Autophagy genes are not only essential for exposing the engulfment signal on apoptotic cells but also function in phagocytes to promote apoptotic cell removal.
Abstract
Apoptotic cell degradation is a fundamental process for organism development, and impaired clearance causes inflammatory or autoimmune disease. Although autophagy genes were reported to be essential for exposing the engulfment signal on apoptotic cells, their roles in phagocytes for apoptotic cell removal are not well understood. In this paper, we develop live-cell imaging techniques to study apoptotic cell clearance in the Caenorhabditis elegans Q neuroblast lineage. We show that the autophagy proteins LGG-1/LC3, ATG-18, and EPG-5 were sequentially recruited to internalized apoptotic Q cells in the phagocyte. In atg-18 or epg-5 mutants, apoptotic Q cells were internalized but not properly degraded; this phenotype was fully rescued by the expression of autophagy genes in the phagocyte. Time-lapse analysis of autophagy mutants revealed that recruitment of the small guanosine triphosphatases RAB-5 and RAB-7 to the phagosome and the formation of phagolysosome were all significantly delayed. Thus, autophagy genes act within the phagocyte to promote apoptotic cell degradation.
Introduction
Apoptosis is an important cellular process that removes excess or damaged cells during organism development and adult tissue maintenance (Lettre and Hengartner, 2006; Conradt, 2009). The failure of apoptotic cell clearance contributes to autoimmune disorders, and excessive apoptosis has been associated with chronic neurodegenerative diseases (Elliott and Ravichandran, 2010). Apoptosis is executed in a highly regulated manner: a cell fated to die initiates an intrinsic program to destroy its intracellular structures and expose phosphatidylserine on the outer membrane as the engulfment signal. Subsequently, a phagocyte internalizes the apoptotic cell corpse and delivers the phagosome to lysosomes for degradation (Conradt and Xue, 2005; Kinchen and Ravichandran, 2008; Conradt, 2009).
Many molecules involved in apoptotic cell removal were identified through genetic screens (Elliott and Ravichandran, 2010). Two evolutionarily conserved signaling pathways (e.g., one by CED-1, CED-6, and CED-7 and the other by CED-2, CED-5, and CED-12 in Caenorhabditis elegans) perceive phosphatidylserine signal and activate a Rac GTPase (CED-10), leading to the reorganization of the actin cytoskeleton for phagocytosis (Reddien and Horvitz, 2004; Elliott and Ravichandran, 2010). Once inside the phagocyte, the endocytic machinery, including Rab GTPases and homotypic fusion and vacuole protein sorting complex components, mediates a series of maturation steps of phagosomes, which culminates lysosome delivery and the final degradation (Kinchen and Ravichandran, 2010). However, the precise molecular events in this clearance pathway are not fully understood.
Previous genetic experiments suggested that autophagy genes are required for apoptotic cell clearance (Qu et al., 2007). Autophagy is an intracellular degradation process, in which a double-membrane structure engulfs a portion of the cytosol and targets it for lysosomal degradation (Levine and Ranganathan, 2010). Yeast and C. elegans genetic screens isolated ∼20 conserved autophagy-related genes (ATGs) and several metazoan-specific autophagy genes (Levine and Ranganathan, 2010; Tian et al., 2010). Among these, genetic deletions of either the Atg5 or Atg6/Beclin1/bec-1 homologue in mouse embryoid bodies caused a failure in the generation of engulfing signals and resulted in the persistence of apoptotic cells (Qu et al., 2007). Recent studies using mammalian cell cultures showed that autophagy proteins were recruited from phagocytes to phagosomes, which contain apoptotic or necrotic cells, or to single-membrane entotic vacuoles, which harbor apoptotic cells and facilitated their elimination (Sanjuan et al., 2007; Florey et al., 2011; Martinez et al., 2011). However, the function of autophagy proteins in apoptotic cell degradation remains unclear in live animals.
Pioneering research on C. elegans postembryonic development reported apoptosis in Q cell lineage using Nomarski optics (Sulston and Horvitz, 1977). In this study, we developed fluorescence protein-based live-cell imaging methodologies to study the role of autophagy genes in Q cell apoptosis. We find that the autophagy proteins ATG-18 and EPG-5 do not function in initiating apoptosis in the Q cell but rather function in the phagocyte to process the engulfed Q cell after their internalization and help to deliver it to lysosomes for degradation.
