Abstract
Genetic information is encoded not only by the linear sequence of DNA, but also by epigenetic modifications of chromatin structure that include DNA methylation and covalent modifications of the proteins that bind DNA. These “epigenetic marks” alter the structure of chromatin to influence gene expression. Methylation occurs naturally on cytosine bases at CpG sequences and is involved in controlling the correct expression of genes. DNA methylation is usually associated with triggering histone deacetylation, chromatin condensation, and gene silencing. Differentially methylated cytosines give rise to distinct patterns specific for each tissue type and disease state. Such methylation-variable positions (MVPs) are not uniformly distributed throughout our genome, but are concentrated among genes that regulate transcription, growth, metabolism, differentiation, and oncogenesis. Alterations in MVP methylation status create epigenetic patterns that appear to regulate gene expression profiles during cell differentiation, growth, and development, as well as in cancer. Environmental stressors including toxins, as well as microbial and viral exposures, can change epigenetic patterns and thereby effect changes in gene activation and cell phenotype. Since DNA methylation is often retained following cell division, altered MVP patterns in tissues can accumulate over time and can lead to persistent alterations in steady-state cellular metabolism, responses to stimuli, or the retention of an abnormal phenotype, reflecting a molecular consequence of gene-environment interaction. Hence, DNA epigenetics constitutes the main and previously missing link among genetics, disease, and the environment. The challenge in oral biology will be to understand the mechanisms that modify MVPs in oral tissues and to identify those epigenetic patterns that modify disease pathogenesis or responses to therapy.
Keywords: epigenetics, DNA methylation, gene regulation, infection, inflammation, field effect
Introduction
Four years after the unveiling of the complete sequence of the human genome in 2003, “Human Genetic Variation” was recognized by the journal Science as the “breakthrough of the year” (Kennedy, 2007). This report emphasized the number of studies in that year that led to a new view of human genetic diversity, with appreciation of the extent to which our genomic sequences differ from person to person, and the implications of these variations for potentially deciphering the complexity of the biological systems in the human body. Undoubtedly, variations in the linear sequence of the genetic code, like single nucleotide polymorphisms (SNPs), may play a key role in explaining inter-individual differences in structure and function, as well as insight into disease susceptibility and resistance. However, the function of our genome is also dependent upon epigenetic mechanisms—which are by definition “beyond the genome”, and include alterations of chromatin structure, involving covalent modification of the central DNA molecule itself, as well as the complex macromolecules that form chromatin. The rapidly evolving field of epigenetics is contributing to our understanding of gene-environment interactions, as epigenetic mechanisms exert an additional layer of transcriptional control that regulates gene expression.
Historical Perspective
Prior to the middle of the twentieth century, before DNA was given a special status in biology, the developmental biologist and evolutionist Conrad H. Waddington (1905-1975) emphasized that genetics and developmental biology were related (Pennisi, 2001), hypothesizing that patterns of gene expression, turning genes on and off, and not the genes themselves, define each cell type, thus linking genes and gene action to development. To denote the dynamic actions leading from the genotype to the phenotype, Waddington coined the term ‘epigenetics’ from the Greek word epigenesis, referring to embryology and genetics as “a gradual coming into being of newly formed organs and tissues out of an initially undifferentiated mass”. In this way, Waddington indicated that an epigenetic landscape underlies each developing organism, referring to the existence of a complex network in which genetic interactions, the feedback and “feedforward” relationships among DNA, proteins, and other internal and external biochemical compounds are highly intermingled (Van de Vijver et al., 2002). In 1975, two papers were published (Holliday and Pugh, 1975; Riggs, 1975) outlining a molecular model for the switching of gene activities, and also the heritability of gene activity or inactivity. This model was based on the enzymatic methylation of cytosine in DNA. The suggestion was that DNA methylation could have strong effects on gene expression, and changes in DNA methylation may therefore explain the switching on and off of genes during development, and that the pattern of methylation could be heritable, persisting through cell divisions. That is, during DNA replication, the methylation patterns in cytosine bases would also be conserved during strand duplication. More recently, the scope of epigenetics extends to heritable modifications of genes, leading to alteration in the expression of specific DNA sequences that vary among different tissues within the same organism and cannot be explained by changes in DNA sequence.
Epigenetics, as the term suggests, can be seen as a major turn away from molecular biology’s Central Dogma, recognizing that there are epigenetic inheritance systems through which non-sequence-dependent DNA variations can be transmitted in cell, tissue, and organismal lineages. Thus, current epigenetics not only offers new insights into gene regulation and heredity, but it also profoundly challenges the way we think about evolution, genetics, and development. Most interestingly, it suggests testable mechanisms whereby environmental factors (ranging from stress to infection) can influence genetic expression. Furthermore, these potential epigenetic modifications can occur throughout the lifetime of the organism, beginning as early as the intra-uterine environment, and can accumulate in tissues and cells over time to modify gene expression patterns and cellular phenotypes.
