Abstract
Human papillomaviruses (HPVs) are small dsDNA tumor viruses, which are the etiologic agents of most cervical cancers and are associated with a growing percentage of oropharyngeal cancers. The HPV capsid is non-enveloped, having a T=7 icosahedral symmetry formed via the interaction among 72 pentamers of the major capsid protein, L1. The minor capsid protein L2 associates with L1 pentamers, although it is not known if each L1 pentamer contains a single L2 protein. The HPV life cycle strictly adheres to the host cell differentiation program, and as such, native HPV virions are only produced in vivo or in organotypic “raft” culture. Research producing synthetic papillomavirus particles—such as virus-like particles (VLPs), papillomavirus-based gene transfer vectors, known as pseudovirions (PsV), and papillomavirus genome-containing quasivirions (QV)—has bypassed the need for stratifying and differentiating host tissue in viral assembly and has allowed for the rapid analysis of HPV infectivity pathways, transmission, immunogenicity, and viral structure.
Keywords: papillomavirus, assembly, structure, cancer, life cycle
Introduction
Over 200 human papillomavirus (HPV) types have been identified. All HPV types are epitheliotropic viruses that can be subdivided based on their ability to infect either mucosal or cutaneous keratinocytes. Mucosal-infecting HPV types can be further subdivided into low-risk and high-risk types. Low-risk HPV types lead to the development of benign neoplasms, such as warts and condyloma acuminatum, whereas high-risk HPV types can lead to malignant neoplasms, such as cervical cancer (de Villiers et al., 2004; Longworth and Laimins, 2004). A very strong association has been made between malignant progression and certain HPV types such as HPV16, HPV18, HPV31, HPV45, etc. (de Villiers et al., 2004; IARC, 2007; Longworth and Laimins, 2004). Infection from any of the high-risk HPV types is considered the most significant risk factor regarding the development of cervical cancer (Franquemont et al., 1989; Morrison et al., 1991; Madsen et al., 2008). In the case of infections of the oral cavity and/or oro-pharynx, HPV6, HPV11, HPV16, and HPV18 are common, with approximately 40-60% of head and neck cancers containing oncogenic HPV types such as HPV16 and HPV18 (zur Hausen, 1996; Herrero et al., 2003; Gillison, 2004; D’Souza et al., 2007; Anaya-Saavedra et al., 2008; Fakhry et al., 2008). While HPV16 is found in approximately 90% of HPV-containing head and neck cancers, several other high-risk HPV types have been detected in these lesions (Hoshikawa et al., 1990; Niedobitek et al., 1990; Ostwald et al., 1994; Shindoh et al., 1995; zur Hausen, 1996; Mellin et al., 2000; Venuti et al., 2000; Mork et al., 2001).
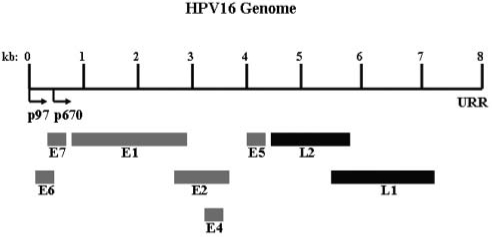