Results and discussion
C. elegans apoptotic Q cells are degraded by an epithelial hyp7 cell
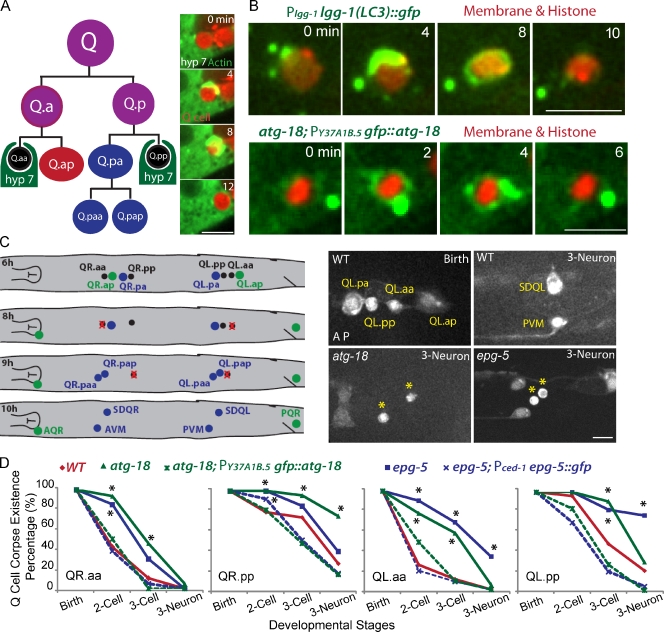
C. elegans Q neuroblast provides an appealing in vivo model system for understanding the contribution of autophagy genes to apoptosis. Q neuroblast at the left (QL) or the right (QR) of C. elegans L1 larva generates two apoptotic cells and three neurons by asymmetric cell divisions (Fig. 1 A; Sulston and Horvitz, 1977; Ou and Vale, 2009; Ou et al., 2010). We developed fluorescence markers and live-cell imaging methodology to follow, for the first time, the cellular events that the Q cell corpse undergoes from birth to its final degradation. Both apoptotic Q cells and the neighboring hyp7 cell can be individually identified using cell type–specific promoters (egl-17 gene promoter for the Q cell; ced-1 or Y37A1B.5 gene promoter for the hyp7 cell; Zhou et al., 2001; Hunt-Newbury et al., 2007). We observed that actin (a GFP-tagged construct expressed from a promoter in the epithelial hyp7 cell) formed an “actin halo” around the Q cell corpse, indicating that the hyp7 cell is the phagocyte for the apoptotic Q cell (Fig. 1 A, Fig. S1 A, and Video 1).
Figure 1.
C. elegans Q neuroblast apoptosis and the function of autophagy genes in Q cell corpse degradation. (A) The left cartoon shows the lineage of C. elegans Q neuroblasts and the phagocyte for Q cell corpse degradation. Three rounds of asymmetric cell divisions generate three neurons and two apoptotic cells (Q.aa and Q.pp). The neighboring epithelial cell, hyp7, engulfs and degrades the apoptotic Q cell. The right images show frames from time-lapse videos in which the GFP::actin cytoskeleton in the hyp7 cell forms an actin halo around the Q cell corpse (marked by cytosolic mCherry). More frames are shown in Fig. S1 A. (B) Still Images from videos show that autophagy markers GFP::LGG-1/LC3 (top) and GFP::ATG-18 (bottom) were recruited onto the outer surface of the Q cell corpse (red). Both markers were expressed under either the endogenous promoter for lgg-1 or hyp7 cell promoter for atg-18. The Q cell plasma membrane (mCherry with a myristoylation signal) and histone (his-24::mCherry) are shown in red. (C, left) The cartoon shows the developmental stage of Q cell at 6 h (birth of apoptotic Q cell), 8 h (two cell), 9 h (three cell), and 10 h (three neuron) after hatching. Definition is in the text (see The loss of autophagy genes delays Q cell corpse degradation section). Right images show Q cell corpse degradation in WT (top) or atg-18 (bottom left) or epg-5 mutant (bottom right). Asterisks show the ectopic Q cell corpses in autophagy mutants. Q cell names are adjacent. Genotypes are given in the top left. Developmental stages are given in the top right. Anterior of the cell is to the left. (D) Quantifications of Q cell corpse degradation in WT, atg-18 mutant, epg-5 mutant, or their rescue animals (dotted lines) at different Q cell developmental stages. *, P < 0.01, χ2 test. For each data point, n = 15–22 from a single experiment. Bars: (A and C) 5 µm; (B) 2.5 µm.