Epigenome and DNA Methylation
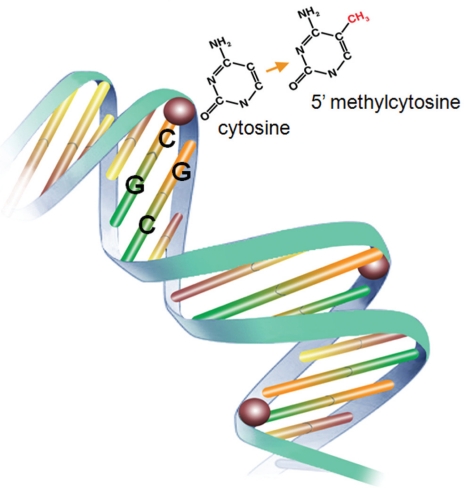
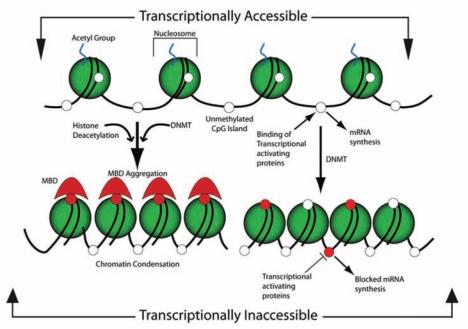
The most-studied epigenetic modification of DNA in mammals is methylation of cytosine in CpG dinucleotides (Bird, 2002). The other main group of epigenetic modifications includes post-translational modification of histones, principally changes in phosphorylation, acetylation, and ubiquitinylation status. Often, these epigenetic mechanisms are coupled and interact to modify chromatin structure and function. DNA methylation is a covalent biochemical modification that, in the mammalian genome, takes place predominantly at cytosine bases that are located 5´ to a guanosine (Fig. 1). CpGs are vastly under-represented in the genome, as compared with what would be expected by chance (0.23 in the human genome). Furthermore, CpG-rich regions are not evenly distributed throughout the genome, but appear most often among the promoter regions and first exons of specific genes (Larsen et al., 1992). Within the structure of chromatin, DNA is complexed with histone proteins to form octamers around which DNA loops to form the nucleosome, the individual packaging unit of genomic DNA. The enzymes involved in DNA methylation include the DNA methyltransferases (DNMTs), which establish and maintain DNA methylation patterns using S-adenosyl methionine (SAM) as the methyl donor. When DNA becomes methylated, those methyl groups protrude from the cytosine nucleotides into the major groove of the DNA to displace transcription factors that normally bind to the DNA (Hark et al., 2000). The exposed methylation sites attract methyl-binding proteins, the methyl-CpG-binding domain proteins (MBDs), which are involved in ‘reading’ methylation marks (Loenen, 2006) to affect chromatin condensation by recruiting histone deacetylases that covalently modify the tails of histone proteins (Vucic et al., 2008). Histone modification results in gene silencing and chromatin compaction (Bird and Wolffe, 1999) (Fig. 2).
Figure 1.
Methylation of DNA occurs at cytosine residues when present as CG dinucleotides. Methylation occurs by the addition of a methyl group at the 5′ site of cytosine (depicted as shaded sphere).
Figure 2.
The shaded sphere depicts the octameric histone complex, which forms the nucleosome with the acetylated tails of histones and the cytosines of the CpG sites in an unmethylated state, shown as open white circles. In this conformation, the chromatin is loosely packed and available for the binding of transcriptional activating proteins, which, by the action of RNA polymerase II, synthesize mRNA. The action of DNA methyl transferase (DNMT) methylates the cytosine residues, depicted as red circles, which provide a docking site for the methyl binding domain proteins (MBD), which aggregate in conjunction with the action of the histone deacetylase, which cleaves the histone acetyl group. Both of these serve to alter the structure of the chromatin by causing a condensation that impedes the access of the transcriptional activating proteins and thereby blocks mRNA synthesis. Alternatively, the normal active structure of chromatin can become inaccessible for the binding of transcriptional activating proteins by the action of CpG methylation at sites that sterically hinder the binding of activating proteins, independent of MBD aggregation.
It is important that methylation patterns be generally conserved following DNA replication carrying epigenetic patterns to cellular progeny, thereby creating a link or communication that conveys the developmental or environmental pressures of preceding cellular generations. In 2004, Fazzari and Greally pointed out that the methylation of CpG-rich promoters is used by mammals to prevent transcriptional initiation and to ensure the silencing of genes on the inactive X chromosome, imprinted genes, and parasitic DNAs. The potential role of DNA methylation in tissue-specific gene expression has been explored more recently, since it was realized that CpG methylation can regulate tissue-specific gene expression and can be influenced by external stressors, environmental toxins, and aging (Dolinoy et al., 2007; Hanson and Gluckman, 2008), potentially increasing or decreasing the level of transcription, depending on whether the methylation inactivates a positive or negative regulatory element.
The ontological role of DNA methylation, through transcriptional silencing, contributes to an epigenetic regulation of the embryonic and morphogenetic developmental gene expression program (Holliday and Pugh, 1975). DNA methylation is also recognized as an ancient host defense system designed to protect against exogenous parasitic nucleic acid sequence elements or deleterious endogenous sequences, which have been evolutionarily incorporated and retained vestigially within our genome (Doerfler, 1991; Yoder et al., 1997). In normal cells, DNA methylation occurs predominantly in repetitive genomic regions, including satellite DNA and parasitic elements (such as long interspersed transposable elements [LINES], short interspersed transposable elements [SINES], and endogenous retroviruses [Yoder et al., 1997]), offering a mechanism by which the environment can stably change gene expression. Changes in methylation status can also regulate microRNA (miRNA) expression, which, in turn, modulates post-transcriptional gene expression and plays important roles in essential processes, such as differentiation, cell growth, and cell death (Miska, 2005; Zamore and Haley, 2005). The key concept is that the epigenome, consisting of chromatin and its modifications, functions as an interface between the inherited genome and the dynamism imposed by the environment, and such interaction promotes epigenetic modifications that are specific DNA methylation patterns, which then result in a relatively stable or homeostatic profile of gene expression. This has been referred to (Feinberg, 2008) as a metastable condition that represents an epigenetic modification, resulting in a new cellular or tissue homeostatic “set-point” with a new range of gene expression patterns that differ from those of the original state. Chromatin modifications, including CpG methylations, allow for sculpting of the epigenome during development, modified by individual environmental exposures, providing absolutely unique identity, even for monozygotic twins (Fraga et al., 2005).
