Abstract
Background
Clostridium difficile infection (CDI) is an important cause of morbidity and mortality internationally, yet there are important regional differences in the epidemiology and microbiology of disease. Most reports have come from North America and Europe, with limited information from other regions, including the Middle East. Given the changes in the epidemiology of CDI in developed countries, particularly associated with the dissemination of hypervirulent epidemic clones, an understanding of the epidemiology and microbiology of CDI in diverse regions is warranted. This study involved collection of stool samples from individuals with diarrhea at the Isfahan University of Medical Sciences Teaching Hospital, Isfahan, Iran, between October 2010 and March 2011. Selective enrichment culture for C. difficile was performed and isolates were characterised using ribotyping, PCR for the detection of tcdA, tcdB and cdtB genes, and tcdC sequence analysis.
Findings
Clostridium difficile was isolated from 19/89 (21%) stool samples of 17/86 (20%) patients. 13/17 (77%) cases of CDI were hospital-associated. Patients with CDI were significantly older (43 ± 28y) than those with non-CDI diarrhea (24, ± 26y)(P = 0.018). All isolates were toxigenic, and possessed genes encoding for toxins A and B. Six (32%) of 19 isolates also possessed cdtB. Twelve ribotypes were identified. Ribotype 078/toxinotype V was most common, accounting for 4 (21%) of isolates. A single isolate of a different toxinotype V ribotype was identified, as was a toxinotype XXIV isolate. The remaining isolates consisted of 9 different toxinotype 0 ribotypes.
Conclusions
CDI is an important cause of diarrhea in patients in this hospital. The diversity of ribotypes was striking, and the number of different types suggests the presence of a broad range of strains in the community, the hospital or both. The predominance of toxinotype V strains, which have been associated with community-associated disease and food animals, was unexpected and possible sources of this type require further investigation.
Keywords: Clostridium difficile, Diarrhea, Nosocomial, Infectious disease, Gastroenterology
Findings
Clostridium difficile is a leading cause of hospital-associated and antimicrobial-associated diarhea, and is of significant concern because of the increasing morbidity, mortality and relapse rates [1], along with the emergence of community-associated disease [2]. Some of these clinical and epidemiological changes have been associated with dissemination of hypervirulent clones, particularly ribotype 027 (toxinotype III, North American pulsotype (NAP)1)[3] and to a lesser degree ribotype 078 (toxinotype V, NAP7/8) [4,5]. Clostridium difficile infection (CDI) has been reported throughout much of the world, but most data come from developed countries in North America and Europe. Limited information is available regarding the role of C. difficile in diarrheic hospital patients in people in Iran or other Middle Eastern countries, or about C. difficile strains that are involved. Therefore, the aim of present study was to determine the prevalence of C. difficile in diarrheic patients in hospital in Isfahan, Iran, and to characterize isolates.
The study was conducted at the Isfahan University of Medical Sciences Teaching Hospital, Isfahan, Iran, the largest tertiary care medical centre in the region with 800 beds, three intensive care units and all major clinical specialties. Aliquots of stool specimens that had been collected from diarrheic patients between October 2010 and March 2011 for other diagnostic testing were studied. Both patients that were admitted to hospital with diarrhea and those who developed diarrhea during hospitalization were included. Diarrhea was defined as watery, loose or unformed stool passed at a frequency of three times or more per 24 hours. Demographic and basic medical information were collected from the medical record. Cases were classified as community associated (CA) if the onset of symptoms occurred prior to, or within 48 h of admission and if they had not been hospitalized in the preceding 3 months [6]. Cases were classified has hospital-associated (HA) if the onset was greater than 48 h after admission. Cases that had diarrhea at admission or developed diarrhea within the first 48 h of admission but had been hospitalized within the preceding 4 weeks were classified as hospital-associated, community-onset (HA-CO). Studies such as this are exempt from ethics board review at Isfahan University and instead require approval of the Isfahan University of Medical Sciences Vice Chancellor for Research, which was obtained.
