Abstract
Background
MicroRNA (miR) expression may have prognostic value for many types of cancers. However, the miR literature comprises many small studies. We systematically reviewed and synthesized the evidence.
Methods
Using MEDLINE (last update December 2010), we identified English language studies that examined associations between miRs and cancer prognosis using tumor specimens for more than 10 patients during classifier development. We included studies that assessed a major clinical outcome (nodal disease, disease progression, response to therapy, metastasis, recurrence, or overall survival) in an agnostic fashion using either polymerase chain reaction or hybridized oligonucleotide microarrays.
Results
Forty-six articles presenting results on 43 studies pertaining to 20 different types of malignancy were eligible for inclusion in this review. The median study size was 65 patients (interquartile range [IQR] = 34–129), the median number of miRs assayed was 328 (IQR = 250–470), and overall survival or recurrence were the most commonly measured outcomes (30 and 19 studies, respectively). External validation was performed in 21 studies, 20 of which reported at least one nominally statistically significant result for a miR classifier. The median hazard ratio for poor outcome in externally validated studies was 2.52 (IQR = 2.26–5.40). For all classifier miRs in studies that evaluated overall survival across diverse malignancies, the miRs most frequently associated with poor outcome after accounting for differences in miR assessment due to platform type were let-7 (decreased expression in patients with cancer) and miR 21 (increased expression).
Conclusions
MiR classifiers show promising prognostic associations with major cancer outcomes and specific miRs are consistently identified across diverse studies and platforms. These types of classifiers require careful external validation in large groups of cancer patients that have adequate protection from bias. –
CONTEXT AND CAVEATS
Prior knowledge
Many small studies have examined associations between tumor expression of microRNAs (miRs)—small, noncoding highly stable nucleic acids that regulate DNA translation to protein—and clinical outcome for a variety of cancers. However, there has been no formal systematic synthesis of the evidence on miR prognostic markers for cancer outcomes.
Study design
A systematic review of 46 articles presenting results on 43 studies pertaining to 20 different cancers that examined associations between miRs and prognosis using tumor specimens for more than 10 patients during classifier development. Included studies used either polymerase chain reaction or hybridized oligonucleotide microarrays to assess associations with a major clinical outcome in an agnostic fashion (ie, without selection for prior credibility).
Contribution
The median study size was 65 patients, the median number of miRs assayed was 328, and the most commonly measured outcomes were overall survival and recurrence. Of the 21 studies that performed external validation, 20 reported at least one nominally statistically significant result for a miR classifier. The median hazard ratio for poor outcome in externally validated studies was 2.52. Two miRs appeared repeatedly among the selected classifiers for different malignancies: let-7 (decreased expression in patients with cancer) and miR 21 (increased expression).
Implications
MiR classifiers show promising prognostic associations with major cancer outcomes. That fact that specific miRs are consistently identified across diverse studies and assay platforms suggests that regulatory pathways coordinated by miRs operate across cancers.
Limitations
The review excluded non–English language articles and studies that analyzed fewer than 20 miRs, cross-sectional studies, studies concerning genetic alterations of a miR, and studies involving oncoviruses. Some biases at the study or outcome level were difficult to assess due to non-standardized reporting in the primary studies.
From the Editors
Insights gained from novel molecular tests may improve the ability to make accurate prognostic assessments for patients with cancer. One of the most promising new biological measurements involves the expression of microRNAs (miRs), which are small, noncoding, highly stable 22-base-pair nucleic acids first discovered in the nematode Caenorhabditis elegans (1). Unlike messenger RNA, which serves as a template for DNA translation to protein, miRs are unique because they directly regulate this translation. To date, investigators have discovered several hundred of these oligomers (2). Furthermore, the ability of these molecules to act as gene suppressors or activators at multiple sites, their relatively limited number, and their remarkable in vivo stability make them appealing to study as clinical biomarkers in cancer and other diseases (3).
Many articles have published promising results for miR-based prognostic classifiers in diverse malignancies. Whereas some studies test only a handful of miRs on an opportunistic basis, there are also many studies that evaluate a large number of miRs in an agnostic fashion, that is, they select the most prognostic miRs from a large initial unselected set. The development of miR-based classifiers requires rigorous methods and validation processes, and it is often challenging to make sense of the evidence accumulated from complex “omics” studies to ensure that results are unbiased and well validated (4,5). This problem applies to the cancer miR literature as well, and it is accentuated by the fact that the literature comprises many, mostly small, studies among different cancer types. Currently, a formal systematic synthesis of the evidence on miR prognostic markers for cancer outcomes is lacking.
For this study, we systematically evaluated and synthesized the data from miR studies that have attempted to identify prognostic classifiers in malignancies. We describe the characteristics of studies published to date, assess study design and limitations thereof—with emphasis on the validation practices—and provide a quantitative synopsis of the performance of these biomarkers for important clinical outcomes in cancer. We further evaluate the specific miRs that are most frequently identified as important prognostic indicators of survival for diverse malignancies.
Methods
Search Strategy and Eligibility Criteria
We searched MEDLINE for English language studies that analyzed associations between miRs and prognosis in cancer patients. The search strategy used the clinical queries prognosis filter, which has a sensitivity of 92% for detecting articles related to prognosis (6): (Prognosis/Broad [filter]) AND ((microRNA OR miRNA OR miR) AND (cancer OR tumor OR neoplas* OR tumour OR malignan* OR metastat* OR metastas* OR leukemia OR lymphoma OR recurren* OR tumor control OR lymph node OR response) AND (Humans[Mesh] AND English[lang])). The search was last updated to include articles published through December 31, 2010.
Studies were iteratively screened for inclusion at the title, abstract, and full-text levels. Titles or abstracts that were not clearly categorized as being eligible or not were reviewed at the next stage until a definitive assessment could be made. Articles were considered eligible if they described miR analyses in samples from patients with any type of malignancy, with the aim of evaluating major clinical outcomes. Eligible outcomes included disease progression, response to therapy, metastasis, recurrence, and overall survival. We only considered studies that analyzed a large number of miRs (>20) without specifying a priori that only a select number of miRs should be tested based on speculation about their biological importance. Studies that included such agnostic testing of multiple miRs were eligible for consideration in this review if they examined associations between these miRs and clinical outcomes in a single dataset or if they also performed validation of these associations (either by cross-validation or by external validation in a separate dataset). Articles that presented the results of studies with selective testing of one or a few miRs were eligible if they evaluated miRs associated with clinical outcomes in an earlier study that had started from the agnostic testing of a large number of miRs; these articles were then considered as efforts to validate the earlier studies. Finally, we only considered studies with a sample size exceeding 10 patients because the ability to evaluate any prognostic marker would be negligible in smaller studies.