Recruitment of autophagy proteins to the engulfed apoptotic Q cell
Mammalian microtubule-associated protein LC3 (light chain 3) has been widely used as a cell biological marker for autophagy (Kabeya et al., 2000; Mizushima et al., 2010). We found that LGG-1::GFP (C. elegans LC3/ATG-8 homologue) was recruited as a ring or puncta on the outer surface of apoptotic Q cell (Fig. 1 B, top, 22% for ring; and Fig. S1 B, 32% for puncta, n = 23). We also examined the localization of two other autophagy proteins in the phagocyte: the WIPI/ATG-18 homologue and the metazoan-specific autophagy protein EPG-5 (Tian et al., 2010; Lu et al., 2011). Both GFP::ATG-18 and EPG-5::GFP appeared on the outer surface of apoptotic Q cells even more prominently than LGG-1::GFP (ATG-18: Fig. 1 B, bottom, 39% for ring; and Fig. S1 C, 39% for puncta, n = 18; and EPG-5: Fig. 4 C, 100% ring, n = 21). Thus, the autophagy proteins LGG-1, ATG-18, and EPG-5 are recruited from the phagocyte to the outer surface of internalized Q cell corpses.
Figure 4.
Sequential action of autophagy proteins during Q cell corpse degradation. (A) Still images show the corecruitment of BFP::RAB-5 with GFP::ATG-18 on the Q cell corpse. Time at 2 min is the first frame of RAB-5 and ATG-18 on the phagosome. (B, top) The PI(3)P binding motif of ATG-18 is required for ATG-18 recruitment on the Q cell corpse. n shows the number of Q.aa corpses examined. (bottom) Quantifications of QR.aa and QL.aa corpse degradation in WT and different genetic backgrounds at three-cell developmental stages. *, P < 0.01, χ2 test (mutant paired with WT). For each data point, n = 15–22 from a single experiment. Data about other cell types and developmental stage are given in Fig. S3. (C) The sequence of BFP::RAB-5 and EPG-5::GFP recruitment onto the Q cell corpse. Time at 0 min (the left column) shows the last frame of EPG-5::GFP on the phagosome, and time at 7 min (right) shows the first frame of GFP::RAB-5 on the Q cell corpse. (D) The sequence of engulfment (actin::BFP) and EPG-5::GFP during Q cell corpse degradation. Actin and EPG-5 were corecruited at 8 min, but EPG-5::GFP lasts longer on the phagosome (frame at 29 or 34 min). Bars, 5 µm.
The loss of autophagy genes delays Q cell corpse degradation
We examined whether the autophagy proteins are important for Q cell corpse degradation. Lgg-1 mutant alleles, like many other autophagy mutations, are embryonically lethal or show severe growth defects (Tian et al., 2010), preventing the examination of Q cell corpse degradation in their mutants. However, deletion alleles of atg-18 (gk378) and epg-5 (tm3425) are viable for phenotype analysis (Zhang et al., 2009; Tian et al., 2010), and we compared their Q cell corpse degradation with wild-type (WT) animals.
Apoptotic QR.aa and QL.aa cells (referred to Q.aa) underwent a similar degradation process (Fig. 1 C). Normally, more than half of Q.aa was eliminated at the two-cell stage of Q cell development (8 h after hatching, two cells, Q.ap and Q.pa, were born, and Q.ap migrates to its final position); ∼90% of Q.aa disappeared at the three-cell stage (Q.ap, Q.paa, and Q.pap were born), and no Q.aa cells were detected at the three-neuron stage (neurites of Q.ap, Q.paa, and Q.pap were visible at this stage; Fig. 1, C and D). In the atg-18 or epg-5 mutant, the clearance of the Q.aa corpse was significantly delayed. At the two-cell stage, >75% of animals retained their corpses. Only 50% of Q.aa was degraded at the three-cell stage, and even at the three-neuron stage, some Q.aa corpses were still visible (e.g., 33% of QL.aa in the epg-5 mutant; Fig. 1, C [bottom right] and D). A similar impairment in Q.pp corpse degradation also was observed in the atg-18 and epg-5 mutants (Fig. 1 D). Thus, autophagy genes atg-18 and epg-5 are required to remove apoptotic Q cells efficiently.
Expression of autophagy genes in the phagocyte rescues the Q cell corpse degradation phenotype
We determined whether the addition of ATG-18 or EGP-5 protein to the phagocyte rescues the delay of Q cell corpse degradation. The hyp7 cell–specific promoter, the Y37A1B.5 gene 5′ untranslated region (UTR; Hunt-Newbury et al., 2007), or the promoter of ced-1, which encodes a phagocytic receptor (Zhou et al., 2001), was used to express GFP::ATG-18 or EPG-5::GFP in the atg-18 or epg-5 mutant, respectively. We found that expression of GFP::ATG-18 or EPG-5::GFP in the hyp7 cell rescued the apoptotic Q cell degradation phenotype (Fig. 1 D, dotted lines). Thus, atg-18 and epg-5 function in the phagocyte to promote Q cell corpse clearance.