Selective enrichment culture was performed. Approximately 5 g of stool was inoculated into 25 ml of C. difficile selective enrichment broth containing 40 g/l proteose peptone, 5.0 g/l disodium hydrogen phosphate, 0.1 g/l magnesium sulphate, 2.0 g/l sodium chloride, 6.0 g/l fructose and 1.0 g/l sodium taurocholate supplemented with cysteine hydrochloride, norfloxacin and moxalactam, and anaerobically incubated at 37 C for 5–7 days. Two ml of each culture were then added to an equal volume of absolute ethanol, mixed and left at room temperature for 1 h. Alcohol shocked cultures were then centrifuged (4000 rpm/10 min) and the pellet was streaked onto C. difficile moxalactam norfloxacin (CDMN) agar (Oxoid) and incubated anaerobically at 37 C for 24-48 h. Suspect colonies were subcultured and identified as C. difficile on the basis of characteristic colony morphology, odour, Gram stain morphology and L-proline aminopeptidase test (Prodisk, Remeb, Lenexa, KS, USA). All isolates were screened for the presence of genes encoding toxin A (tcdA), toxinB (tcdB), the binding component of CDT (cdtB) and triose phosphate isomerase (tpi) as have been previously described [7-9]. Isolates were also subjected to PCR ribotyping [10], toxinotyping[11] and tcdC sequence analysis [12]. For ribotyping, ribotypes were assessed visually and compared to an internal collection of ribotypes from over 3000 isolates from humans and animals. When a ribotype pattern was known to be a recognized international ribotype through previous typing of reference strains from the HPA Anaerobic Reference Laboratory (http://www.webcitation.org/64HSI8B56) (Cardiff, UK), the appropriate numerical designation (i.e. 078) was used. Otherwise, internal nomenclature was used.
Categorical comparisons were performed using Fisher’s exact test while t-test was used for continuous data. A P value of <0.05 was considered significant.
Eighty-six diarrheic patients were enrolled. Clostridium difficile was isolated from 19/89 (21%) of stool samples from 17/86 (20%) patients; 8/36 (22%) women and 9/50 (18%) men (P = 0.78). 13/17 (76%) cases were HA-CDI while the other 4 (24%) were CA-CDI. Patients with CDI were significantly older (43 ± 28y) than those with non-CDI diarrhea (24, ± 26y)(P = 0.018). Five (29%) CDI patients were over the age of 65 compared to 6/69 (8.7%) without non-CDI diarrhea (P = 0.037). Fifteen (88%) of CDI patients had received antimicrobials prior to the onset of diarrhea, compared to 58 (84%) others (P = 1.0).
Two patients had separate episodes of diarrhea 62 and 33 days apart, and the same ribotype was isolated from each episode. Follow-up data were limited but this represents a minimal recurrence rate of 12%. Two (12%) patients with CDI died during hospitalization, however death was only attributed to CDI in 1 (5.9%) individual.
All isolates possessed genes encoding for toxins A and B, and 12 different ribotypes were identified (Table 1). In addition to tcdA and tcdB, six (32%) isolates also possessed cdtB. Four of these were ribotype 078 and toxinotype V, and contained a 39 bp deletion and C184T nonsense mutation in tcdC. One ribotype was a toxinotype V isolate that differed from ribotype 078 but had the same tcdC deletion and mutation. Patients carrying these five isolates were admitted over a period of approximately 2 months, with no more than one per week (Figure 1). The other cdtB positive isolate was a toxinotype XXIV strain that contained an 18-bp tcdC deletion in tcdC but not the upstream truncating mutation that is characteristic of the toxinotype III hypervirulent ribotype 027/NAP1. The remaining 13 isolates from 11 patients were toxinotype 0 strains consisting of 9 different ribotypes, including ribotype 014 and 8 ribotypes not previously encountered in this laboratory.
Table 1.