HPV virions are non-enveloped, consisting of dsDNA-containing, icosahedral capsids. HPV viral genomes are approximately 8 kb, circular, and are replicated within the nuclei of host cells. Within the capsids, viral genomes are associated with cellular histones, forming chromatin-like structures (Larsen et al., 1987; Doorbar, 2005). Viral genomes from all HPV types harbor an average of 8 open reading frames (ORFs), and these ORFs are expressed from polycistronic mRNAs which are transcribed from a single DNA strand (Favre et al., 1975; Zheng and Baker, 2006). The 8 ORFs can be subdivided into early and late genes, as well as a non-coding region called the upstream regulatory region (URR) (Fig. 1) (Zheng and Baker, 2006; Buck et al., 2008). The early (E) ORFs encode the E1, E2, E4, E5, E6, and E7 proteins (Doorbar et al., 1986; Zheng and Baker, 2006; Buck et al., 2008). E1 and E2 proteins have been shown to regulate viral replication as well as the coordinated expression of the other early viral genes (Cripe et al., 1987; Gloss and Bernard, 1990; Mohr et al., 1990; Meyers et al., 1992; Conger et al., 1999). The spliced E1^E4 protein may affect the replication of viral genomes through the inhibition of the G2-to-M transition, while also interacting with cellular keratin networks, causing their collapse, thus allowing mature virions to escape from the cornified envelope (White et al., 1999; Bryan and Brown, 2000; Brown et al., 2006; Knight et al., 2006; Mach et al., 2006; Gambhira et al., 2007). E5, E6, and E7 proteins that are coded by the high-risk HPVs are oncogenic, since they are able to transform and stimulate the growth of cells (Androphy et al., 1987; Phelps et al., 1988; Bedell et al., 1991). The late (L) ORFs encode the L1 and L2 proteins. L1 serves as the major capsid protein, while L2 serves as the minor capsid protein (Favre, 1975; Doorbar and Gallimore, 1987; Chen et al., 2000; Buck et al., 2008).
Figure 1.
Genomic organization of high-risk HPV16 in linear fashion. Early ORFs are indicated in grey, while late ORFs are indicated in black. The early (p97) and late (p670) promoters are designated by black arrows. The upstream regulatory region (URR) is depicted.
High-risk HPV types initiate the majority of transcription via 2 major viral promoters (Smith et al., 2007). The first, or early, promoter initiates upstream from the E6 ORF, and synthesizes transcripts that are translated early in the viral life cycle. Since the early promoter functions independently from differentiation, it can initiate prior to the productive phase of the viral life cycle. In HPV16 the early promoter is referred to as p97, in HPV31 the early promoter is referred to as p99, and in HPV18 it is referred to as p105 (Fig. 1) (Hirochika et al., 1987; Gloss and Bernard, 1990; Romanczuk et al., 1990; Kyo et al., 1995; Kanaya et al., 1997; Cumming et al., 2004; Smith et al., 2007). The second, or late, promoter is initiated in a differentiation-dependent manner, and thus is activated only when cells are grown in the host’s stratifying/differentiating tissue, or in vitro through the use of methylcellulose or organotypic culture techniques. Once activated, the late promoter directs transcription from a heterogeneous set of start sites which are clustered around nucleotide 742 (p742) in HPV31 (Ozbun and Meyers, 1998, 1999; Smith et al., 2007). A late promoter has been identified in HPV16 (p670), and evidence suggests that a late promoter exists in HPV18 as well (Hummel et al., 1992; Meyers et al., 1992; Grassmann et al., 1996; Parker et al., 1997). The late promoter specifically serves to produce a set of transcripts that facilitate the translation of L1 and L2 proteins.