Apoptotic Q cells are engulfed normally but are not efficiently degraded in autophagy mutants
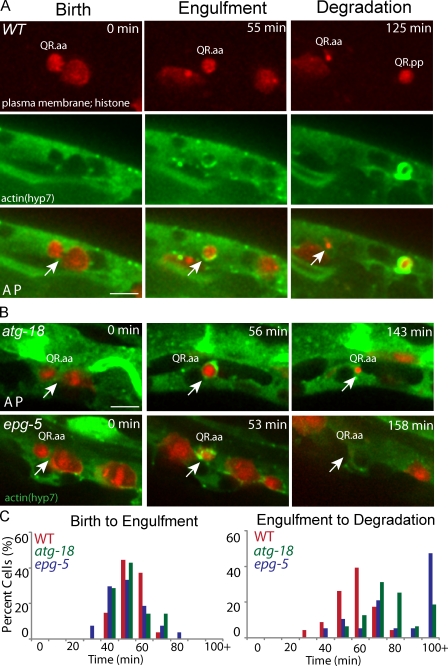
We performed kinetic analysis of Q cell corpse clearance in autophagy mutants to examine which step is defective. We first examined whether the engulfment of the Q cell corpse by the hyp7 cell is abnormal in autophagy mutants. To measure the time interval from the birth of apoptotic Q cells to their engulfment or final degradation, we constructed a C. elegans strain in which mCherry marked the Q cell plasma membrane and chromosomes, and GFP::actin in the hyp7 cell revealed the actin halo formation during engulfment.
In WT animals, Q.aa cells were engulfed 58 ± 8 min (mean ± SD; n = 54) after birth (Fig. 2, A and C; and Video 1), whereas Q.pp cell corpse degradation displays great variation in this timing as reported previously (Sulston and Horvitz, 1977). Our study thus focused on Q.aa cells. Strikingly, the C. elegans autophagy mutants did not exhibit any delay in actin halo formation in the hyp7 cell. Q cell corpse was internalized within 57 ± 10 min (n = 14) in the atg-18 mutant or 55 ± 11 min (n = 27) in the epg-5 mutant after birth (Fig. 2, B and C; and Fig. S2 A for statistical analysis). This result suggests that the death signal exposed by the apoptotic Q cell and the downstream reorganization of actin cytoskeleton in the phagocyte during the internalization process were all normal.
Figure 2.
Q cell corpse engulfment is normal, but its degradation in phagocytes is defective. (A) Still images of the birth, engulfment, and degradation of the Q cell corpse in WT. (top) Q cell plasma membrane and chromosome are imaged by mCherry. (middle) The actin in a hyp7 cell is labeled by GFP. The merged images are shown on the bottom. (B) Q cell corpse in the atg-18 (top) or epg-5 (bottom) mutant. The birth of the apoptotic Q cell was the end of cytokinesis of their mother (arrows; the cleavage furrows in the left of A and B). The engulfment of the apoptotic Q cell was the last frame with actin halo (arrows in the middle of A and B). The degradation was the last frame showing Q cell corpse (arrows in the right of A and B). Anterior of the cell is to the left. (C) Quantifications of the time for Q cell corpse engulfment and degradation. n = 10–29 from a single experiment. Statistical analysis was shown in Fig. S2 (A and B). Bars, 2.5 µm.
We continued to measure the time interval from Q cell internalization to the final clearance. WT animals took 62 ± 10 min (n = 23) to degrade apoptotic cell corpses (time from actin halo formation to complete loss of fluorescence from membranes and chromosomes). However, this time was significantly longer in the atg-18 mutant (84 ± 16 min; n = 16) and the epg-5 mutant (98 ± 35 min; n = 19; Fig. 2 and Fig. S2 B). Thus, the degradation of apoptotic Q cells inside the phagocyte was delayed in autophagy mutants.