Ribotype and toxinotype data for Clostridium difficile isolated from diarrheic individuals in an Iranian hospital
| Ribotype | n | Toxin gene profile | Toxinotype |
|---|---|---|---|
| 078 |
4 |
tcdA, tcdB, cdtB |
V |
| IR1 |
1 |
tcdA, tcdB, cdtB |
V |
| 014 |
1 |
tcdA, tcdB |
0 |
| IR2 |
1 |
tcdA, tcdB, cdtB |
XXIV |
| IR3 |
1 |
tcdA, tcdB |
0 |
| IR4 |
3 |
tcdA, tcdB |
0 |
| IR5 |
1 |
tcdA, tcdB |
0 |
| IR6 |
1 |
tcdA, tcdB |
0 |
| IR7 |
1 |
tcdA, tcdB |
0 |
| IR8 |
1 |
tcdA, tcdB |
0 |
| IR9 |
1 |
tcdA, tcdB |
0 |
| IR10 | 1 | tcdA, tcdB | 0 |
Figure 1.
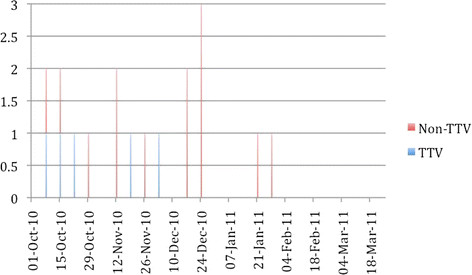
Temporal distribution of Clostridium difficile infection caused by toxinotype V (TT V) and non toxinotype V (Non TT V) isolates
All 4 patients from which ribotype 078 was isolated were from rural areas, but had hospital-associated diarrhea. The mean age of patients with ribotype 078 was 29.2y compared to 49.1y for other ribotypes, but this was not statistically significant (P = 0.25)
The prevalence of CDI (21%) was consistent with many studies of hospitalized individuals in other regions, albeit higher than two recent Iranian studies that reported isolation of toxigenic C. difficile from 6.1-6.8% of stool samples [13,14]. Other regional studies have reported 4.6-9.5% prevalence in two studies from Saudi Arabia [15], 10.5% in Kuwait [16], and 13.7% from Jordan [17]. However, limited emphasis should be placed on comparisons of prevalence because of differences in study methodology. The key finding is that CDI appears to be common in patients in Iranian hospitals, and may be underdiagnosed since testing appears to be rarely performed, in part because of limited access to commercial assays.
As is consistent with other reports, CDI was associated with older individuals. However, in this study, the CDI patients were younger (mean 43y) compared to most other reports [1,18] and less than 30% of CDI patients were over the age of 65. Reasons for this difference are not apparent. Classically, CDI has been considered a disease predominantly of elderly individuals, however there are increasing reports of CDI in younger individual, including people that would be considered at low risk of infection [19-21]. Only limited investigation of risk factors was performed here based on the available data. There was no association between antimicrobial use and CDI in these patients, however antimicrobial use was very common in both CDI and non-CDI groups, and the role of antimicrobials in CDI can certainly not be dismissed by these data.
Community-associated CDI is being more commonly reported in some regions, often associated with milder disease in younger individuals and people with little or no antimicrobial exposure [2,20,22]. In this study, 23% of cases were classified as CA-CDI, and all involved different toxinotype 0 strains. While the majority of cases were therefore associated with hospitalization, it cannot be stated with certainty that exposure occurred in hospital since testing of patients at admission, prior to the onset of disease, would be required to ensure that they were not carrying C. difficile at the time of admission. Since there was a wide diversity in types and little evidence of temporal clustering (Figure 1), a point-source of hospital exposure was not apparent. Hospital-associated CDI could have occurred from exposure of different patients to different strains in hospital, but also could have been the result of development of disease from C. difficile that was resident in the intestinal tract at the time of admission.
The wide diversity of ribotypes was interesting. Toxinotype 0 strains predominated, yet only 1 isolate (ribotype 014) was consistent with major reported human types. The diversity of ribotypes within toxinotype 0 strains and the differences between common toxinotype 0 ribotypes in this versus other studies was interesting. The relatively high prevalence of ribotype 078 was consistent with two studies from Kuwait that identified ribotype 078 as one of the predominant types [23,24]. Additionally, the one other toxinotype V isolate is likely closely related to ribotype 078, and the overall prevalence of toxinotype V strains was 29%. Recent studies have reported increases in the prevalence of ribotype 078 or toxinotype V strains in humans, particularly in community-associated disease [19,25]. This strain can also be found commonly in food animals and food [26-28], leading to concern that it might be foodborne or zoonotic [29]. While all patients with ribotype 078 were from rural regions, disease onset was in hospital and the origin of infection cannot be discerned. Further, no information about animal or food contact is available, and no data regarding C. difficile in animals or food are available for this region. Therefore no assumptions can be made regarding potential sources of this strain. The four patients with ribotype 078 were admitted over a 37 day period (Figure 1), and the potential that these represent a small hospital-associated cluster cannot be dismissed. Ribotype 027, a toxinotype III strain associated with hypervirulent disease and epidemics internationally [30], was not identified in any patient. The diversity in ribotypes is consistent with the only available Iranian study, which reported 28 different ribotypes amongst 178 isolates [14]. That study did not compare strains to internationally recognized reference strains and did not perform other typing methods, so detailed comparison with these data is impossible.