The life cycle of HPV is strictly linked to the differentiation program of the host keratinocyte, whereby the assembly of mature virions is restricted to terminally differentiated suprabasal cells, limited in part by the differentiation-dependence of the late promoter (Bedell et al., 1991; Hummel et al., 1992; Meyers et al., 1992; Barksdale and Baker, 1993; Grassmann et al., 1996; Ozbun and Meyers, 1997; Smith et al., 2007). Initial infection by HPVs is thought to occur through micro-abrasions of the epithelial tissue, thus allowing entry of the HPV particle into cells of the basal layer (Belnap et al., 1996). Basal layer cells consist mainly of stem cells and transit-amplifying cells, and it is the epithelial stem cells that must be infected for a lesion to be maintained (Kaur and Li, 2000; Egawa, 2003). These cells continuously divide and replenish cells that are lost due to desquamation (Kaur and Li, 2000). Controversies exist as to the primary and/or secondary receptors for HPV entry, with alpha integrin and heparin sulfate as key possibilities, in addition to a role for laminin 5 (Giroglou et al., 2001; Yoon et al., 2001; Culp and Christensen, 2004; Doorbar, 2005; Patterson et al., 2005). Heparin sulfate appears important for adsorption and infection of a variety of HPV-typed VLPs, PsV, and native virions in non-host and transformed cell lines, whereas HPV31b organotypic culture-derived native virions (OTNV) do not require heparin sulfate for infection of HaCat or N/Tert-1 human keratinocytes, but do require it for infection of non-host and transformed cell lines (Giroglou et al., 2001; Yoon et al., 2001; Culp and Christensen, 2004; Doorbar, 2005; Patterson et al., 2005). Post-adsorption to the cell surface, internalization of virions has been shown to take many hours, and, depending on the papillomavirus type, virions can enter via clathrin-coated pits or caveolae (Fligge et al., 2001; Bousarghin et al., 2003; Day et al., 2003; Hindmarsh and Laimins, 2007; Smith et al., 2007). Recent studies involving entry pathways (e.g., clathrin- vs. caveolar-mediated endocytosis) and entry kinetics (e.g., few vs. many hours) of HPV have suffered due to a lack of consistency (Bousarghin et al., 2003; Culp and Christensen, 2004; Hindmarsh and Laimins, 2007; Smith et al., 2007; Laniosz et al., 2008). This may be due to the use of multiple types of synthetic papillomavirus particles and cell lines, in addition to the use of native virions, and cross-talk between clathrin and caveolar pathways (Fligge et al., 2001; Bousarghin et al., 2003; Hindmarsh and Laimins, 2007; Schelhaas et al., 2007; Laniosz et al., 2008). Recent discoveries with polyomaviruses suggest that controversies over papillomavirus entry may be from the initial usage of clathrin-mediated endocytosis and later exploitation of caveolar endocytic machinery within the cell (Bousarghin et al., 2003; Querbes et al., 2006; Laniosz et al., 2008).
While many details are lacking regarding the delivery of the viral genome into the nucleus, it is thought that the N-terminus of L2 is cleaved within the endosomal compartment via the cellular protease, furin, thus releasing an L2/genome complex into the cytosol (Day et al., 2003; Richards et al., 2006). The L2/genome complex may then interact with syntaxin 18, which ferries the complex to a perinuclear site (Bossis et al., 2005; Richards et al., 2006; Laniosz et al., 2008). L2 may then translocate the genome into the nucleus through its NLS (Fay et al., 2004; Bordeaux et al., 2006). Once in the nucleus, a sophisticated cascade of viral gene expression occurs which serves to maintain a minimum number of viral DNA copies per cell (Doorbar, 2005).
E1 and E2 are among the first early proteins that are expressed, assisting in the establishment of 20 to 100 episomal copies per basal cell (Mohr et al., 1990; Meyers et al., 1992; Conger et al., 1999; Smith et al., 2007). These 2 proteins form a complex with the viral origin of replication and recruit cellular polymerases and the necessary accessory proteins to facilitate replication (Mohr et al., 1990). E2 is a DNA-binding protein that recruits E1 to the origin of replication, but also serves to regulate transcription of E6 and E7 from the early promoter (Cripe et al., 1987; Gloss and Bernard, 1990; Mohr et al., 1990; Meyers et al., 1992). The E6 and E7 proteins from high-risk, but not low-risk, HPV types are oncoproteins. High-risk E6 is able to bind the tumor suppressor protein, p53, forming a trimeric complex along with the cellular ubiquitin ligase, E6AP. The ubiquitin ligase activity of E6AP leads to the rapid turnover of p53 (Rolfe et al., 1995). High-risk E7 is able to bind to and modulate a variety of proteins, such as the retinoblastoma (Rb) family of tumor suppressors, and cell-cycle regulatory proteins, leading to enhanced cellular replication (Dyson et al., 1989; Neary and DiMaio, 1989).