Autophagy mutants show delayed recruitment of RAB-5, RAB-7, and lysosomal markers onto the apoptotic Q cell corpse
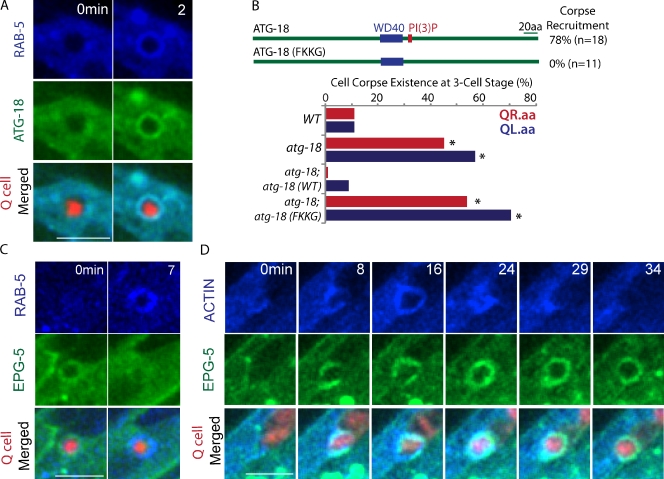
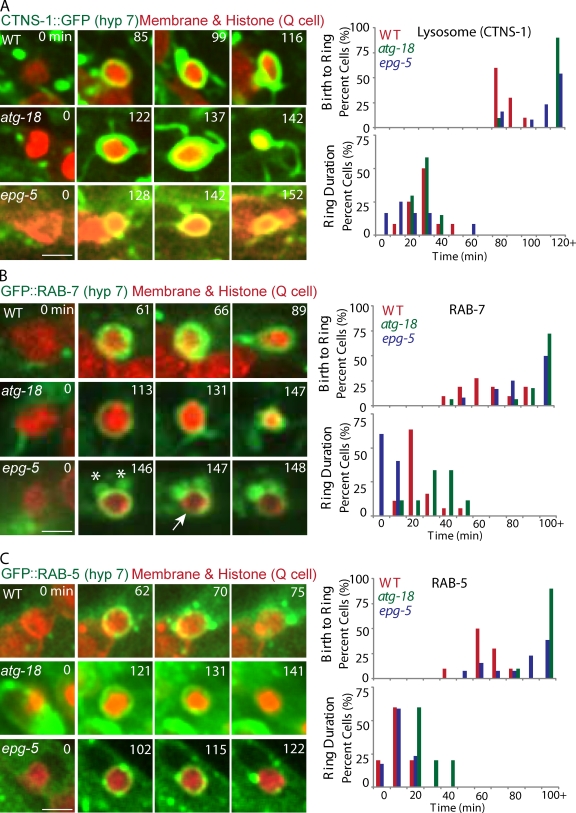
We investigated how autophagy proteins regulate Q cell corpse degradation in the phagocyte. After internalization, the phagosome undergoes a series of compositional changes that culminates in its fusion with lysosomes (Kinchen and Ravichandran, 2010). We first assessed whether the formation or stability of phagolysosomes containing Q cell corpses is affected in autophagy mutants. In WT animals, the lysosome marker CTNS-1::GFP was recruited onto Q cell corpses at 91 ± 8 min (n = 10) after Q cell birth, and CTNS-1::GFP rings lasted for another 32 ± 10 min (n = 12) until the corpse was completely degraded (Fig. 3 A and Video 2). We found that CTNS-1::GFP was recruited at 147 ± 27 min in the atg-18 mutant (n = 12) or 134 ± 49 min in the epg-5 mutant (n = 15) after Q cell birth, and we did not observe abnormality in the stability of the CTNS-1::GFP ring (Fig. 3 A and Fig. S2 C). Thus, recruitment of the lysosomal marker to the internalized Q cell corpse was significantly delayed in autophagy mutants.
Figure 3.
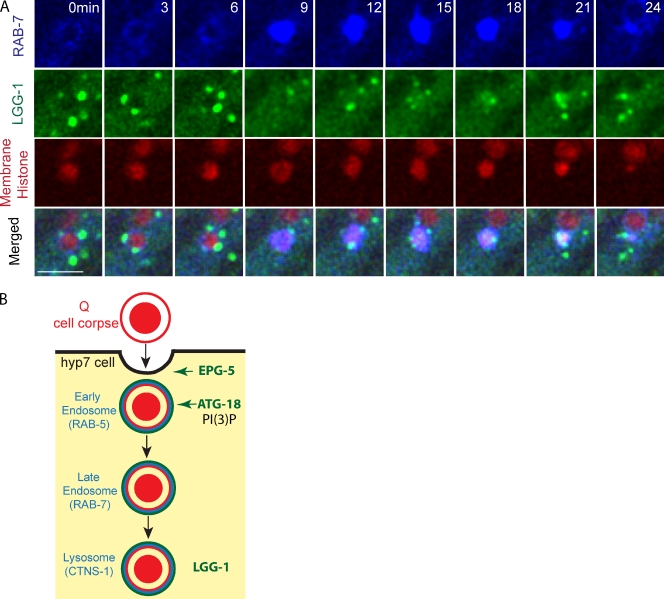
The recruitment and stability of lysosome, RAB-7, and RAB-5 onto Q cell corpse. (A–C) The left images are representative frames from time-lapse videos showing lysosome (labeled by CTNS-1::GFP; A), GFP::RAB-7 (B), and GFP::RAB-5 (C) recruitment and stability on the Q cell corpse in WT (top), atg-18 (middle), and epg-5 (bottom) mutants. The first column with time 0 shows the birth of apoptotic Q cells (mCherry), the second column shows the first frame of GFP signal on the Q cell corpse, and the right column shows the last frames of GFP signal on the Q cell corpse. The right top shows quantifications of the time interval from Q cell birth to the recruitment of lysosome, RAB-7, or RAB-5, and the bottom shows GFP signal duration time. n = 10–18 from a single experiment. Statistical analysis was shown in Fig. S2 (C–E). The asterisks show that the RAB-7 vesicles are associated with the Q cell corpse. The arrow shows the breakage of the RAB-7 ring on the Q cell corpse. Bars, 2.5 µm.