This study indicates that C. difficile might be an important enteric pathogen in patients in Iranian hospitals. Efforts must be undertaken to diagnose CDI to allow for targeted therapy and to provide a better understanding of this important disease. While some aspects identified here are consistent with reports of CDI in other areas, further study of the epidemiology and microbiology of CDI in this region is required to explore some apparent differences, such as the younger age distribution and the predominance of ribotype 078. While ribotype 027 was not identified, further surveillance is required to determine whether this strain is indeed present in the country and to monitor for its emergence.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MJ, JSW and KW designed the study. FK coordinated sample and medical record data collection. MJ performed the laboratory tests under the direction of JSW. All authors contributed to writing and reviewing the final manuscript. All authors read and approved the final manuscript.
Contributor Information
Mohammad Jalali, Email: mjalali@uoguelph.ca.
Farzin Khorvash, Email: khorvash@med.mui.ac.ir.
Keith Warriner, Email: kwarrine@uoguelph.ca.
J Scott Weese, Email: jsweese@uoguelph.ca.
Acknowledgements
The authors thank Joyce Rousseau for technical assistance.
References
- Pépin J, Valiquette L, Alary M, Villemure P, Pelletier A, Forget K, Pépin K, Chouinard D. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004;171(5):466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Severe Clostridium difficile-associated disease in populations previously at low risk–four states, 2005. MMWR Morb Mortal Wkly Rep. 2005;54(47):1201–1205. [PubMed] [Google Scholar]
- Goorhuis A, Van der Kooi T, Vaessen N, Dekker F, Van den Berg R, Harmanus C, van den Hof S, Notermans D, Kuijper E. Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin Infect Dis. 2007;45(6):695–703. doi: 10.1086/520984. [DOI] [PubMed] [Google Scholar]
- Mulvey MR, Boyd DA, Gravel D, Hutchinson J, Kelly S, Mcgeer A, Moore D, Simor A, Suh KN, Taylor G, Weese JS, Miller M. Hypervirulent Clostridium difficile strains in hospitalized patients. Canada. Emerging Infect Dis. 2010;16(4):678–81. doi: 10.3201/eid1604.091152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik M, Widmer A, Zimmermann O, Eckert C, Barbut F. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J Clin Microbiol. 2008;46:2146. doi: 10.1128/JCM.00598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L, Coignard B, Dubberke E, Song X, Horan T, Kutty P. Unknown: Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28(2):140–145. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim S, Chong Y, Wasito E. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36(8):2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186(2):307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- Lemee L, Dhalluin A, Testelin S, Mattrat M, Maillard K, Lemeland J, Pons J. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42(12):5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175(2):261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmée M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36(8):2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry SR, Marsh JW, Muto CA, O’Leary MM, Pasculle AW, Harrison LH. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45(1):215–221. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghifard N, Salari MH, Ghassemi MR, Eshraghi S, Amin Harati F. The incidence of nosocomial toxigenic Clostridium difficile associated diarrhea in Tehran tertiary medical centers. Acta Med Iran. 2010;48(5):320–325. [PubMed] [Google Scholar]
- Sadeghifard N, Salari MH, Ranjbar R, Ghafouryan S, Raftari M, Abdulamir AS, Fatimah AB, Kazemi B. The clinical and environmental spread and diversity of toxigenic Clostridium difficile diarrheic in the region of the Middle East. Rev Infection. 2010;1:180–187. [Google Scholar]