As HPV-infected basal cells divide, each new daughter cell is replete with a set of viral genomes as the genomes are equally partitioned during mitosis. Recent studies suggest that both Brd4-dependent and independent partitioning of viral genomes into daughter cells can occur, depending on the papillomavirus type studied (You et al., 2004; McPhillips et al., 2006). Post-mitosis, one cell remains attached to the basal layer, while the other cell is detached from the basal layer and begins to migrate up through the suprabasal layers (Lehman and Botchan, 1998; Bastien and McBride, 2000). During migration of the cell up through the strata, the cell begins a process of terminal differentiation (Fig. 2) (Pang et al., 1993). In non-HPV-infected epithelia, cells normally exit the cell cycle once they detach from the basal layer, and this is often accompanied by the loss of nuclei in differentiating suprabasal cells (Pang et al., 1993). In the case of HPV-infected epithelia, detached cells remain mitotically active due to the oncogenic properties of the E7 protein (Morris et al., 1993). Because of this, infected cells that have already differentiated can re-enter the S phase and enhance the expression of cellular replication factors that are required for concomitant viral genome amplification, and late gene expression (Bedell et al., 1991; Barksdale and Baker, 1993; Swindle et al., 1999; Banerjee et al., 2006). While designated ‘early proteins’, E1^E4 and E5 are both translated in a differentiation-dependent manner, both of which may affect the replication of viral genomes through the inhibition of G2-to-M transition, and stimulation of cell-cycle progression, respectively (Doorbar et al., 1997; White et al., 1999; Fehrmann et al., 2003; Knight et al., 2006). Similarly, the L1 and L2 capsid proteins are expressed only in terminally differentiated keratinocytes (Barksdale and Baker, 1993). Studies with both papillomaviruses and polyomaviruses suggest that, once expressed, capsid proteins assemble into icosahedral capsids via assistance from chaperone proteins (Buck et al., 2005; Chromy et al., 2003, 2006; Bird et al., 2008). It is unknown if encapsidation of the viral genome takes place during capsid assembly or after; however, encapsidation is assisted by L2 and may be facilitated by E2 proteins (Heino et al., 2000; Gu et al., 2004; Holmgren et al., 2005). In the cornified envelope, E1^E4 proteins are thought to interact with cellular keratin networks, causing their collapse, thus allowing mature virions to escape from the cornified cells (Doorbar et al., 1986; Bryan and Brown, 2000; Brown et al., 2006; Mach et al., 2006; Gambhira et al., 2007). Mechanistic details concerning the host cell differentiation program that leads to the productive phase of the HPV life cycle are unclear. It is also unclear as to the regulated molecular interactions that must take place for individual capsid proteins to assemble into higher-ordered structures over the time-course of stratification and differentiation of the host keratinocyte.
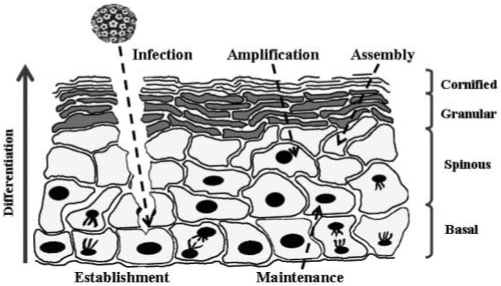
Figure 2.
HPV virions establish an infection in basal epithelial cells by gaining access through micro-abrasions within the tissue. Upon establishment, viral genomes are maintained at approximately 20 100-episomal copies per cell. Once maintained, viral genomes transit up through the differentiating strata as the mitotically active basal cells divide. In the suprabasal layers, concomitant late gene expression and viral genome amplification allow for assembly of progeny virions. Progeny virions escape with the highly keratinized cornified envelope, allowing the viral life cycle to continue.
To study aspects of the HPV life cycle, including HPV infectivity pathways, transmission, immunogenicity, and viral structure, recent research predominantly utilizes synthetic papillomavirus particles, such as virus-like particles (VLPs), papillomavirus-based gene transfer vectors, known as pseudovirions (PsV), and papillomavirus genome-containing quasivirions (QV), which bypasses the need for stratifying and differentiating tissue in the production of native HPV and facilitates rapid scientific discovery.