In a landmark publication, Fraga and colleagues (2005) reported, in a large cohort of monozygotic twins, that DNA methylation increased over time within different tissues and cell types, including oral epithelial, lymphocytic, muscle, and fat. Most importantly, tissues from identical twins who were 3 years of age displayed low levels of DNA methylation and virtually identical gene expression profiles for the different tissues obtained from each twin, reflecting the high concordance of gene expression patterns associated with monozygosity, i.e., identical DNA sequences. However, when they compared DNA methylation patterns in identical twins at the age of 50, there were higher DNA methylation levels in general. The gene expression patterns in the twins were significantly different, and those differences were mapped to the alterations in DNA methylation status. Thus, the authors suggested that the “distinct profiles of DNA methylation and histone acetylation patterns that among different tissues (that) arise during the lifetime of monozygotic twins may contribute to the explanation of some of their phenotypic discordances and underlie their differential frequency/onset of common diseases”, dependent upon the cumulative history of external exposures (i.e., environmental, nutritional, toxins, and infectious) that reprogram epigenetic status.
Imprinting
Diploid organisms carry two copies of every autosomal gene, one from each parent. In the great majority of cases, the two copies are either repressed or transcribed identically, but this is not the case for genes that exhibit the phenomenon of parental imprinting. Observations through the centuries have suggested that the genes passed on by each parent had somehow been permanently marked—or “imprinted”, as it eventually came to be known—so that the expression patterns of the maternal and paternal genes differ in their progeny. Genomic imprinting in mammals represents a situation where there is non-equivalence in the expression of alleles at certain gene loci, dependent on the parent of origin (Reik and Walter, 1998).The expression of either the paternally or maternally inherited allele is consistently repressed, resulting in mono-allelic expression of a particular gene. Thus, imprinted genes show markedly different behavior, depending on their parental origin. The same pattern of mono-allelic expression is faithfully transmitted to daughter cells following cell division. Imprinting is not a phenomenon entirely unique to mammals, since it also happens in plants, where most commonly the paternal genes are imprinted (Grant-Downton and Dickinson, 2005).
However, it is only since 1991 that researchers have begun to isolate a variety of genes whose expression depended upon their parents of origin. That year, researchers identified two genes, Igf2r [insulin-like growth factor-2 receptor] and H19, that are active only when inherited from the mother; a third, called Igf2 [insulin-like growth factor-2], is turned on only when inherited from the father. Those findings raised essential questions on how genomes become marked differentially during gametogenesis, and how this marking is maintained on the gene throughout development, as well as prompting a broader search for other imprinted genes (Bartolomei et al., 1991; DeChiara et al., 1991; Constancia et al., 1998). DNA methylation appears to be the key mechanism by which one copy of a gene is preferentially silenced according to parental origin and maintained during cell division by 5-cytosine DNA methyltransferase-1 (DNMT1) (Okano et al., 1999; Miranda and Jones, 2007). DNMT1 preferentially methylates hemi-methylated CpG sites, thus copying established methylation patterns to the newly synthesized DNA strands. Reik and Walter (1998) proposed that at least a subset of ~ 100 genes of the ~20,000-25,000 genes in the mammalian genome are thought to be imprinted, and a list of known imprinted genes can be accessed at http://www.geneimprint.com/site/genes-by-status. Although it has not been demonstrated, it appears likely that many small RNA sequences that arise from non-coding regions, and are also involved in gene regulation and metabolism, might be candidates for epigenetic marks and possibly demonstrate parent-of-origin imprinting properties. This possibility is suggested by the presence of CpG-rich regions that are occasionally present in non-coding regions, including miRNAs. The concept of imprinting is an interesting example of interactions between heterologous genomes (maternal and paternal) that seek to establish some sort of mutually acceptable state of equilibrium. The role of imprinting in IGF2 regulation in placental and fetal growth is a fascinating example. This insulin-like growth hormone is the major somatic growth factor for the fetus, since it enhances placental nutrient exchange of glucose for fetal growth, and it serves as a pluripotent tissue growth hormone for the fetus. IGF2 impairment restricts fetal growth. It is perhaps no surprise that the paternal genome serves to enhance IGF-II secretion to stimulate the transfer of nutrients from mother to fetus to make the baby as large and healthy as possible, whereas the maternal genome seeks to attenuate this response by controlling the expression level of the receptor for this hormone. Thus, one can appreciate that the epigenetic response that mediates IGF2 gene expression strikes a balance between competing genetic programs. It now appears that epigenetic mechanisms serve to mediate genomic conflict that can occur between host and exogenous genomes such as those provided by viral infections or even the commensal microbiome.
Development
Differences in programmed gene expression that result in the development of organs, tissues, and cell lineages generally occur without changes to the sequence of our DNA (with one or two exceptions, e.g., immunoglobulin synthesis), indicating that development is, by definition, epigenetic (Reik, 2007). Certain cell lineages are dependent upon epigenetic programs that sequentially regulate gene expression patterns to direct differentiation, maturation, and function effectively. The helper T-cell population of the immune system is an example of this epigenetic programming (Ansel et al., 2003). During the differentiation of CD4+ T-cells, there is an epigenetic activation of the interferon gamma gene (IFNG) and a silencing of the interleukin 4 (IL4) gene. This results in a progressive polarization of T-cell responsiveness as the epigenetic modifications are further modified by antigenic and cytokine actions via sequential divisions within the lineage. Thus, different T-helper cells emerge and maintain a polarized phenotype based upon epigenetic modifications that are retained following cell divisions, but additionally polarized by antigenic and environmental cytokines, which add to the epigenetic modulation. In mice, naïve T-cells have hypermethylated CpG islands within the Il4 locus. Extended demethylation within this region results in Il4 gene activation and is coupled to Ifng silencing, leading to a TH2 phenotype commitment. Retention of Il4 methylation and Ifng activation results in a TH1 phenotype. Thus, epigenetic memory of the T-cells and the lineage modulates immune responses via TH2 (which up-regulates IL4, IL5, and IL13) or TH1 (with enhanced IFN-γ and IL-2) cytokine responses (Fields et al., 2002).