Studies that used either quantitative real-time polymerase chain reaction (qRT-PCR) array or hybridized oligonucleotide microarray (oligoarray) platforms were eligible for inclusion. We excluded cross-sectional studies (eg, those that addressed associations with tumor stage), studies concerning genetic alterations of an miR (eg, polymorphisms or methylation patterns), and studies involving oncoviruses. Studies that defined associations with tumor histology, tumor size, tumor differentiation from normal tissue, or malignant potential without specifically examining associations with a clinical outcome were likewise excluded. Two reviewers (V. S. Nair and L. S. Maeda) identified eligible studies, and contested articles were adjudicated by a third reviewer (J. P. A. Ioannidis). The reference lists from included articles were examined to ensure that relevant studies were not missed.
Data Extraction
We used a worksheet to record information about all studies that qualified for final inclusion. The worksheet documented relevant metrics identified during full-text retrieval of the included studies based on previous empirical evaluations of “omics” studies and information that we thought would be important in identifying systematic variation for this genre of literature (4,5,7). More specifically, the worksheet included information on the journal in which the study was published, demographics, study characteristics, array platform characteristics, technical validation procedures, statistical methods for selecting miRs, validation techniques, adjustment for other potential predictors, and clinical outcomes. The performance of miRs for prognosis was captured separately for training, cross-validation, and independent external validation settings whenever applicable.
Statistical Methods
Descriptive characteristics for studies were described by the median value with interquartile range (IQR) for continuous variables and by frequencies with percentages for categorical variables. MiR classifiers were defined by the authors of each study; some classifiers consisted of a single miR, whereas other classifiers combined information from multiple miRs. We abstracted the miR threshold published by the original investigators to define the association between miR expression and outcome along with the threshold metric used for this purpose. To quantify the risk of poor clinical outcome associated with each classifier, we extracted the hazard ratio (HR) from time-to-event analyses along with the 95% confidence interval for the defined miR categories. Whenever the 95% confidence interval was not directly provided, we approximated it using the hazard ratio and the P value from the log-rank test, or we estimated the variance of the natural logarithm of the hazard ratio using the formula [(n1 + n2)2/(n1 + n2) × t1 × t2], where n was the number of events per group delineated by miR expression levels and t was the total number of subjects per group (8). When the hazard ratio was not reported and it was not possible to approximate it with these calculations, we used the inverse of the ratio of the median survival times in groups that were defined by miR expression levels to approximate the hazard ratio. For consistency, all hazard ratios were expressed as values greater than 1 to denote increased risk (ie, HRs <1 were presented as the inverted ratio). No formal quantitative synthesis (meta-analysis) of these data was performed because they pertained to heterogeneous cancers, miRs, validation settings, and adjustments.
Testing for Small-Study Effects and Excess of Statistically Significant Results
For estimates of effects derived from validation studies, we evaluated whether smaller studies yielded different effects compared with larger studies by creating a funnel plot with sample size on the vertical axis and log10 hazard ratio on the horizontal axis. We used sample size for the funnel plot because there were no complete data on standard errors for all estimates. We used linear regression (9) to assess whether the sample size was associated with the log10 hazard ratio. We also calculated the expected number of positive classifiers based on the power of each study to find nominally statistically significant (P < .05) results given the number of events and assuming an effect equal to the median effect found in these analyses. The expected number was then compared with the observed number using a χ2 test to determine whether an excess of statistically significant findings existed (10,11).
Specific MiRs Associated With Survival
We also identified which specific miRs were identified most frequently across all cancers regardless of expression level direction and those with increased or decreased expression levels. This analysis required adjusting for how many times each miR was assayed because not all known miRs were assessed up front in all studies. We therefore counted how many opportunities each miR had for selection, or in other words, how many times it was assayed in the original agnostic platform, and also counted how many times each miR was identified in training classifiers used for survival analysis. The null hypothesis is that all miRs had the same probability of being selected in a prognostic classifier across any study. This expected probability is equal to pexp= Ncl/Npl, where Ncl is the sum of the number of statistically significant miRs across all survival classifiers and Npl is the sum of the number of miRs across all platforms evaluated in survival studies. We used a binomial test to determine whether the observed frequency of selection for each miR across all classifiers differed from the expected probability. In a sensitivity analysis, we also assessed variants of the same “parent” miR as separate opportunities for classifier prediction. The parent is defined here as the root number assigned to each miR without additional annotation according to the Welcome Trust Sanger Institute’s miRBase registry (12) and represents the fundamental unique biological miR. As a hypothetical example, the parent miR of miR 0*, 0a, 0-1, and 0-3p would be coded as miR 0. We limited our analysis to overall survival because recurrence, progression, and metastasis were often studied in concert with survival and were typically not studied independently (recurrence was examined in nine studies, progression in four studies, and metastasis in one study). MiR platform data were recovered from the supplementary material of the relevant studies, the Gene Expression Omnibus of the National Center for Biotechnology Information, or the European Bioinformatics Institute Array Express Database.
All calculations were performed using Excel software (for Mac 2011, Microsoft Corp, Seattle, WA) or SAS statistical software (v9.2; SAS Institute Inc, Cary, NC). Forest plots were developed using GraphPad Prism (v5; GraphPad Software, Inc, La Jolla, CA). P values are two-sided.
Results
Eligible Studies
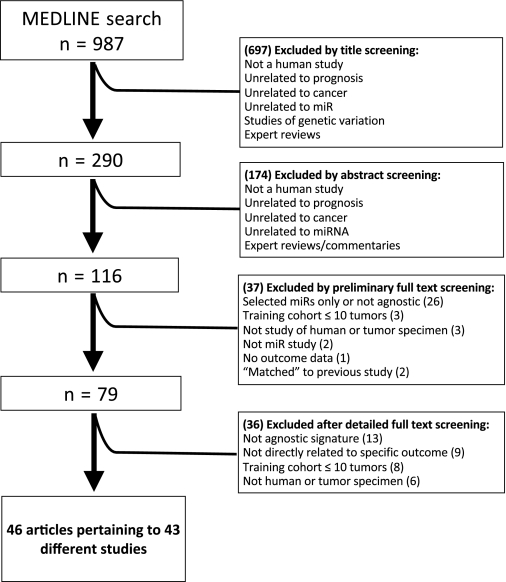
Of the 987 items retrieved by the MEDLINE search, 908 were excluded based on screening of the title or abstract and 79 were evaluated in full text. Of these, ultimately 46 articles (13–58) pertaining to 43 different studies (ie, unique miR classifiers associated with a clinical outcome) were considered eligible for inclusion in this review (Figure 1). One article (52) described a validation study of a classifier that had been developed but not externally validated in a previous article (18), another article (15) described a new validation study of a classifier that had already been externally validated in a previous article (14), and two other articles from the same research group (39,40) described the development and training of the same classifier and were counted as one study. We refer to these combined classifiers from here as “Calin–Visone” (18,52), “Bray–Buckley” (14,15), and “Navarro” (39,40) in the accompanying tables.
Figure 1.
Flow chart of study inclusion.