Structure
Background
HPV virions are non-enveloped, icosahedral particles approximately 50 to 60 nm in diameter (Hagensee et al., 1994; Chen et al., 2000; Modis et al., 2002). Each particle is composed of 72 pentamers of the L1 capsid protein. L1 pentamers are thought to form a network of intra- and interpentameric disulfide interactions which serve to stabilize the capsid (Sapp et al., 1998; Fligge et al., 2001; Buck et al., 2005; Kondo et al., 2007). In addition to L1, an unknown amount of the minor capsid protein, L2, exists within the virion. Previous cryoelectron microscopy studies of native bovine papillomavirus type 1 (BPV1) virions suggested that one L2 protein occludes the center of each pentavalent capsomere, for a total of 12 L2 proteins per virion (Trus et al., 1997). In support of this, SDS-PAGE analyses of L1/L2 VLPs have depicted a 30:1 ratio of L1 to L2, for a total of 12 L2 proteins per virion (Hagensee et al., 1993; Kirnbauer et al., 1993; Volpers et al., 1994; Bossis et al., 2005). However, SDS-PAGE analyses of native HPV1 virions, biochemical analyses of HPV11 L1/L2 proteins, and recent cryoelectron microscopy images of HPV16 pseudovirions (PsV) suggest that papillomavirus particles can contain up to 36 and as much as 72 L2 proteins per particle (Doorbar and Gallimore, 1987; Finnen et al., 2003; Buck et al., 2008). While L1 is sufficient to produce synthetic papillomavirus particles, L2 has been shown to affect the final structure of the virion, in addition to enhancing infectivity and DNA encapsidation of the particle (Kirnbauer et al., 1992, 1993; Holmgren et al., 2005; Kondo et al., 2007). Also housed within the virion is a histone-associated, 8-kb, circular viral genome which has been shown to alter the final structure of the virion (Sapp et al., 1998; Fligge et al., 2001). In addition to L2 and viral DNA, a minority population of unknown viral and/or cellular proteins/factors such as molecular chaperones and karyopherins may also exist which influence the final structure of native virions (Chromy et al., 2003, 2006; Bird et al., 2008).
Due to previous difficulties in propagating native virions in organotypic culture (i.e., technical constraints and low productivity), methods have been utilized to make synthetic virus in vitro (Kirnbauer et al., 1992, 1993; Gambhira et al., 2007). The most efficient synthetic techniques involve the overexpression of L1 with or without L2 via codon-optimized plasmids within monolayers of insect or mammalian cells, although less efficient techniques have also been developed to assemble virus-like particles (VLPs) and PsV in vitro (Kirnbauer et al., 1992; Fligge et al., 2001; Gambhira et al., 2007). L1 can spontaneously assemble into VLPs alone, or in tandem with L2. Expression plasmids or full-length HPV genomes can be incorporated into VLPs to produce PsV and quasivirions (QV), respectively (Buck et al., 2004; Culp and Christensen, 2004; Pyeon et al., 2005; Gambhira et al., 2007). Assembly within the reducing environment of the cytosol/lysates of these cells is insufficient to form the much more extensive disulfide bonding network that is formed in native virions (Sapp et al., 1998). In fact, overnight incubation of PsV allows for the formation of additional disulfide bonds in a redox-dependent and cellular lysate-dependent fashion (Buck et al., 2005). Similarly, stabilization of VLPs has been reported through the removal of reducing agents, lowering of pH, and increasing of ionic strength (Mach et al., 2006). These experiments suggest that differences in the redox states of monolayer vs. organotypic culture may play a role in the structural discrepancies noted between immature synthetic papillomavirus particles and native virions.