During development, imprinting of fetal tissues includes the placenta, principally targeting trophoblast cells. Abnormalities in placental imprinting have been recently implicated as a cause of preeclampsia (Van Dijk et al., 2005) and fetal growth restriction (McMinn et al., 2006). Imprinting patterns have been associated with congenital disorders affecting growth and neurodevelopment that persist into adulthood, including Prader-Willi and Angelman syndromes, which are two clinically distinct diseases associated with abnormal imprinting on chromosome 15q11-q13. Loss of maternal imprinting is responsible for the Angelman syndrome, which is characterized by mental retardation, ataxia, and social disposition. In Prader-Willi syndrome, loss of paternal imprinting in the same region is characterized by learning difficulties, hypogonadism, short stature, and small hands and feet. Beckwith-Weidemann syndrome, another imprinting disorder characterized by macrosomia, hemihypertrophy, abdominal wall defects, organomegaly, and susceptibility to Wilm’s tumor, is the result of loss of imprinting of insulin-like growth factor 2 (IGF2) on chromosome 11p15 (Reik and Maher, 1997; Tycko and Morison, 2002). We recently demonstrated, in a pregnant murine model, that infection with Campylobacter rectus could induce an alteration in placental Igf2 methylation patterns that resulted in reduced insulin-like growth factor II mRNA expression with an associated fetal growth restriction (Bobetsis et al., 2007). This suggests that external stimuli or stressors like infection can modify imprinting patterns in utero.
Environmental Stressors as Epigenetic Modifiers
Alterations in DNA methylation status as a result of environmental stressors have been documented to begin before birth. For example, the methylation of fetal DNA that occurs in utero as a result of low dietary levels of folate, methionine, or selenium can change epigenetic programming that can persist into adulthood (Post et al., 1999; Lund et al., 2004; Zaina et al., 2005). Although many epigenetic marks are potentially reversible, the mechanisms for reversal remain to be clearly elucidated, and many epigenetic changes appear to persist throughout the cell lineage and life of the organism. This provides perhaps an explanation for the Barker hypothesis (Barker et al., 2002), which posits that intra-uterine exposures can result in fetal programming that persists into adulthood and may contribute to the risk for adult-onset diseases such as cardiovascular disease and type 2 diabetes.
Intra-uterine nutrition can determine epigenetic programming of the fetus. For example, methyltetrahydrofolate (folate) is a critical methyl donor for S-Adenosyl Methionine (SAM), which is used by the enzyme DNA methyltransferase (DNMT) to methylate CpG residues selectively during embryonic development (Razin and Shemer, 1995; Carlone and Skalnik, 2001; Hershko et al., 2003). Maternal folate deficiency during pregnancy leads to inadequate levels of SAM, a critical substrate for DNMT-dependent methylation (Okano et al., 1999). Folate deficiencies can thereby result in DNA hypomethylation, which can contribute to improperly elevated expression of certain genes, as well as genetic instability facilitating abnormal chromosomal re-arrangements (Zaina et al., 2005). These folate deficiencies often result in abnormalities in placental development and function and alter patterns in fetal DNA methylation that can result in growth defects and birth anomalies, including neural tube defects (Blom et al., 2006). Thus, gross nutritional deficiencies can lead to impairment of normal epigenetic programming, resulting in abnormal ontological outcomes.
Such dietary or other exogenous environmental factors also appear to modulate chromatin epigenetic marks throughout life. For example, hyperhomocysteinemia (a marker for low levels of methyl donors) is associated with global hypomethylation and has been reported in atherosclerosis models and in humans, which supports an emerging view that alterations in global methylation patterns are characteristic of early stages of cardiovascular disease. In advanced stages of atherosclerosis, smooth-cell proliferation or monocytic clonal expansion within the atheroma may be associated with altered DNA methylation patterns (Castro et al., 2006). The first findings linking DNA methylation of CpG islands to cardiovascular disease identified increased methylation of the CpG region of the estrogen receptor-α (ERα) gene seen in coronary atherectomy or carotid endarterectomy samples (Post et al. 1999).
It has also been proposed that sensitivity to diet or to environmental toxins may vary among individuals, due to pre-existing genetic variants that can challenge methyl metabolism and predispose individuals to epigenetic changes (Lund and Zaina, 2007). Other environmental stimuli that may potentially function as epigenetic modifiers are exposures to metals and aromatic hydrocarbons (e.g., benzopyrene), found in occupational chemicals, fossil fuel emissions, contaminated drinking water, cigarette smoke, and infection (Risch and Plass, 2008). A recent study on the influence of smoking on global DNA methylation indicated that smoking induces generalized alterations in DNA methylation across multiple tissues and organ systems, also showing an association of the offspring’s DNA methylation with paternal DNA methylation that was strongest if both had never smoked (p = 0.02) (Hillemacher et al., 2008). The authors also found that the association completely vanished if descendants smoked themselves or had ever smoked, suggesting an association between smoking behavior and global DNA methylation, which may be of importance for a wide range of diseases. Smoking may also be linked to oncogenesis by inducing specific epigenetic modifications (Tessema et al., 2008). For example, promoter methylation of several tumor suppressor genes has frequently been reported in a high percentage (20-100%) of human lung cancers (Zochbauer-Muller et al., 2002). Methylation of the tumor suppressor p16 gene has been suggested to play a critical role in lung cancer survival rate pathogenesis (Kim et al., 2001; von Zeidler et al., 2004; Dammann et al., 2005).