The median impact factor of the journals in which the eligible studies were published was 7.5 (IQR = 4.7–11). The first eligible article was published in the New England Journal of Medicine in 2005 and 21 different journals published this research (Table 1). The median study size was 65 patients (IQR = 34–129 patients), most studies analyzed solid tumors, and more than three times as many studies used frozen tumor tissue vs formalin-fixed paraffin-embedded tumor tissue. Hybridized oligoarrays were used by 81% of the studies to identify statistically significant miRs, whereas PCR arrays were used by 19% of the studies. The median number of miRs assayed across studies was 328 (IQR = 250–470).
Table 1.
Descriptive characteristics for included studies*
| General study characteristics | |
| Journal impact factor† | 7.5 (4.7–11) |
| Year of publication | |
| 2005–2007 | 3 (7) |
| 2008 | 12 (28) |
| 2009 | 13 (30) |
| 2010 | 15 (35) |
| Sample size, all studies | 65 (34–129) |
| Sample size, internal validation studies only | 76 (53–142) |
| Sample size, external validation studies only | 129 (69–182) |
| Tumor characteristics | |
| Tumor type | |
| Solid tumor | 35 (81) |
| Hematological | 8 (19) |
| Tissue preservation method | |
| Frozen | 22 (51) |
| Paraffin-embedded | 7 (16) |
| Frozen and paraffin-embedded | 2 (5) |
| Other | 5 (12) |
| Not reported | 7 (16) |
| Assay characteristics | |
| Array type | |
| Oligoarray | 35 (81) |
| Polymerase chain reaction | 8 (19) |
| MiRs assayed | 328 (250–470) |
| Net miRs‡ | 208 (167–298) |
| Predictive miRs | 6 (3.5–9.5) |
| Increased expression | 3 (0–6) |
| Decreased expression | 1 (0–3.3) |
| Validation | |
| Internal validation | 20 (47) |
| Sample size | 62 (47–92) |
| External validation | 21 (49) |
| Sample size | 55 (32–79) |
| Internal and external validation | 8 (19) |
| Sample size | 186 (110–241) |
| Validation method§ | 33 (77) |
| Cross-validation | 20 (47) |
| K means | 5 (12) |
| Leave-one-out | 1 (2) |
| Bootstrapping | 2 (5) |
| Prediction of analysis microarray | 2 (5) |
| Other | 10 (21) |
| External validation§‖ | 21 (49) |
| Split samples | 7 (16) |
| Independent samples | 6 (13) |
| Combined cohorts | 3 (7) |
| Not clearly defined | 5 (12) |
| Outcome¶ | |
| Response to therapy | 1 (2) |
| Metastasis | 2 (5) |
| Disease progression | 6 (14) |
| Recurrence | 19 (44) |
| Overall survival | 30 (59) |
Continuous variables are displayed as the median (interquartile range) and categorical variables as No. (%). Data are based on 43 studies, except where explicitly enumerated and for the variables net miRs, miRs with increased expression, and miRs with decreased expression for which information was available from 17, 41, and 41 studies, respectively. miR = microRNA.
Based on 2009 data.
After data processing and filtration and before classifier development.
For prespecified outcomes as stated in the inclusion criteria.
Split-pair sampling was considered external validation. For one study, both split-sample and independent external cohort testing was performed. In that case, we considered only the external cohort as the external validation.
All outcomes in training or test cohorts are tabulated per study.
Array Data Processing
In general, most studies that used oligoarray analysis reported data normalization procedures consisting of: 1) filtering probe signal intensity by mean normalization across replicates to adjust for sample variability, 2) median centering across arrays to adjust for interarray variability, and 3) log normalization to accommodate further statistical analyses (Supplementary Table 1, available online). By contrast, studies that used qRT-PCR arrays performed mean normalization for replicates based on the amplification threshold cycle (Ct) with normalization to an endogenous control to account for between-sample variability. Among the 43 included studies, 33 of the 35 oligoarray studies defined a normalization procedure for microarray data processing, with global median normalization being the most common method. Thirty-two oligoarray studies validated signal intensity by PCR, and none of the PCR array studies provided additional signal validation. Normalization using endogenous miR controls to account for between-sample variability for PCR arrays was performed in all cases, and the most commonly used endogenous miR controls were U6B and let-7. Comparative expression analysis was the dominant method used for quantifying PCR transcript expression (59).
Analytic Methods Used for Classifier Development
Although the included studies used diverse statistical methods, the majority (31 [72%] of 43) provided methods to account for multiple comparisons, either by reporting a false discovery rate (Q value) (60) or an adjusted P value. The most commonly used Q value was 0.05 (n = 9) and the median adjusted P value was .005 (n = 8).
Thirty-three studies (13–19,22–28,32,33,35–37,39–50,53–55,58) performed either cross-validation or external validation, and seven studies (16,18,28,37,47,48,55) performed both cross-validation and external validation (Table 1). Among the 20 studies that performed cross-validation (14,16–19,23,26–28,36,37,41,42,45,47,48,53–55,58), the most common methods were K-means clustering with bootstrapping, leave-one-out cross-validation, and prediction analysis of microarrays (61–63). Two studies (23,24) used multiple cross-validation methods, and six studies (22,35,43,46,47,49) validated results using different model building methods for the same cohort. Of the 21 studies that carried a developed miR classifier to an external validation stage (14–19,23,27,28,32,36,37,41,42,47,48,53–55,58), 17 used a distinct previously unexamined group of patients, whereas the other four studies (16,28,37,58) that claimed to externally validate actually combined the training and test groups for final classifier modeling.
Outcomes
The most commonly measured outcomes were overall survival (30 studies) and recurrence (19 studies), and 10 studies examined both (Table 1). Metastasis, disease progression, and response to therapy were not commonly studied endpoints.
Characteristics of Training Processes
The included studies addressed outcomes for 20 different malignancies, with the most common being lung cancer (five studies), hepatocellular carcinoma (four studies), leukemia (four studies), and ovarian cancer (four studies) (Table 2). Several common cancers (ie, prostate, colon, and breast cancer) were evaluated in comparatively few studies. Training set sample size varied considerably. The largest training set was in a study of hepatocellular carcinoma (n = 131), and studies that addressed outcomes for gynecologic cancers generally had the smallest training cohorts. Sixteen studies assessed clinical variables in the miR training cohort, and 14 of these studies stated that these potentially prognostic variables were incorporated in multivariable adjusted analyses in the training set during miR classifier development (Table 2). Stage (n = 10 studies), age (n = 6 studies), grade (n = 5 studies), and sex (n = 5 studies) were the most commonly used adjustment factors.
Table 2.