Structural studies of synthetic papillomavirus particles may or may not elucidate the native interactions that are necessary for the assembly of monomeric/pentameric L1. In fact, the crystal structure of an HPV16, 12-pentamer VLP lacks disulfide bonds altogether (Chen et al., 2000). The potential differences between synthetic and native virions stress the need for caution in the analysis of HPV structure and life-cycle strategies, and call for the development of novel techniques to produce native virions efficiently for comparative studies.
Production of Synthetic and Native Papillomavirus Particles
Virus-like Particles (VLPs)
VLPs have been produced by a variety of similar methods (Kirnbauer et al., 1992, 1993; Hagensee et al., 1993; Rose et al., 1993; Zhang et al., 1998; Chen et al., 2000; Querbes et al., 2006) (Fig. 3). Generally, expression plasmids or viral vectors that express L1 alone, or L1 plus L2, are transfected or used to infect the following eukaryotic or prokaryotic cell types: 293T, BSC-1, E. coli, yeast, or Sf9 cells (Kirnbauer et al., 1992, 1993; Hagensee et al., 1993, 1994; Rose et al., 1993; Belnap et al., 1996; Zhang et al., 1998; Chen et al., 2000; Querbes et al., 2006). Upon expression of L1, structures which retain the native T=7 icosahedral symmetry of the native virions are intracellularly self-assembled. VLPs are antigenically similar to native virions, suggesting gross structural similarities. In fact, VLPs can induce high titers of type-specific neutralizing antibodies, which made them good candidates for first-generation HPV vaccines (Rose et al., 1994; Belnap et al., 1996; Harper et al., 2004). While not necessary for the production of VLPs, one advance in VLP technology incorporates L2 into the particle, which has been suggested to increase their overall stability (Yang et al., 2005; Gambhira et al., 2007; Kondo et al., 2007). Novel findings into the cross-neutralization potential of the N-terminal peptide of L2 highlight the potential utility of L1/L2 chimeric VLPs in eliciting an optimized immune response (Gambhira et al., 2007; Kondo et al., 2007). Advancements in VLP technology have also increased the disulfide-bonding-mediated stabilization of VLPs via the manipulation of buffers, by removing reducing agents, lowering pH, and increasing ionic strength (Mach et al., 2006). Most novel VLP production techniques seek to produce VLPs which appear more like native virions.
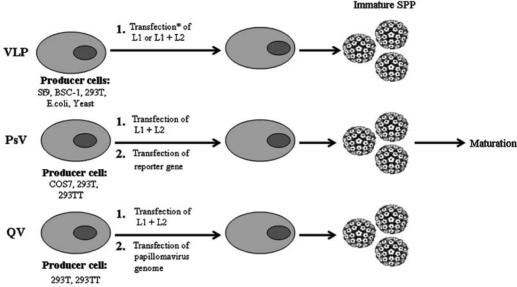
Figure 3.
Virus-like particles (VLPs), pseudovirions (PsV), and quasivirions (QV) are all synthetically derived papillomavirus particles which are made via transfection and/or infection-based techniques that overexpress L1 alone, or in tandem with L2. The differentiating factor among the various forms of synthetic papillomavirus particles is the presence and/or absence of plasmid-based reporter genes or full-length papillomavirus genomes. VLPs are empty, while PsV contain plasmid-based reporter genes, and QV contain full-length papillomavirus genomes. A maturation step has been characterized with PsV which make them appear more like native virions via TEM and biochemical assays. *Transfection is used interchangeably with co-transfections and infection, since all permutations of transfections, co-transfections, and infections with viral vectors have been utilized to produce synthetic papillomavirus particles.
Pseudovirions (PsV)
While many protocols exist, with various degrees of efficacy, to produce PsV, the most efficient PsV production protocol utilizes transfection of codon-optimized, L1- and L2-expression plasmids, in addition to reporter-gene-expressing plasmids into 293T or 293TT cells, which allows for efficient intracellular production of papillomavirus gene transfer vectors (Buck et al., 2005; Gambhira et al., 2007) (Fig. 3). PsV are self-assembled within the transfected producer cells, allowing for the production of several billion transducing units per milliliter when a 5.9-kb plasmid is encapsidated, although 10-fold fewer transducing units per milliliter are obtained when an 8-kb plasmid is encapsidated (Buck et al., 2004).