Epigenetics, Cancer, and Inflammation
The best-studied epigenetic alteration in cancer is DNA methylation. During tumorigenesis, methylation is usually decreased genome-wide, with selective hypermethylation of CpG sites within promoters of tumor-suppressor genes, leading to their silencing and subsequent tumor progression (Breivik and Gaudernack, 1999). This suggests that oncogenesis may also occur through epigenetic dysregulation. Feinberg (2007) has recently reviewed the epigenetic mechanisms involved with oncogene activation or tumor suppressor gene silencing in cancer initiation and progression, discussing the new idea that epigenetic modifications may play a role in cancer predisposition, and that such changes should be considered as targets for preventive oncology.
The role of host inflammation on modification of epigenetic patterns is still unknown, but the activation of the immune response involving potential epigenetic changes has been suggested (Adcock and Lee, 2006). Inflammatory signals that activate NF-κB have been shown to alter histone methylation patterns and activate gene expression (Ito, 2007). Thus, inflammation has some potential to modify chromatin structure via histone structure; however, the role of inflammation in modulating CpG methylation patterns, which are more likely to be conserved following cell replication, remains unclear. Recent reports suggest that loss of epigenetic control over this complex process contributes to autoimmune disease (Yung and Julius, 2008). Logically, this may involve abnormal function and maturation in T-cell lineages as a consequence of aberrant epigenetic patterns.
Epigenetic mechanisms may also explain, in part, the linkage between inflammation and oncogenesis, and the relationship between CpG island methylation phenotype in tumors and inflammation has been discussed (Shaw et al., 2007). For example, gastric inflammation due to bacterial infection with H. pylori has been linked to alteration in DNA methylation patterns of tumor suppressor genes (Tsuji et al., 2006; Ushijima, 2007). H. pylori is an etiologic gastric carcinogen, with about 80% of gastric cancers being H. pylori-related (Forman et al., 1991). However, the cancer risks are different among H. pylori-infected individuals, which probably reflects the diversity of H. pylori strains, and differences in host susceptibility or other environmental factors (Uemura et al., 2001). There exists, however, a close anatomical relationship in cancerous lesions, whereby H. pylori is in direct contact with gastric cells that display altered MVP methylation patterns. One specific MVP target in gastric cancer appears to be cyclo-oxygenase 2 (COX-2). Low levels of gastric secretion of prostaglandin E2 (a product of cyclo-oxygenase 2) and COX-2 suppression have long been known to be associated with gastric cancer. Recent studies (Huang et al., 2006; Perri et al., 2007) have demonstrated that cancerous gastric cells exhibit abnormal PTGS2 (Prostaglandin G/H Synthase-2) promoter hypermethylation patterns. Non-cancerous regions of the gastric lesion do not demonstrate the presence of H. pylori or the abnormal PTGS2 promoter hypermethylation and COX-2 suppression. Thus, the evidence supports the concept that the H. pylori infection promotes oncogenesis by epigenetic modification that includes the PTGS2 promoter.
In 2007, we were the first group to report alteration in DNA methylation status of the Igf2 gene in murine placental tissues due to maternal infection with the periodontopathogenic bacteria Campylobacter rectus (Bobetsis et al., 2007). By analogy, H. pylori and C. rectus are close phylogenetic neighbors that share, for example, GroEL protein (HSP60 family) expression, which can stimulate IL-6 production (Tanabe et al., 2003). We were able to demonstrate, in placentas from growth-restricted fetuses, that the hypermethylation found in the promoter region (P0) of the Igf2 gene was related to the C. rectus placental exposure in pregnant mice. This appears causal, since Igf2 gene function involves growth and development. Population studies have shown that, among humans, prematurity and impaired fetal growth have been associated with adult-onset diabetes and cardiovascular disease (Barker et al., 2002). If abnormalities in IGF2 methylation were to occur in utero among humans, it is possible that these epigenetic marks could persist into adulthood and may be associated with the observed abnormalities in IGF2 metabolism in adults with cardiovascular disease and diabetes. Thus, the role of infection as an intra-uterine modifier of epigenetic programming should be considered as a possible link to adult disease. This may be particularly relevant in humans, since C. rectus exposure of the fetus has been found to be associated with preterm delivery (Madianos et al., 2001).
Following the observation and establishment of a connection between oral bacteria altering placental DNA methylation, we asked the question, Could the oral biofilm epigenetically modify the local adjacent periodontal tissues? We have conducted a pilot survey using CpG Island Microarray analysis (data not shown) comparing genomic-wide MVP methylation status of periodontally diseased gingival tissues with healthy gingival tissue. We could preliminarily identify a list of genes that were differentially methylated in gingival tissues from individuals with periodontal disease, and these results will be forthcoming shortly. Thus, the role of bacterial infection and chronic inflammation as a potential stimulus for altering local periodontal tissue DNA methylation patterns provides a fertile area for further investigation. Furthermore, the link between inflammation and oral cancer is well-established, and a connection between bacterial infection and inflammation is evident. Thus, epigenetic influences may serve as a plausible potential mechanism that connects all three pathways and should be further explored, especially as it relates to mucosal cancers, which emerge in the presence of high microbial burdens.