Signature development characteristics for microRNA classifiers*
| First author (reference) | Cancer type | Outcome* | Training set sample size | No. of selected miRs | Expression increased | Expression decreased | Change in expression not stated | Variables assessed in training model | No. of miRs carried forward† /No. of selected miRs | Reason for not further testing all miRs |
| Zhi (58) | Astrocytoma | OS | 84 | 7 | 2 | 5 | 0 | NR | 3/7 | Not stated |
| Catto (20) | Bladder | P | 52 | 4 | 4 | 0 | 0 | Stage, T, grade | NA | NA |
| Dyrskjot (24) | Bladder | P | 106 | 4 | 4 | 0 | 0 | Stage, grade | NA | NA |
| Veerla (51) | Bladder | OS | 14 | 2 | 2 | 0 | 0 | NR | NA | NA |
| Foekens (26) | Breast | M | 38 | 4 | 1 | 0 | 3 | Age, menopausal status, tumor size, grade, mRNA levels | 4/4 | NR |
| Schepeler (46) | Colon | R | 37 | 2 | 0 | 2 | 0 | Age, sex, T, grade, histology | NA | NA |
| Schetter (47) | Colon | OS | 84 | 5 | 5 | 0 | 0 | Stage | 5/5 | NA |
| Cohn (22) | Endometrial | P, OS | 20 | 1 | 0 | 1 | 0 | NR | NA | NA |
| Hiroki (30) | Endometrial | R, OS | 21 | 6 | 0 | 6 | 0 | Stage, invasion | NA | NA |
| Guo (28) | Esophageal | R, OS | 30 | 2 | 2 | 0 | 0 | Age, sex, stage, tobacco, alcohol, T, N‡ | 2/2 | NA |
| Mathe (37) | Esophageal | OS | 76 | 1 | 1 | 0 | 0 | NR | 1/5 | Not stated |
| Ogawa (41) | Esophageal | OS | 30 | 6 | 6 | 0 | 0 | T, N | 1/6 | Not stated |
| Ueda (50) | Gastric | P, R, OS | 106§ | 14 | 6 | 5 | 3 | Stage | NA | NA |
| Haller (29) | GIST | P, R | 49 | 2 | 0 | 2 | 0 | NR | NA | NA |
| Budhu (16) | Hepatocellular | M, R, OS | 131 | 20 | 4 | 16 | 0 | NR | 20/20 | NA |
| Chung (21) | Hepatocellular | R | 23 | 7 | 6 | 1 | 0 | NR | NA | NA |
| Jiang (31) | Hepatocellular | OS | 25 | 19 | 0 | 0 | 19 | NR | NA | NA |
| Li (35) | Hepatocellular | OS | 75 | 1 | 0 | 1 | 0 | Cirrhosis | NR | NA |
| Cairo (17) | Hepatoblastoma | OS | 19 | 4 | 0 | 0 | 4 | NR | 4/4 | NA |
| Navarro (39,40) | HL | R | 84 | 1 | 0 | 1 | 0 | Age, sex, WHO status, disease extent | NA | NA |
| Calin (18) | Leukemia | P | 94 | 9 | 8 | 1 | 0 | NR | 9/9 | NA |
| Garzon (27) | Leukemia | R, OS | 122 | 5 | 5 | 0 | 0 | NR | 4/5 | PCR validation‖ |
| Marcucci (36) | Leukemia | R | 64 | 9 | 7 | 2 | 0 | NR | 9/9 | NA |
| Zhang (56) | Leukemia | OS | 29 | 7 | 3 | 4 | 0 | NR | NA | NA |
| Landi (32) | Lung | OS | 107 | 5 | 0 | 5 | 0 | Age, sex, histology, stage, smoking‡ | NA | NA |
| Patnaik (43) | Lung | R, OS | 77 | 6 | 0 | 5 | 1 | Sex | NA | NA |
| Raponi (44) | Lung | OS | 54 | 20 | 9 | 11 | 0 | NR | NA | NA |
| Yanaihara (53) | Lung | OS | 32 | 5 | 3 | 2 | 0 | Age, sex, stage, smoking | 2/5 | Not stated |
| Yu (55) | Lung | R, OS | 112 | 5 | 2 | 3 | 0 | NR | 5/5 | NA |
| Caramuta (19) | Melanoma | OS | 16 | 6 | 5 | 1 | 0 | NR | 2/6 | PCR validation‖, P value |
| Segura (49) | Melanoma | OS | 59 | 18 | 18 | 0 | 0 | Stage | NA | NA |
| Pass (42) | Mesothelioma | R, OS | 37 | 1 | 0 | 1 | 0 | NR | 1/1 | NA |
| Bray (14) | Neuroblastoma | R, OS | 96 | 15 | 15 | 0 | 0 | NR | 2/15 | Stratified by MYC status |
| Schulte (48) | Neuroblastoma | R, OS | 50 | 42 | 0 | 0 | 42 | NR | 42/42 | NA |
| Di Lisio (23) | NHL | OS | 22 | 12 | 1 | 0 | 11 | NR | 12/12 | NA |
| Lawrie (33) | NHL | R | 52 | 12 | 3 | 9 | 0 | NR | NA | NA |
| Zhao (57) | NHL | OS | 30 | 3 | 0 | 3 | 0 | NR | NA | NA |
| Eitan (25) | Ovarian | Rx, R, OS | 30§ | 5 | 4 | 1 | 0 | NR | NA | NA |
| Lee (34) | Ovarian | R | 33 | 2 | 0 | 2 | 0 | NR | NA | NA |
| Nam (38) | Ovarian | OS | 20 | 10 | 7 | 3 | 0 | NR | NA | NA |
| Yang (54) | Ovarian | R | 72 | 9 | 6 | 3 | 0 | NR | 1/9 | Plausibility, P value |
| Bloomston (13) | Pancreas | OS | 65 | 6 | 6 | 0 | 0 | NR | NA | NA |
| Schaefer (45) | Prostate | R | 75 | 15 | 5 | 7 | 3 | Stage, grade | 1/15 | P value |
GIST = gastrointestinal stromal tumor; HL = Hodgkin lymphoma; M = metastasis; miR = microRNA; N = node stage; NA = not applicable; NHL = non-Hodgkin lymphoma; NR = not reported; OS = overall survival; P = progression; PCR = polymerase chain reaction; PSA = prostate-specific antigen; R = recurrence; Rx = response to therapy; T = tumor stage; WHO = World Heath Organization.
Number of miRs carried forward to a test set of samples after classifier development.
Only examined univariaby.
Averaged across outcome.
Not specified why only this many were validated using PCR.
Most studies assayed hundreds of miRs (range = 73–911), and the number of miRs that were found to be statistically significantly associated with an outcome of interest ranged from 1 to 42 (Table 2). Of the 43 studies, 14 used continuous miR expression levels to assess outcome without stratifying, whereas the remaining 29 stratified miR levels to assess outcomes (Supplementary Table 2, available online). When we summed the number of miRs that were statistically significantly associated with outcomes across all developed classifiers, the total number of miRs selected was 339, and of those, 150 displayed increased expression in samples of patients with cancer, 103 displayed decreased expression, and for 86, it was not clearly stated whether their expression was increased or decreased in patients with cancer.