- Al-Tawfiq JA, Abed MS. Clostridium difficile-associated disease among patients in Dhahran, Saudi Arabia. Travel Med Infect Dis. 2010;8(6):373–376. doi: 10.1016/j.tmaid.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Jamal W, Rotimi VO, Brazier J, Duerden BI. Analysis of prevalence, risk factors and molecular epidemiology of Clostridium difficile infection in Kuwait over a 3-year period. Anaerobe. 2010;16(6):560–565. doi: 10.1016/j.anaerobe.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Nasereddin LM, Bakri FG, Shehabi AA. Clostridium difficile infections among Jordanian adult hospitalized patients. Am J Infection Control. 2009;37(10):864–866. doi: 10.1016/j.ajic.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Barbut F, Corthier G, Charpak Y, Cerf M, Monteil H, Fosse T, Trévoux A, De Barbeyrac B, Boussougant Y, Tigaud S. et al. Prevalence and pathogenicity of Clostridium difficile in hospitalized patients. A French multicenter study. Arch Intern Med. 1996;156(13):1449–1454. doi: 10.1001/archinte.1996.00440120107012. [DOI] [PubMed] [Google Scholar]
- Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, Bergwerff AA, Dekker FW, Kuijper EJ. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47(9):1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Surveillance for community-associated Clostridium difficile--Connecticut, 2006. MMWR Morb Mortal Wkly Rep. 2008;57(13):340–343. [PubMed] [Google Scholar]
- Vindigni SM, Shane AL. Clostridium difficile infections among hospitalized children, United States, 1997–2006. Emerging Infect Dis. 2010;16(10):1651–1652. doi: 10.3201/eid1610.100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltansing S, van den Berg RJ, Guseinova RA, Visser CE, van der Vorm ER, Kuijper EJ. Characteristics and incidence of Clostridium difficile-associated disease in The Netherlands, 2005. Clin Microbiol Infect. 2007;13(11):1058–1064. doi: 10.1111/j.1469-0691.2007.01793.x. [DOI] [PubMed] [Google Scholar]
- Jamal WY, Rotimi VO, Grubesic A, Rupnik M, Brazier JS, Duerden BI. Correlation of multidrug resistance, toxinotypes and PCR ribotypes in Clostridium difficile isolates from Kuwait. J Chemother. 2009;21(5):521–526. doi: 10.1179/joc.2009.21.5.521. [DOI] [PubMed] [Google Scholar]
- Rotimi VO, Jamal WY, Mokaddas EM, Brazier JS, Johny M, Duerden BI. Prevalent PCR ribotypes of clinical and environmental strains of Clostridium difficile isolated from intensive-therapy unit patients in Kuwait. J Med Microbiol. 2003;52(Pt 8):705–709. doi: 10.1099/jmm.0.05207-0. [DOI] [PubMed] [Google Scholar]
- Jhung M, Thompson A, Killgore G, Zukowski W, Songer G, Warny M, Johnson S, Gerding D, McDonald L, Limbago B. Toxinotype V Clostridium difficile in Humans and Food Animals. Emerging Infect Dis. 2008;14(7):1039–1045. doi: 10.3201/eid1407.071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Zwartkruis-Nahuis A, Heuvelink AE, Harmanus C, Kuijper EJ. Prevalence of Clostridium difficile in retailed meat in the Netherlands. Int J Food Microbiol. 2011;144(3):561–564. doi: 10.1016/j.ijfoodmicro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. Clostridium difficile in retail meat products, USA, 2007. Emerging Infect Dis. 2009;15(5):819–821. doi: 10.3201/eid1505.081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weese JS, Avery BP, Rousseau J, Reid-Smith R. Detection and enumeration of Clostridium difficile in retail beef and pork. Appl Environ Microbiol. 2009;75(15):5009–5011. doi: 10.1128/AEM.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik M. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin Microbiol Infect. 2007;13(5):457–459. doi: 10.1111/j.1469-0691.2007.01687.x. [DOI] [PubMed] [Google Scholar]
- McDonald L, Killgore G, Thompson A, Owens R, Kazakova S, Sambol S, Johnson S, Gerding D. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]