We next examined RAB-7 and RAB-5 recruitment during phagosome maturation. RAB-7 mediates the fusion of lysosome to phagosome, and RAB-5 acts upstream of RAB-7 by tethering early endosomes and nascent phagosomes (Guo et al., 2010; Kinchen and Ravichandran, 2010). We found that the recruitment of RAB-7 onto the phagosome was delayed from 70 ± 15 min in WT (n = 11) to 145 ± 66 min in atg-18 (n = 18) and 102 ± 30 min in epg-5 (n = 12) mutants (Fig. 3 B, Fig. S2 D, and Video 3), and GFP::RAB-5 recruitment was prolonged from 67 ± 9 min in WT (n = 11) to 140 ± 31 min in the atg-18 (n = 10) and 92 ± 23 min in the epg-5 (n = 13) mutant (Fig. 3 C, Fig. S2 E, and Video 4). The GFP::RAB-7 ring was less stable in the epg-5 mutant than that of WT or the atg-18 mutant (Fig. 3 B and Fig. S2 D). Thus, the delay of RAB-5 and RAB-7 recruitment or RAB-7 stability onto phagosomes is an early cause for the delay of phagolysosome formation in autophagy mutants.
The sequential recruitment of autophagy proteins on the Q cell corpse
We visualized the sequential recruitment of autophagy proteins and phagosomal markers on Q cell corpse. We first developed triple fluorescence protein labeling and imaging techniques in C. elegans. We modified the DNA coding sequence of a mammalian BFP, TagBFP, by C. elegans codon optimization and artificial intron insertions to facilitate its expression in nematodes.
Using time-lapse analysis, we found that GFP::ATG-18 was recruited on the Q cell corpse at approximately the same time as BFP::RAB-5 recruitment (n = 10; Fig. 4 A and Video 5). To elucidate how ATG-18 was recruited onto phagosomes, we examined whether phosphatidylinositol 3-phosphate (PI(3)P) binding motif in ATG-18 protein is essential for its recruitment and function. PI(3)P is generated on phagosomal membranes to promote apoptotic cell degradation by recruiting PI(3)P-binding effectors (Elliott and Ravichandran, 2010; Guo et al., 2010). FRRG (amino acids 227–230) in ATG-18 was identified as the PI(3)P binding site, and the mutation from FRRG to FKKG abolished its binding activity in vitro (Lu et al., 2011). We constructed transgenic C. elegans expressing GFP::ATG-18 (FKKG) and found that GFP::ATG-18 (FKKG) protein failed to be recruited to Q cell corpse and did not rescue the cell corpse degradation phenotype in the atg-18 mutant (Fig. 4 B and Fig. S3), suggesting that PI(3)P may recruit ATG-18 onto the cell corpse and that this recruitment is essential for Q cell corpse degradation.
The EPG-5 protein was reported to evenly distribute in the cytosol (Tian et al., 2010), but we found that EPG-5::GFP was recruited on the Q cell corpse 6 ± 4 min (n = 10) earlier than BFP::RAB-5 with a duration time of 11 ± 4 min (n = 21; Fig. 4 C). In support of this observation, we found that EPG-5::GFP was recruited onto the Q cell corpse at 59 ± 9 min (n = 19) after its birth, comparable with the time of actin halo formation in phagocytes. Our triple labeling analysis indeed showed that the EPG-5::GFP ring forms at the same time of actin::BFP halo formation on the Q cell corpse (n = 12; Fig. 4 D and Video 6). Thus, EPG-5 is recruited at a very early step of phagosome maturation and may play a role in RAB-5 recruitment.
LGG-1 was proposed to function at a late stage of autophagy (Zhang et al., 2009; Tian et al., 2010). We expressed a lgg-1::gfp transgene in the hyp7 cell and found that LGG-1::GFP was initially recruited to the Q cell corpse 11 ± 7 min (n = 10) after BFP::RAB-7 recruitment (Fig. 5 A and Video 7). The late recruitment of LGG-1::GFP on phagosomes was confirmed using the lysosome marker CTNS-1::BFP. In 50% of animals, LGG-1::GFP and CTNS-1::BFP were recruited at the same time onto the Q cell corpse; for the other animals, LGG-1::GFP was recruited 4 ± 3 min (n = 10) later than CTNS-1::BFP (Video 8).
Figure 5.