Selection of MiRs for External Validation
Of the 21 studies that performed external validation, 11 carried forward to the test set all of the statistically significant miRs in the training set, whereas the remaining 10 carried forward only part of the developed classifier for external validation testing. Four of the studies that dropped part of the prognostic classifier in the test cohort did not provide a clear reason for doing so (Table 2).
External Validation Studies
The median sample size for training and test cohorts for the 21 studies that performed external validation was 62 (range = 20–131) and 55 (range = 16–147), respectively. These studies externally validated their data on a total of 1188 patients by using disease progression (one study), metastasis (two studies), recurrence (10 studies), or overall survival (14 studies) as an outcome measure (Table 3). Six of 21 studies provided externally validated data for more than one outcome. Nine studies used a classifier for outcome analysis that combined information for more than one miR, and 12 studies used only a single miR for outcome analysis. Six of these 12 studies presented separate analyses for several miRs.
Table 3.
Hazard ratios for microRNA classifiers by cancer type and outcome in test cohorts*
| First author (reference) | Cancer type | Outcome | Test set sample size | Prognostic miR(s)† | Unadjusted HR (95% CI) | Adjusted HR (95% CI) | Variables adjusted for in test set |
| Zhi (58)‡ | Astrocytoma | OS | 40 | 21 | 1.84 (1.05 to 3.22) | 1.88 (1.07 to 3.31) | Sex, age, WHO grade |
| Zhi (58) | Astrocytoma | OS | 40 | 106a | 1.72 (0.99 to 2.99) | 1.63 (0.90 to 2.95) | Sex, age, WHO grade |
| Zhi (58) | Astrocytoma | OS | 40 | 181b | 1.87 (1.07 to 3.26) | 1.86 (1.03 to 3.37) | Sex, age, WHO grade |
| Foekens (26) | Breast | M | 147 | 57, 128a, 210, 516-3p | NR | 5.4 (2.4 to 11.8) | Age, menopausal status |
| Schetter (47) | Colon | OS | 103 | 21 | 2.4 (1.4 to 3.9) | 2.4 (1.4 to 4.1) | Stage |
| Guo (28)‡ | Esophageal | OS | 22 | 103, 107 | 3.62 (1.53 to 8.57) | 2.60 (1.01 to 6.70) | Stage, N |
| Ogawa (41) | Esophageal | OS | 19 | 129 | 5.78 (1.49 to 22.4) | NR | NR |
| Mathe (37)‡ | Esophageal | OS | 69 | 223 | 2.33 (1.03 to 5.26) | 4.00 (1.45 to 11.1) | Study center, neoadjuvant therapy, N |
| Mathe (37) | Esophageal | OS | 69 | 21 | 1.17 (0.54 to 2.51) | NR | NR |
| Budhu (16)‡ | Hepatocellular | OS | 110 | 20 miR composite | 2.1 (1.2 to 3.7) | 2.1 (1.2 to 3.6) | Stage, tumor encapsulation |
| Budhu (16) | Hepatocellular | M | 110 | 20 miR composite | 1.85§ | NR | NR |
| Cairo (17) | Hepatocellular | OS | 46 | 371–3, 100, let-7a-2, 125b-1 | 6.74 (2.02 to 22.5) | NR | NR |
| Calin–Visone (18,52)‖ | Leukemia | P | 16 | 181 | 2.00§ | NR | NR |
| Garzon (27) | Leukemia | OS | 60 | 191 | NR | 2.35 (1.18 to 4.75) | Allelic ratio, WBC |
| Garzon (27) | Leukemia | OS | 60 | 199 | NR | 2.81 (0.96 to 8.23) | Allelic ratio, WBC |
| Garzon (27) | Leukemia | R | 60 | 191 | NR | 2.29 (1.15 to 4.55) | Allelic ratio, WBC |
| Garzon (27) | Leukemia | R | 60 | 199 | NR | 2.57 (0.93 to 3.51) | Allelic ratio, WBC |
| Marcucci (36) | Leukemia | R | 55 | 9 miR composite | 1.99 (1.07 to 3.69) | Age, WBC, cytogenetics | |
| Yanaihara (53) | Lung | OS | 32 | let-7a | 2.22 (0.91 to 5.71) | 2.97 (1.07 to 8.23) | Age, sex, stage, smoking |
| Yanaihara (53) | Lung | OS | 32 | 155 | 2.52 (1.10 to 7.45) | 3.77 (1.32 to 10.6) | Age, sex, stage, smoking |
| Yu (55) | Lung | R | 62 | 137, 372, 182, 221, let-7a | NR | 2.86 (1.20 to 6.82) | Stage, age |
| Yu (55) | Lung | OS | 62 | 137, 372, 182, 221, let-7a | NR | 3.65 (1.29 to 10.4) | Stage, age |
| Caramuta (19) | Melanoma | OS | 16 | 191 | 8.82 (2.00 to 38.8) | NR | NR |
| Caramuta (19) | Melanoma | OS | 16 | 193b | 8.43 (1.92 to 37.1) | NR | NR |
| Pass (42) | Mesothelioma | R | 92 | 29c | 2.31§ | NR | Stage, sex, N |
| Pass (42) | Mesothelioma | OS | 92 | 29c | 2.39 (1.26 to 3.11) | NR | NR |
| Bray (14) | Neuroblastoma | OS | 48 | 15 miR composite | 4.31 (1.66 to 11.2) | NR | NR |
| Bray (14) | Neuroblastoma | R | 48 | 15 miR composite | 4.50 (1.74 to 11.7) | NR | NR |
| Bray–Buckley (14,15)‖ | Neuroblastoma | OS | 37 | 15 miR composite | 2.26 (1.07 to 4.78) | NR | NR |
| Bray–Buckley (14,15)‖ | Neuroblastoma | R | 37 | 15 miR composite | 2.22 (1.04 to 4.75) | NR | NR |
| Schulte (48) | Neuroblastoma | R | 19 | 42 miR composite | 13.8 (2.62 to 72.4) | NR | NR |
| Schulte (48) | Neuroblastoma | OS | 19 | 42 miR composite | 19.7 (2.99 to 130) | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 20b | 1.39§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 130b | 1.15§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 454 | 1.15§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 99b | 0.96§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 181c | 0.98§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | let-7 | 1.06§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 532 | 0.99§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 362 | 1.00§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 363 | 1.15§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 625 | 1.11§ | NR | NR |
| Di Lisio (23) | NHL | OS | 54 | 660 | 1.07§ | NR | NR |
| Yang (54) | Ovarian | R | 62 | let-7i | 2.47 (1.39 to 4.38) | NR | NR |
| Schaefer (45) | Prostate | R | 79 | 96 | NR | 3.55 (0.95 to 13.3) | Grade |
CI = confidence interval; HR = hazard ratio; MiR = microRNA; N = node stage; M = metastasis; NHL = non-Hodgkin lymphoma; NR = Not reported or calculations not possible for deriving metric; OS = overall survival; P = progression; R = recurrence; WBC= white blood cell count; WHO = World Health Organization.
Classifiers comprising more than five miRs are presented in Supplementary Table 2 (available online).