Epigenetic Marks in Carcinogenesis
Epigenetic changes set the stage for alterations in gene expression and have been identified as important components of carcinogenesis. As previously mentioned, global DNA hypomethylation is a general feature of genomic DNA derived from solid and hematologic tumors as isolated from animal models and human tumors (Gaudet et al., 2003; Fraga et al., 2005; Holm et al., 2005). Hypomethylation is consistent with the overall increased transcriptional activity seen in most tumors. However, loss of DNA methylation, which often occurs at sequences which are unstable, is likely related to increased tumor frequency due to chromosomal instability, and it has been considered as the earliest epigenetic change from a normal to a pre-malignant cell. However, the expression of certain oncogenes appears to be directly activated by hypomethylation, whereas hypermethylation of tumor suppressor genes can also be seen. Examples of promoter DNA hypermethylation and chromatin hypoacetylation, which result in the silencing of tumor suppressor genes, include p16 (also known as cyclin-dependent kinase inhibitor 2A) and MutL protein homologue 1 (MLH1) (Herman and Baylin, 2003; Feinberg and Tycko, 2004).
In a recent review, Choi and Myers (2008) emphasized the genetic and epigenetic alterations in the molecular pathogenesis of oral squamous cell carcinoma (OSCC), discussing the role of oncogenes like Ras oncogene, Cyclin D1, AP-1 complex, and tumor suppressor genes like p53, p16, and p21. These investigators reported aberrant hypermethylation patterns in the promoter region of p16 and E-cadherin which influence cell division and cell-cell adhesion, respectively. p16 was one of the first genes to be found associated with aberrant DNA methylation patterns in head and neck cancer (Reed et al., 1996). p16 inhibits G1 to S phase passage by binding cyclin-dependent kinase, preventing formation of its complex with cyclin D. Methylation of p16 promoter has been considered as a predictive marker for malignant transformation, since the methylation depicts uncontrolled cell division (Hall et al., 2008). In OSCC, p16 methylation has been reported to vary between 31% (Maruya et al., 2004) and 67% (Kulkarni and Saranath, 2004). E-cadherin plays a role in cell-cell adhesion, and, when underexpressed, may affect tumor invasion by leading to a greater probability of tumor invasion or metastasis. E-cadherin was found silenced by hypermethylation in other studies of oral cancer (Hasegawa et al., 2002; Kudo et al., 2004; Maruya et al., 2004).
Other genes have also been investigated for aberrant DNA methylation in oral squamous cell carcinomas. The epigenetic silencing of the MGMT (O6-methylguanine–DNA methyltransferase) DNA-repair gene which, by promoter methylation, compromises DNA repair, has been considered an early event in the development of oral cancer and is associated with 25-52% of primary oral squamous-cell carcinomas (Viswanathan et al., 2003; Kulkarni and Saranath, 2004; Maruya et al., 2004). The death-associated protein kinase 1(DAPK1) gene, a tumor-suppressor gene involved in apoptosis, is also found methylated between 7% and 68% in oral cancers (Li, 2002; Ogi et al., 2002; Maruya et al., 2004).
It has also been shown that miRNAs, which modulate post-transcriptional gene expression, can be aberrantly expressed or mutated in cancers, suggesting that they may also function as oncogenes or tumor suppressor genes. More recently, studies have shown that miRNA genes may be regulated also by epigenetic mechanisms and have even been identified embedded within CpG islands (Lujambio and Esteller, 2007). The promoter of an miRNA methylated in normal tissues can be maintained in cancer, as in the case of two putative tumor suppressor miRNAs—miR-127 and miR-124a—which are transcriptionally inactivated by CpG island hypermethylation (Saito et al., 2006; Lujambio et al., 2007), whereas in lung cancer, the overexpression of let-7a-3 seems to be due to DNA hypomethylation (Brueckner et al., 2007; Weber et al., 2007). In both cases, the functional significance is the opposite. miR127 and miR-124a seem to act as tumor suppressors, and so methylation is maintained in cancer, while let7a3 is thought to act as an oncogene, so its demethylation would contribute to the tumoral phenotype. Clearly, the role of epigenetic influence on miRNA function is a new area of investigation and represents just one area in which the role of epigenetics and oral disease needs further exploration.
Future research will help us understand, for example, how systemic exposures, like smoking, may alter global epigenetic patterns to affect the expression of oral conditions such as oral cancer or advanced periodontitis. We need to understand how the oral microbiome and local biofilm may create an epigenetic “footprint” in the adjacent mucosa and periodontal tissues, and potentially modify the local inflammatory response and oncogenic potential. Biofilm-induced epigenetic patterns may influence local tissue metabolism to alter the microbial ecology and alter local healing responses of the periodontal tissues. Recently, Park and colleagues (2008) were able to reprogram somatic human cells to a pluripotent state, which is in essence a reversal of differentiation to a more embryonic state, by inducing the ectopic expression of four transcriptional regulatory factors (Oct4, Sox2, Klf4, and Myc). This resulted in significant epigenetic remodeling, which was sufficient to result in cellular reprogramming to a pluripotent state. This suggests that epigenetic reprogramming might prove to be a mechanism to create new wound-healing or tissue-regenerative potential, and agents which modify epigenetic patterns are a fertile area for new drug development strategies. These are just a few important questions and opportunities that will await further studies that explore the role of epigenetics in oral biology. Thus, epigenetic codes, which are just becoming revealed, can help us better understand the biological phenotype that arises from the interaction of the human genome with the environment in health and in disease.
Footnotes
This work is supported by grant DE-01243 from NIDCR and grant RR-00046 from NCRR to SO.