Recruitment of LGG-1 and RAB-7 on Q cell corpse and proposed model. (A) Still images show the sequence of BFP::RAB-7 and LGG-1::GFP recruitment onto the Q cell corpse (red). Bar, 5 µm. (B) Proposed model of the function of autophagy genes in Q cell corpse degradation. EPG-5 was recruited onto the phagosome during engulfment. ATG-18 was corecruited with RAB-5, and ATG-18 recruitment requires its association with PI(3)P on apoptotic corpses. EPG-5 and ATG-18 are required for RAB-5 recruitment. LGG-1 recruitment occurs at the phagolysosome stage.
Conclusion
Our study on C. elegans Q cell apoptosis showed that autophagy proteins ATG-18 and EPG-5 function sequentially to facilitate apoptotic cell degradation in the phagocyte by promoting phagosome maturation (Fig. 5 B). As ATG-18 and EPG-5 are two among >20 autophagy genes, a systematical examination of other autophagy genes is required for understanding how the autophagy pathway is involved in apoptotic cell degradation.
RNAi of the autophagy protein BEC-1, Beclin1 homologue, led to the delay of RAB-5 recruitment on phagosomes in C. elegans embryos and germlines (Florey et al., 2011; Martinez et al., 2011), which is consistent with our findings of ATG-18 and EPG-5 on Q cell corpse clearance. As many cell death genes play similar roles in different cell types (Conradt and Xue, 2005; Elliott and Ravichandran, 2010), we expected that ATG-18 and EPG-5 proteins might play roles for the apoptotic cell clearance in other cell lineages. ATG-5 and Beclin1 were reported to be required for apoptotic cell removal by generating ATP-dependent engulfment signals during mouse developmental programmed cell death (Qu et al., 2007), whereas loss of C. elegans atg-18 and epg-5 only affects degradation, suggesting that autophagy proteins may function differently in mammals and C. elegans apoptosis.
ULK1/ATG-1 was known to be associated with ATG-18 to early autophagosomal structures and is required for the autophagosome formation (Polson et al., 2010). However, we did not detect any defects of Q cell corpse clearance in the C. elegans ATG-1/unc-51 mutant (Fig. S3), which is consistent with the recent findings from Toll-like receptor–mediated phagosomes or entotic cells (Florey et al., 2011; Martinez et al., 2011). Analysis of LGG-1::GFP during cell corpse clearance showed that LGG-1 is directly recruited onto corpses (Fig. 1 B), forming autophagosome-like puncta near the phagosome (Fig. S1 B), or is not associated with corpses, reflecting a complex behavior of LGG-1 in cell corpse degradation. The lipidation machinery regulates the recruitment of LGG-1 onto autophagosomes (Mizushima et al., 2010). However, we did not observe any association of GFP-tagged ATG-5 or ATG-7 with the Q cell corpse (unpublished data), and in atg-7 mutant, no apoptotic Q cell corpse degradation phenotype was detected (Fig. S3). Thus, the role of some autophagy proteins in phagocytosis might be different from macroautophagy.
Autophagy machinery was recently identified to be involved in Toll-like receptor–regulated phagolysosomal pathways (Sanjuan et al., 2007) and the degradation of phagocytosed microorganisms, such as virus, bacteria, and protozoa (Levine et al., 2011). Our experiments provide in vivo evidence for the function of autophagy genes in the apoptotic cell removal in C. elegans. Recent work on mammalian cells showed that autophagy proteins are required for the degradation of necrotic cells or entotic cells (Florey et al., 2011; Martinez et al., 2011). Thus, the involvement of autophagy proteins in the clearance of engulfed dead cells may be a conserved function across species.
Materials and methods
C. elegans strains, genetics, and DNA manipulations
C. elegans strains were raised on a nematode growth medium plate seeded with the Escherichia coli strain OP50 at 20°C. Strains are listed in the Table S1. PCR templates and primers are listed in Table S2, and plasmid constructs are given in Table S3. DNA at the concentration of 10–30 ng/µl were transformed by germline microinjection. We reengineered TagBFP for expression in C. elegans by a similar strategy of optimizing mCherry (Green et al., 2008).
Live-cell imaging
C. elegans L1 larva were anesthetized with 0.1 mmol/liter levamisole in M9 buffer, mounted on 3% agarose pads, and maintained at 20°C. Our imaging system includes a microscope (Axio Observer.Z1; Carl Zeiss) equipped with a 100×, 1.45 NA objective, an EM charge-coupled device camera (iXon+; Andor), and the 405-, 488-, and 561-nm lines of a laser system (CW CDRH USB; Sapphire) or an argon and krypton laser attached to a spinning-disk confocal scan head (CSU-X1 Spinning Disk Unit; Yokogawa). Time-lapse images were acquired with exposure time of 200–300 ms at every 60 s by µManager or Focus Image software (developed by X. Zhang at the Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). We used ImageJ software (National Institutes of Health) to process the images.