These studies combined training and testing cohorts and the resulting data are presented.
CI not given and/or unable to be imputed.
Separate study providing external validation to parent study.
To assess the effect of miR expression levels on patient outcome, studies performed survival analysis by regression modeling or Kaplan–Meier plots against a threshold miR expression level in most test sets except for one study where the threshold was not explicitly stated (52). Single miR classifiers used the median (n = 10 studies), mean (n = 1 study), or some other (n = 1 study) expression threshold to dichotomize expression levels. Composite classifiers used a “summary” score for further analysis and selected various cutoffs (Supplementary Table 2, available online).
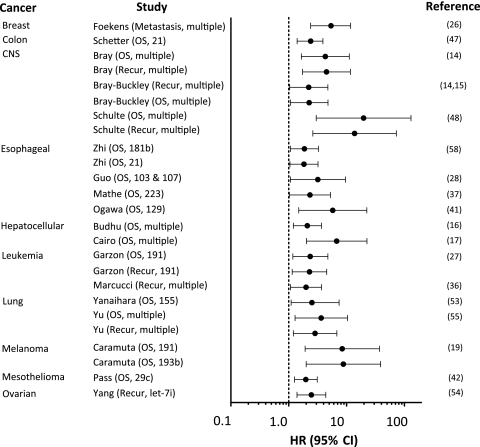
For these 21 externally validated studies, 45 cancer-specific outcomes were assessed (Table 3). We imputed hazard ratios for 16 of the 45 outcomes reported in these 21 studies and 95% confidence intervals for 19 of the 45 outcomes for which this information was not explicitly provided. For 14 of the 45 outcomes, we could extract point estimates only. Examining unadjusted hazard ratios (unless only an adjusted value was available) revealed that 16 of the 45 miR classifiers were not statistically significantly associated with outcome (Table 3). The median hazard ratio in external validation studies that reported statistically significant results at the P = .05 level for which we were able to extract or approximately calculate a hazard ratio (n = 25) was 2.52 (IQR = 2.26–5.40; range = 1.39–19.7) (Figure 2). Fifteen of these 25 estimates had a P value less than .01. For all 45 reported hazard ratios, regardless of statistical significance, the median hazard ratio was 2.29 (IQR = 1.39–3.28) (Table 3).
Figure 2.
Nominally statistically significant hazard ratios of miR classifiers in test cohorts. Prognostic impact of miRs by cancer type and study are shown for statistically significant results. The point estimate (denoted by a circle) is bounded by a 95% confidence interval (CI) (indicated by error bars), and the dashed line represents no increased risk (unity) for the outcome. Each study is referred to by author name followed by the measured outcome and the miR(s) included in the classifier. Classifiers with many miRs per classifier are denoted as “multiple” (see Supplementary Table 2 [available online] for full details). Unadjusted hazard ratios (HRs) and 95% CIs are presented for all studies except for those by Garzon and Yu (27,55) for which only adjusted hazard ratios were available. CNS = central nervous system; miR = microRNA; OS = overall survival; recur = recurrence.
Twelve studies adjusted for potentially prognostic clinical and pathological variables during multivariable regression in test cohorts (Table 3). Of the six studies that presented hazard ratios and 95% confidence intervals for both unadjusted and adjusted classifiers, only one (28) reported a substantially smaller hazard ratio with adjustment, whereas in the other studies (16,37,47,53,58), there was no substantial change or an increase in the hazard ratio estimate with adjustment. Only three studies performed adjusted and unadjusted analysis for their miR classifier in both training and test cohorts: two studies (47,53) adjusted for the same variables in both the test and training cohorts, and one study (45) adjusted for different variables.
Testing for Small-Study Effects and Excess of Statistically Significant Results
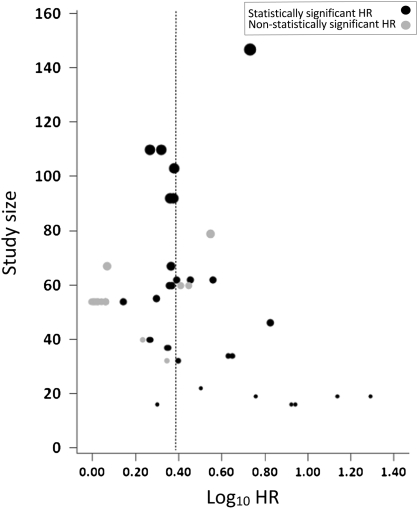
As shown in Figure 3, there was a suggestion that large effect sizes were seen predominantly in smaller studies (P = .08 for correlation between log10HR and sample size). Moreover, we estimated the statistical power required for 40 of the 45 measured outcomes to achieve the median hazard ratio of 2.29 (we excluded five study outcome analyses for which the number of events was missing and could not be approximated). Power calculations showed that the expected number of measured outcomes with nominally statistically significant results among the 40 available was 17.9 vs the 25 observed (P = .02 for an excess statistical significance of expected vs observed positive classifiers).
Figure 3.
Association between sample size and effect size. Hazard ratios (HRs) for studies reporting statistically significant and non-statistically significant findings are shown by study size (P = .08 for correlation between log10HR and sample size). The relative sizes of the gray and black circles indicate the study size and the median log10HR is shown by the dashed line.
Specific MiRs Associated With Overall Survival
Thirty studies evaluated overall survival as a clinical outcome in a training cohort, and for 27 of these studies, we were able to extract data for the full miR profiles tested (Supplementary Tables 3 and 4, available online). A total of 257 prognostic miRs from the original 339 miRs were identified in these 27 classifiers, and 20 miRs (7.8%) were identified statistically significantly more frequently than expected at an alpha level of 5%. Four miRs (let-7, 21, 100, and 125) were statistically significantly associated with overall survival at an alpha level of 1% (Table 4). In addition, four miRs (20, 21, 155, and 193) appeared more frequently than expected as miRs with increased expression and four miRs (let-7, 29, 30, and 7039) appeared more frequently than expected as miRs with decreased expression at an alpha level of .01. Let-7 and miR 21 were commonly part of classifiers associated with poor outcome, both overall and when miRs were grouped by direction of expression level. Expression of let-7 was decreased in a variety of tumors, including gastric, lung, liver, and ovarian carcinomas. Expression of miR 21 was increased consistently in lung, ovarian, and colon cancer as well as in astrocytoma. Additional sensitivity analyses for fully annotated miRs are presented in Supplementary Tables 5–8 (available online).
Table 4.