References
- Adcock IM, Lee KY. (2006). Abnormal histone acetylase and deacetylase expression and function in lung inflammation. Inflamm Res 55:311-321, erratum in Inflamm Res 55:572, 2006 [DOI] [PubMed] [Google Scholar]
- Ansel KM, Lee DU, Rao A. (2003). An epigenetic view of helper T cell differentiation. Nat Immunol 4:616-623 [DOI] [PubMed] [Google Scholar]
- Barker DJ, Eriksson JG, Forsen T, Osmond C. (2002). Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 31:1235-1239 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman MS. (1991). Parental imprinting of the mouse H19 gene. Nature 351:153-155 [DOI] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev 16:6-21 [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. (1999). Methylation-induced repression: belts, braces, and chromatin. Cell 99:451-454 [DOI] [PubMed] [Google Scholar]
- Blom HJ, Shaw GM, den Heijer M, Finnell RH. (2006). Neural tube defects and folate: case far from closed. Nat Rev Neurosci 9:724-731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy D, Jirtle RL, et al. (2007). Bacterial infection promotes DNA hypermethylation. J Dent Res 86:169-174 [DOI] [PubMed] [Google Scholar]
- Breivik J, Gaudernack G. (1999). Genomic instability, DNA methylation, and natural selection in colorectal carcinogenesis. Semin Cancer Biol 9:245-254 [DOI] [PubMed] [Google Scholar]
- Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, et al. (2007). The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res 67: 1419-1423 [DOI] [PubMed] [Google Scholar]
- Carlone DL, Skalnik DG. (2001). CpG binding protein is crucial for early embryonic development. Mol Cell Biol 22:7601-7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R, Rivera I, Blom HJ, Jakobs C, Tavares de, Almeida I. (2006). Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: an overview. J Inherit Metab Dis 29:3-20 [DOI] [PubMed] [Google Scholar]
- Choi S, Myers JN. (2008). Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res 87:14-32 [DOI] [PubMed] [Google Scholar]
- Constancia M, Pickard B, Kelsey G, Reik W. (1998). Imprinting mechanisms. Genome Res 8: 881-900 [DOI] [PubMed] [Google Scholar]
- Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, et al. (2005). CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer 41:1223-1236 [DOI] [PubMed] [Google Scholar]
- DeChiara M, Robertson EJ, Efstratiadis A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859 [DOI] [PubMed] [Google Scholar]
- Doerfler W. (1991). Patterns of DNA methylation—evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol Chem Hoppe Seyler 372:557-564 [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. (2007). Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 104:13056-13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. (2004). Epigenomics: beyond CpG islands. Nat Rev Genet 5:446-455 [DOI] [PubMed] [Google Scholar]
- Feinberg AP. (2007). Phenotypic plasticity and the epigenetics of human disease. Nature 447:433-440 [DOI] [PubMed] [Google Scholar]
- Feinberg AP. (2008). Epigenetics at the epicenter of modern medicine. J Am Med Assoc 299:1345-1350 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. (2004). The history of cancer epigenetics. Nat Rev Cancer 4:143-153 [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. (2002). Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol 169:647-650 [DOI] [PubMed] [Google Scholar]
- Forman D, Newell DG, Fullerton F, Yarnell JW, Stacey AR, Wald N. et al. (1991). Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from prospective investigation. BMJ 302:1302-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. (2005). Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391-400 [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, et al. (2003). Induction of tumors in mice by genomic hypomethylation. Science 300:489-492 [DOI] [PubMed] [Google Scholar]
- Grant-Downton RT, Dickinson HG. (2005). Epigenetics and its implications for plant biology. The epigenetic network in plants. Ann Bot (Lond) 96:1143-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall GL, Shaw RJ, Field EA, Rogers SN, Sutton DN, Woolgar JA, et al. (2008). p16 promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiol Biomarkers Prev 17:2174-2179 [DOI] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. (2008). Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol 102:90-93 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. (2002). Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 21:4231-4236 [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. (2003). Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042-2054 [DOI] [PubMed] [Google Scholar]
- Hershko AY, Kafri T, Fainsod A, Razin A. (2003). Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene 302:65-72 [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, Kornhuber J, et al. (2008). Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol 18:295-298 [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. (1975). DNA modification mechanisms and gene activity during development. Science 187:226-232 [PubMed] [Google Scholar]
- Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, 3rd, Jaenisch R. (2005). Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8:275-285; erratum in Cancer Cell 8:433, 2005, and Cancer Cell 9:69, 2006 [DOI] [PubMed] [Google Scholar]
- Huang L, Zhang KL, Li H, Chen XY, Kong QY, Sun Y, et al. (2006). Infrequent COX-2 expression due to promoter hypermethylation in gastric cancers in Dalian, China. Hum Pathol 37:1557-1567 [DOI] [PubMed] [Google Scholar]
- Ito K. (2007). Impact of post-translational modifications of proteins on the inflammatory process. Biochem Soc Trans 35:281-283 [DOI] [PubMed] [Google Scholar]
- Kennedy D. (2007). Breakthrough of the year. Science 318:1833 [DOI] [PubMed] [Google Scholar]
- Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, et al. (2001). p16 (INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 61:3419-3424 [PubMed] [Google Scholar]
- Kudo Y, Kitajima S, Ogawa I, Hiraoka M, Sargolzaei S, Keikhaee MR, et al. (2004). Invasion and metastasis of oral cancer cells require methylation of E-cadherin and/or degradation of membranous beta-catenin. Clin Cancer Res 10:5455-5463 [DOI] [PubMed] [Google Scholar]
- Kulkarni V, Saranath D. (2004). Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncol 40:145-153 [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. (1992). CpG islands as gene markers in the human genome. Genomics 13:1095-1107 [DOI] [PubMed] [Google Scholar]