The utility of PsV is evidenced by the shear volume of recent publications that report on the utilization of PsV for structural and neutralization analyses, and studies of papillomavirus entry pathways (Fligge et al., 2001; Day et al., 2003; Gu et al., 2004; Buck et al., 2005, 2006, 2008; Yang et al., 2005; Gambhira et al., 2007; Kondo et al., 2007). In addition to the enhanced efficiency of PsV production/purification in comparison with native virions, PsV are structurally similar by cryoelectron microscopy, and have similar immunoreactive profiles against conformation-dependent neutralizing antibodies, suggesting that some, if not all, surface conformational epitopes are retained in PsV (Day et al., 2003; Embers et al., 2004; Yang et al., 2005; Gambhira et al., 2007; Buck et al., 2008).
While the efficient production of PsV allows for rapid analysis of many aspects of the papillomavirus life cycle, a thorough structural comparison between PsV and native virions is lacking; thus, experimental results should be verified with native virions (Sapp et al., 1998; Fligge et al., 2001; Buck et al., 2005). While the incorporation of plasmids into VLPs has been shown to enhance the amount of disulfide interactions that occur within each particle, it is apparent that further maturation events are required for further stabilization of PsV particles (Buck et al., 2005). Such stabilization, which has been depicted via TEM and through biochemical studies, would predict global conformational changes throughout the PsV which may lead to altered immunoreactivities with neutralizing antibodies. Maturation of PsV takes advantage of a simple overnight incubation at 37°C. This step further orders these particles so they appear more like native virions by TEM and cryoelectron microscopy, presumably by exposing these particles to an oxidizing environment for extended periods of time (Buck et al., 2005, 2008). At this point, it is unknown if the correct disulfide interactions are formed during PsV and VLP assembly/maturation. It is also unclear if additional factors such as the native, 8-kb, histone-associated genome, in addition to unknown cellular factors within stratified/differentiating tissue may further enhance the similarities among mature PsV, VLPs, and native virions.
Quasivirions (QV)
QV utilize technology similar to that of PsV, in that transfection of L1 and L2 codon-optimized expression plasmids, in addition to full-length, recircularized HPV genomes, into 293T or 293TT cells allows for efficient intracellular production of the native virion-like particles (Culp and Christensen, 2004; Pyeon et al., 2005; Smith et al., 2007) (Fig. 3). Optiprep-purified cottontail rabbit papillomavirus (CRPV) QV have been shown to have infectious titers similar to those of native virions in vitro and in vivo, are neutralizable by anti-L1 antibodies, and induce papilloma formation (Culp and Christensen, 2004; Pyeon et al., 2005). These attributes suggest that QV are similar, if not identical, to native virions. The most beneficial aspect of the production of QV is that it employs a simple DNA preparation and transfection protocol, utilizing a single 10-cm dish of 293T or 293TT cells; however, similar yields of QV and native virions can be obtained with the more labor-intensive, organotypic culture. While the QV technique is an enormous advance in papillomavirus research, it remains uncertain whether QV are structurally and functionally identical to native virions. Replication strategies of papillomavirus genomes, for instance, have revealed a differentiation-dependent switch from theta replication in monolayer culture to rolling-circle replication in stratifying and differentiating tissue (Flores and Lambert, 1997). Rolling-circle replication has been shown to be intimately involved in the formation of mature capsids in a variety of virus systems, and this possibility has yet to be explored in HPV (Koths and Dressler, 1980; Vlazny et al., 1982; Flores and Lambert, 1997). Until the details of QV and native virions have been studied comprehensively, it is premature to treat QV and native virions identically, and thus interpretation of future studies that utilize QV should consider the potential structural and functional differences.