MicroRNAs selected most frequently in classifiers compared with expected frequency*
| MiR | No. of times assessed, npl (%)† | No. of times selected in classifier, ncl (%)‡ | P§ | Cancer type |
| All miRs in patients with poor outcome | ||||
| let-7 | 244 (2.46) | 16 (6.23) | <.001 | Gastric, HCC, leukemia, lung, melanoma, NHL, ovarian |
| 21 | 34 (0.34) | 5 (1.95) | .0018 | Astrocytoma, colon, lung, ovarian |
| 100 | 26 (0.26) | 4 (1.56) | .0043 | Gastric, HCC, lung |
| 125 | 71 (0.72) | 6 (2.33) | .0084 | HCC, gastric, neuroblastoma |
| MiRs with increased expression in patients with poor outcome | ||||
| 21 | 34 (0.34) | 5 (4.72) | <.001 | Astrocytoma, colon, lung, ovarian |
| 20 | 47 (0.47) | 4 (3.77) | .0046 | Colon, leukemia, NHL |
| 155 | 26 (0.26) | 3 (2.83) | .0063 | Lung, melanoma |
| 193 | 55 (0.56) | 4 (3.77) | .0078 | Melanoma |
| MiRs with decreased expression in patients with poor outcome | ||||
| let-7 | 244 (2.46) | 7 (9.59) | .0017 | Gastric, lung, HCC, ovarian |
| 30 | 135 (1.36) | 5 (6.85) | .0028 | Astrocytoma, HCC, lung |
| 29 | 94 (0.95) | 4 (5.48) | .0046 | Endometrial, NHL |
| 7039‖ | 1 (0.01) | 1 (1.37) | .0073 | Ovarian |
For studies assessing overall survival as an outcome with platform data available (n = 27). HCC = hepatocellular carcinoma; miR = microRNA; NHL = non-Hodgkin lymphoma.
From 9900 total miRs across 27 platforms.
Percentages calculated for 257, 146, and 73 of the selected miRs in the all, increased expression, and decreased expression sets, respectively (for 38 selected miRs, it was not specified whether their expression was increased or decreased in patients with cancer).
Two-sided, using the binomial distribution to assess a statistically significant difference between npl and ncl.
This is a proprietary miR from Ambion (Foster City, CA) used by Nam et al. (38).
Discussion
In this systematic review, we found that several miR classifiers were associated with prognosis above and beyond traditional clinical and pathological metrics for a diverse group of cancers. The median hazard ratio for statistically significant classifiers that were developed and tested in separate groups of patients was 2.52, suggesting a modestly strong discriminatory ability. Moreover, we identified specific miRs that appeared repeatedly among the selected classifiers for different malignancies. This finding suggests that miR-coordinated regulatory pathways are common to many cancers.
Although these results are promising, this overview suggests several caveats and areas for improvement in miR-related translational research. There are many critical steps in the appraisal of miRs for prognostic purposes, and errors or biases can appear at all steps (64), including data acquisition, which is subject to preanalytical imprecision (65), and the large variety of statistical methods that are applied for classifier generation, which allows for a range of different results and creates opportunities for selective analysis and other reporting biases. We found that adjustment for important prognostic factors in both training and test cohorts was neither common nor standardized. Moreover, miRs that were considered in datasets for external validation were often a selected subset of those found in the training sets and were sometimes chosen without clear justification. Furthermore, most test cohorts were small, and although a large number of classifiers reached nominally statistically significant results in external validation, many had wide 95% confidence intervals, and most P values were close to the .05 threshold. A “winner’s curse” has been demonstrated in other biomarker fields: small studies are typically performed first and have large effect sizes (66–68), whereas subsequent larger replication studies report smaller effects or no effect at all (69).
We also found some evidence in the available data that smaller studies tended to report the largest effect estimates and that the number of statistically significant studies was probably excessive given the generally small sample sizes and relatively limited statistical power of many of these investigations. These observations are consistent with the possibility of publication bias, although other explanations such as genuine heterogeneity of effects cannot be excluded, especially given the large diversity of cancers and markers analyzed (70).
An interesting question that remains poorly defined for the field is the number of features required for a robust classifier. Most studies in this review developed an miR classifier and carried it forward to an external test set using only a few miRs. Patnaik et al. (43) suggested that at least four miRs were required for optimal test performance when examining the area under the receiver operating characteristic curve during cross-validation, whereas Bray et al. (14) found very marginally, albeit statistically significantly, improved test performance when expanding their classifier from 10 to 42 miRs. Previous work in the field (71) suggests that differences in expression levels across genes necessarily define the size of a classifier and that a 1.5-fold increase in expression across seven different features (eg, miRs) will result in a small classification error rate between classes or populations (eg, patients with cancer who have different outcomes). Given that the majority of studies to date have externally validated classifiers with only one or a few selected miRs, it is likely that larger studies and a larger number of validated miRs will provide substantial improvement in discriminatory ability.
The miR field is evolving rapidly. The first database of miR sequences (miRBase) contained 506 entries for six organisms in 2002; by 2010, it contained 1424 entries for humans alone (2). Furthermore, miR terms are dynamic, and some miRs are “deceased” based on misclassifications due to high sequence similarities between preprocessed miRs, actual miRs, and minor variations among different miRs (72). Because platform arrays are made from probes that are specific for sequences and annotated to the most recent registry, each change can render current or previous studies inaccurate. For these reasons, we chose to use an ontological approach commonly used in gene enrichment analysis to account for some of these issues and to understand the expression of miRs across diverse cancers (73).
Although previous reviews (74,75) have illustrated the importance of certain cancer-associated miRs, including let-7 and miR 21, our analysis augments this literature by systematically pooling prognostic associations for classifiers across cancers. By using an ontological approach, we demonstrate that let-7 and miR 21 remained statistically significantly associated with outcome across all classifiers as well as when analyzed separately for miRs with increased (miR 21) or decreased (let-7) expression.
Specific miRs could be selected more frequently in cancer prognosis classifiers if either they have large average expression differences in patients with good vs poor prognosis and/or if the values of expression within each of these two groups have small variability. There are not sufficient data to tell whether the variances of the distributions of miR values consistently tend to be higher for some miRs than for others across very heterogeneous malignancies. One possibility is that miRs that are more frequently found to be differentially expressed have smaller variances within compared groups and thus have higher statistical power to detect differences. Alternatively, it is possible that some miRs indeed tend to be more important determinants of cancer survival than others.
Limitations of this systematic review are as follows: The review excluded non–English-language articles and studies that analyzed fewer than 20 miRs, cross-sectional studies, studies concerning genetic alterations of a miR, and studies involving oncoviruses. Some of the outcome data were missing (eg, HR estimates were not available or possible to calculate for all studies). Similarly, information was not always provided on each analyzed miR separately. Finally, some of the primary studies did not provide full details on their design and some selection or information biases cannot be excluded, whereas adjustments for other factors, including potential confounders were not standardized. This systematic review suggests that these are fronts where standardized reporting may improve the quality of this research literature in the future.
In conclusion, imperfect methodology and potential biases hinder the clinical application of miR expression testing as a biomarker of prognosis. However, the availability of many promising results suggests that larger scale standardized investigations with robust external validation practices for the most promising miRs are likely to better define the true potential of these novel cancer biomarkers and help select miR classifiers for further successful clinical translation.
Funding
This work was supported by the National Institutes of Health (5T32HL00794 to V.S.N.).