Statistical analysis
Student’s t test and χ2 analysis were used to examine statistical differences of Q cell corpse duration, protein recruitment, and stability on the phagosome between WT and autophagy gene mutants as indicated in the figure legends.
Online supplemental material
Fig. S1 shows actin halo formation and LGG-1 and ATG-18 recruitment onto the Q cell corpse. Fig. S2 shows the statistical analysis of Q cell corpse degradation in WT and autophagy mutants. Fig. S3 shows quantifications of Q cell corpse degradation in WT and autophagy mutants. Video 1 shows the engulfment and degradation of the apoptotic Q cell. Videos 2–4 show the recruitment of CNTS-1 (Video 2), RAB-7 (Video 3), and RAB-5 (Video 4) onto the Q cell corpse. Videos 5–8 show the recruitment order of RAB-5 and ATG-18 (Video 5), EPG-5 and actin (Video 6), RAB-7 and LGG-1 (Video 7), and CTNS-1 and LGG-1 (Video 8) onto the Q cell corpse. Table S1 shows C. elegans strains. Table S2 shows PCR templates and primers. Table S3 shows plasmid constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111053/DC1.
Acknowledgments
We thank Dr. H. Zhang and the Caenorhabditis Genetics Center for strains and Dr. T. Xu, Dr. W. Ji, and Mr. X. Zhang for their support with microscopy setup.
This work was supported by grants from the National Basic Research Program of China to W. Li and G. Ou (973 Program; 2012CB966800 and 2012CB945002), the National Natural Science Foundation of China to W. Li, Y. Yang, and G. Ou (31101002, 31100972, 31171295, and 31190063), Junior Thousand Talents Program of China to G. Ou, a Ministry of Science and Technology grant to X. Wang (2010CB835201), the National Institutes of Health, and the Howard Hughes Medical Institute to R.D. Vale.
Footnotes
Abbreviations used in this paper:
- ATG
- autophagy-related gene
- PI(3)P
- phosphatidylinositol 3-phosphate
- UTR
- untranslated region
- WT
- wild type
References
- Conradt B. 2009. Genetic control of programmed cell death during animal development. Annu. Rev. Genet. 43:493–523 10.1146/annurev.genet.42.110807.091533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., Xue D. 2005. Programmed cell death. WormBook. 10.1895/wormbook.1.32.1 (accessed March 7, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.R., Ravichandran K.S. 2010. Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189:1059–1070 10.1083/jcb.201004096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O., Kim S.E., Sandoval C.P., Haynes C.M., Overholtzer M. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13:1335–1343 10.1038/ncb2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Audhya A., Pozniakovsky A., Dammermann A., Pemble H., Monen J., Portier N., Hyman A., Desai A., Oegema K. 2008. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 85:179–218 10.1016/S0091-679X(08)85009-1 [DOI] [PubMed] [Google Scholar]
- Guo P., Hu T., Zhang J., Jiang S., Wang X. 2010. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc. Natl. Acad. Sci. USA. 107:18016–18021 10.1073/pnas.1008946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., et al. 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5:e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., Ravichandran K.S. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9:781–795 10.1038/nrm2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., Ravichandran K.S. 2010. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 464:778–782 10.1038/nature08853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G., Hengartner M.O. 2006. Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 7:97–108 10.1038/nrm1836 [DOI] [PubMed] [Google Scholar]
- Levine B., Ranganathan R. 2010. Autophagy: Snapshot of the network. Nature. 466:38–40 10.1038/466038a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., Lin L., Kovács A.L., Yu L., Zhang H. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 21:343–357 10.1016/j.devcel.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Martinez J., Almendinger J., Oberst A., Ness R., Dillon C.P., Fitzgerald P., Hengartner M.O., Green D.R. 2011. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 108:17396–17401 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mizushima N., Yoshimori T., Levine B. 2010. Methods in mammalian autophagy research. Cell. 140:313–326 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Vale R.D. 2009. Molecular signatures of cell migration in C. elegans Q neuroblasts. J. Cell Biol. 185:77–85 10.1083/jcb.200812077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Stuurman N., D’Ambrosio M., Vale R.D. 2010. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science. 330:677–680 10.1126/science.1196112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson H.E., de Lartigue J., Rigden D.J., Reedijk M., Urbé S., Clague M.J., Tooze S.A. 2010. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 6:506–522 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R.N., Gilpin C., Levine B. 2007. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 128:931–946 10.1016/j.cell.2006.12.044 [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Horvitz H.R. 2004. The engulfment process of programmed cell death in caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20:193–221 10.1146/annurev.cellbio.20.022003.114619 [DOI] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 450:1253–1257 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110–156 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E., Horvitz H.R. 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 104:43–56 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]