- Li E. (2002). Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev Genet 3:662-673 [DOI] [PubMed] [Google Scholar]
- Loenen WA. (2006). S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans 34(Pt 2):330-333 [DOI] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. (2007). CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 6:1455-1459 [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67:1424-1429 [DOI] [PubMed] [Google Scholar]
- Lund G, Zaina S. (2007). Atherosclerosis, lipids, inflammation and epigenetics. Curr Opin Lipidol 18:699-701 [DOI] [PubMed] [Google Scholar]
- Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, et al. (2004). DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem 279:29147-29154 [DOI] [PubMed] [Google Scholar]
- Madianos PN, Lieff S, Murtha AP, Boggess KA, Auten RL, Jr, Beck JD, et al. (2001). Maternal periodontitis and prematurity. Part II: Maternal infection and fetal exposure. Ann Periodontol 6:175-182 [DOI] [PubMed] [Google Scholar]
- Maruya S, Issa JP, Weber RS, Rosenthal DI, Haviland JC, Lotan R, et al. (2004). Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin Cancer Res 10:3825-3830 [DOI] [PubMed] [Google Scholar]
- McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, et al. (2006). Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27:540-549 [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. (2007). DNA methylation: the nuts and bolts of repression. J Cell Physiol 213:384-390 [DOI] [PubMed] [Google Scholar]
- Miska EA. (2005). How microRNAs control cell division, differentiation, and death. Curr Opin Genet Dev 5:563-568 [DOI] [PubMed] [Google Scholar]
- Ogi K, Toyota M, Ohe-Toyota M, Tanaka N, Noguchi M, Sonoda T, et al. (2002). Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res 8:3164-3171 [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. (1999). DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell 99:247-257 [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141-146 [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2001). Behind the scenes of gene expression. Science 293:1064-1067 [DOI] [PubMed] [Google Scholar]
- Perri F, Cotugno R, Piepoli A, Merla A, Quitadamo M, Gentile A, et al. (2007). Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol 102:1361-1371 [DOI] [PubMed] [Google Scholar]
- Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, et al. (1999). Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res 43:985-991 [DOI] [PubMed] [Google Scholar]
- Razin A, Shemer R. (1995). DNA methylation in early development. Hum Mol Genet 4:1751-1755 [DOI] [PubMed] [Google Scholar]
- Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, et al. (1996). High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 56:3630-3633 [PubMed] [Google Scholar]
- Reik W. (2007). Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425-432 [DOI] [PubMed] [Google Scholar]
- Reik W, Maher ER. (1997). Imprinting in clusters: lessons from Beckwith-Wiedemann syndrome. Trends Genet 13:330-334 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. (1998). Imprinting mechanisms in mammals. Curr Opin Genet Dev 8:154-164 [DOI] [PubMed] [Google Scholar]
- Riggs AD. (1975). X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14:9-25 [DOI] [PubMed] [Google Scholar]
- Risch A, Plass C. (2008). Lung cancer epigenetics and genetics. Int J Cancer 123:1-7 [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. (2006). Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9:435-443 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Lowe D, Bowers NL, Liloglou T, Field JK, et al. (2007). CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol 43:878-886 [DOI] [PubMed] [Google Scholar]
- Tanabe S, Hinode D, Yokoyama M, Fukui M, Nakamura R, Yoshioka M, et al. (2003). Helicobacter pylori and Campylobacter rectus share a common antigen. Oral Microbiol Immunol 18:79-87 [DOI] [PubMed] [Google Scholar]
- Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, et al. (2008). Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res 68:1707-1714 [DOI] [PubMed] [Google Scholar]
- Tsuji S, Tsujii M, Murata H, Nishida T, Komori M, Yasumaru M, et al. (2006). Helicobacter pylori eradication to prevent gastric cancer: underlying molecular and cellular mechanisms. World J Gastroenterol 12:1671-1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B, Morison IM. (2002). Physiological functions of imprinted genes(review). J Cell Physiol 192:245-258 [DOI] [PubMed] [Google Scholar]
- Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. (2001). Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784-789 [DOI] [PubMed] [Google Scholar]
- Ushijima T. (2007). Epigenetic field for cancerization. J Biochem Mol Biol 40:142-150 [DOI] [PubMed] [Google Scholar]
- Van de, Vijver G, Van Speybroeck L, De Waele D. (2002). Epigenetics: a challenge for genetics, evolution, and development? Ann NY Acad Sci 981:1-6 [DOI] [PubMed] [Google Scholar]
- Van Dijk M, Mulders J, Poutsma A, Könst AA, Lachmeijer AM, Dekker GA, et al. (2005). Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat Genet 37:514-519 [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Tsuchida N, Shanmugam G. (2003). Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer 105:41-46 [DOI] [PubMed] [Google Scholar]
- von Zeidler SV, Miracca EC, Nagai MA, Birman EG. (2004). Hypermethylation of the p16 gene in normal oral mucosa of smokers. Int J Mol Med 14:807-811 [DOI] [PubMed] [Google Scholar]
- Vucic EA, Brown CJ, Lam WL. (2008). Epigenetics of cancer progression. Pharmacogenomics 9:215-234 [DOI] [PubMed] [Google Scholar]
- Weber B, Streseman C, Brueckner B, Lyko F. (2007). Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 6:10011005. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. (1997). Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335-340 [DOI] [PubMed] [Google Scholar]
- Yung RL, Julius A. (2008). Epigenetics, aging, and autoimmunity. Autoimmunity 41:329-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaina S, Lindholm MW, Lund G. (2005). Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? J Nutr 135:5-8 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. (2005). Ribo-gnome: the big world of small RNAs. Science 309:1519-1524 [DOI] [PubMed] [Google Scholar]
- Zochbauer-Muller S, Minna JD, Gazdar AF. (2002). Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist 7:451-457 [DOI] [PubMed] [Google Scholar]