Supplementary Material
Footnotes
The authors take full responsibility for the study design, the data collection, the analysis and interpretation of the data, the decision to submit the article for publication, and the writing of the article.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 5.Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362(9394):1439–1444. doi: 10.1016/S0140-6736(03)14686-7. [DOI] [PubMed] [Google Scholar]
- 6.Haynes RB, McKibbon KA, Wilczynski NL, Walter SD, Werre SR. Optimal search strategies for retrieving scientifically strong studies of treatment from Medline: analytical survey. BMJ. 2005;330(7501):1179. doi: 10.1136/bmj.38446.498542.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castaldi PJ, Dahabreh IJ, Ioannidis JP. An empirical assessment of validation practices for molecular classifiers. Brief Bioinform. 2011;12(3):189–202. doi: 10.1093/bib/bbq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyrgiou M, Salanti G, Pavlidis N, Paraskevaidis E, Ioannidis JP. Survival benefits with diverse chemotherapy regimens for ovarian cancer: meta-analysis of multiple treatments. J Natl Cancer Inst. 2006;98(22):1655–1663. doi: 10.1093/jnci/djj443. [DOI] [PubMed] [Google Scholar]
- 9.Sterne JA, Sutton AJ, Ioannidis JP, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;342:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 10.Ioannidis JP. Excess significance bias in the literature on brain volume abnormalities. Arch Gen Psychiatry. 2011;68(8):773–780. doi: 10.1001/archgenpsychiatry.2011.28. [DOI] [PubMed] [Google Scholar]
- 11.Ioannidis JP, Trikalinos TA. An exploratory test for an excess of significant findings. Clin Trials. 2007;4(3):245–253. doi: 10.1177/1740774507079441. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 14.Bray I, Bryan K, Prenter S, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One. 2009;4(11):e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckley PG, Alcock L, Bryan K, et al. Chromosomal and microRNA expression patterns reveal biologically distinct subgroups of 11q-neuroblastoma. Clin Cancer Res. 2010;16(11):2971–2978. doi: 10.1158/1078-0432.CCR-09-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 17.Cairo S, Wang Y, de Reynies A, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107(47):20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 19.Caramuta S, Egyhazi S, Rodolfo M, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130(8):2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 20.Catto JW, Miah S, Owen HC, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69(21):8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung GE, Yoon JH, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23(1):113–119. [PubMed] [Google Scholar]
- 22.Cohn DE, Fabbri M, Valeri N, et al. Comprehensive miRNA profiling of surgically staged endometrial cancer. Am J Obstet Gynecol. 2010;202(6):656e1–8. doi: 10.1016/j.ajog.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Lisio L, Gomez-Lopez G, Sanchez-Beato M, et al. antle cell lymphoma: transcriptional regulation by microRNAs. Leukemia. 2010;24(7):1335–1342. doi: 10.1038/leu.2010.91. [DOI] [PubMed] [Google Scholar]
- 24.Dyrskjot L, Ostenfeld MS, Bramsen JB, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69(11):4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 25.Eitan R, Kushnir M, Lithwick-Yanai G, et al. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol Oncol. 2009;114(2):253–259. doi: 10.1016/j.ygyno.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105(35):13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68(1):26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 29.Haller F, von Heydebreck A, Zhang JD, et al. Localization- and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32.31. J Pathol. 2010;220(1):71–86. doi: 10.1002/path.2610. [DOI] [PubMed] [Google Scholar]
- 30.Hiroki E, Akahira J, Suzuki F, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101(1):241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14(2):419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrie CH, Chi J, Taylor S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13(7):1248–1260. doi: 10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CH, Subramanian S, Beck AH, et al. MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS One. 2009;4(10):e7314. doi: 10.1371/journal.pone.0007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123(7):1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 36.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 37.Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 39.Navarro A, Diaz T, Martinez A, et al. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood. 2009;114(14):2945–2951. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- 40.Navarro A, Gaya A, Martinez A, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111(5):2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa R, Ishiguro H, Kuwabara Y, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol. 2009;42(2):102–109. doi: 10.1007/s00795-009-0443-1. [DOI] [PubMed] [Google Scholar]
- 42.Pass HI, Goparaju C, Ivanov S, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70(5):1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70(1):36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 44.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 45.Schaefer A, Jung M, Mollenkopf HJ, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 46.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 47.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulte JH, Schowe B, Mestdagh P, et al. Accurate prediction of neuroblastoma outcome based on miRNA expression profiles. Int J Cancer. 2010;127(10):2374–2385. doi: 10.1002/ijc.25436. [DOI] [PubMed] [Google Scholar]
- 49.Segura MF, Belitskaya-Levy I, Rose AE, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16(5):1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11(2):136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veerla S, Lindgren D, Kvist A, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124(9):2236–2242. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 52.Visone R, Rassenti LZ, Veronese A, et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114(18):3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 54.Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Luo XQ, Zhang P, et al. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4(11):e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhi F, Chen X, Wang S, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer. 2010;46(9):1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 61.Sauerbrei W, Schumacher M. A bootstrap resampling procedure for model building: application to the Cox regression model. Stat Med. 1992;11(16):2093–2109. doi: 10.1002/sim.4780111607. [DOI] [PubMed] [Google Scholar]
- 62.Verweij PJ, Van Houwelingen HC. Cross-validation in survival analysis. Stat Med. 1993;12(24):2305–2314. doi: 10.1002/sim.4780122407. [DOI] [PubMed] [Google Scholar]
- 63.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kavvoura FK, McQueen MB, Khoury MJ, Tanzi RE, Bertram L, Ioannidis JP. Evaluation of the potential excess of statistically significant findings in published genetic association studies: application to Alzheimer's disease. Am J Epidemiol. 2008;168(8):855–865. doi: 10.1093/aje/kwn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ioannidis JP. Excess significance bias in the literature on brain volume abnormalities. Arch Gen Psychiatry. 2011;68(8):773–780. doi: 10.1001/archgenpsychiatry.2011.28. [DOI] [PubMed] [Google Scholar]
- 68.Pfeiffer T, Bertram L, Ioannidis JP. Quantifying selective reporting and the Proteus phenomenon for multiple datasets with similar bias. PLoS One. 2011;6(3):e18362. doi: 10.1371/journal.pone.0018362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA. 2011;305(21):2200–2210. doi: 10.1001/jama.2011.713. [DOI] [PubMed] [Google Scholar]
- 70.Lau J, Ioannidis JP, Terrin N, Schmid CH, Olkin I. The case of the misleading funnel plot. BMJ. 2006;333(7568):597–600. doi: 10.1136/bmj.333.7568.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobbin KK, Zhao Y, Simon RM. How large a training set is needed to develop a classifier for microarray data? Clin Cancer Res. 2008;14(1):108–114. doi: 10.1158/1078-0432.CCR-07-0443. [DOI] [PubMed] [Google Scholar]
- 72.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 73.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 74.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nature reviews. Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 75.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157(4):216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.