOI] [PubMed] [Google Scholar]
- David A., Dolan B.P., Hickman H.D., Knowlton J.J., Clavarino G., Pierre P., Bennink J.R., Yewdell J.W. 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 197:45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Turris V., Nicholson P., Orozco R.Z., Singer R.H., Mühlemann O. 2011. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA. 17:2094–2107 10.1261/rna.02918111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C.L., Randal-Whitis L., Ellis S.R. 1999. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 59:704–710 [PubMed] [Google Scholar]
- Goidl J.A., Canaani D., Boublik M., Weissbach H., Dickerman H. 1975. Polyanion-induced release of polyribosomes from HeLa cell nuclei. J. Biol. Chem. 250:9198–9205 [PubMed] [Google Scholar]
- Iborra F.J., Jackson D.A., Cook P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science. 293:1139–1142 10.1126/science.1061216 [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Escargueil A.E., Kwek K.Y., Akoulitchev A., Cook P.R. 2004. Molecular cross-talk between the transcription, translation, and nonsense-mediated decay machineries. J. Cell Sci. 117:899–906 10.1242/jcs.00933 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324:218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis E.H., Zhu X., Singer R.H. 1997. β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136:1263–1270 10.1083/jcb.136.6.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.J., Beemon K.L. 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 78:5139–5146 10.1128/JVI.78.10.5139-5146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 282:2082–2085 10.1126/science.282.5396.2082 [DOI] [PubMed] [Google Scholar]
- Nathanson L., Xia T., Deutscher M.P. 2003. Nuclear protein synthesis: a re-evaluation. RNA. 9:9–13 10.1261/rna.2990203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D.W., Nicchitta C.V. 2012. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287:5518–5527 10.1074/jbc.M111.312280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J.A., Plant J.J., Ohsumi T.K., Borowsky M., Blower M.D. 2011. Functional analysis of the microtubule-interacting transcriptome. Mol. Biol. Cell. 22:4312–4323 10.1091/mbc.E11-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S.A., Warner J.R. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412–1416 [PubMed] [Google Scholar]