Native Virions
Native virions are produced only in stratified and differentiated epithelia and are thus synthesized only during a natural infection or in organotypic culture (Meyers et al., 1992; Ozbun and Meyers, 1997; McLaughlin-Drubin et al., 2003, 2004; Gu et al., 2004; McLaughlin-Drubin and Meyers, 2005). While direct structural comparisons are lacking between organotypic culture-derived native virions (OTNV) and native virions derived from xenograft tissue or natural infection, the strict differentiation-dependence of OTNV highlights their novelty. For years, the ability to synthesize native virions in vitro was hindered due to the lack of a reproducible tissue culture system which allowed for virion morphogenesis in a differentiation-dependent manner. Our laboratory has established a system utilizing organotypic “raft” cultures which allows for the completion of the HPV life cycle and allows us to examine all aspects of this life cycle, including, but not limited to, viral DNA amplification, late gene expression, virion morphogenesis, and infectivity. We have successfully depicted propagation of HPV16, HPV18, HPV31, HPV33, HPV39, HPV45, HPV11, and a variety of chimeric and mutant OTNV (Meyers et al., 1992; Ozbun and Meyers, 1997; McLaughlin-Drubin et al., 2003, 2004; Gu et al., 2004; McLaughlin-Drubin and Meyers, 2004, 2005). Previously, the low productivity of organotypic culture garnered too few virions for many low-sensitivity analyses. However, advances in PCR, qPCR, mass spectroscopy, and purification techniques have afforded novel opportunities to examine aspects of the viral life cycle that were previously too difficult to explore.
Structural Details of Papillomavirus Particles
High-resolution Structural Analyses of Synthetic Capsids
Unfortunately, the low productivity of organotypic culture previously made obtaining enough OTNV impossible for high-resolution, crystallographic studies. The production of synthetic papillomavirus particles has bypassed this setback. However, VLPs composed of full-length, HPV16 L1 are unable to be crystallized. Because of this, VLPs containing N-terminally truncated forms of HPV16 L1, which can be crystallized, have provided the only atomic resolution structures of HPV particles (Chen et al., 2000). N-terminally truncated forms of L1 efficiently self-assemble into VLPs, as can full-length L1; however, deletion of 10 amino acids starting from the second N-terminal methionine leads to the assembly of T=1 icosahedral lattices made from 12 L1 pentamers, rather than the native T=7 icosahedral lattice made from 72 L1 pentamers (Fig. 4B) (Chen et al., 2000). Deleting fewer than 10 amino acids starting from the second N-terminal methionine retains the T=7 icosahedral lattice made from 72 L1 pentamers, but this native structure does not crystallize (Chen et al., 2000). As mentioned above, atomic resolution has been obtained only from the T=1, 12-pentamer HPV16 capsids (Chen et al., 2000). This structure is quite an accomplishment and allows our field to make further hypotheses regarding the structure of HPV; however, the ostensible small size of the T=1 particle and lack of disulfide bonds mean that this model may not be representative of all the detailed interactions that occur between and within L1 pentamers of native virions (Chen et al., 2000).
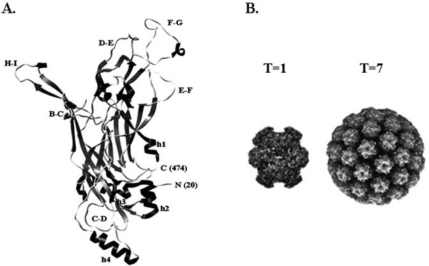
Figure 4.
Atomic structure of HPV16 major capsid protein L1. (A) Atomic resolution of monomeric HPV16 L1 from an N-terminally truncated form of L1. No disulfide bonds were formed in this particle. (B) Comparison of an HPV16, T=1, 12-pentamer particle composed of N-terminally truncated forms of L1, and a native, T=7, 72-pentamer, particle composed of full-length L1. Interpentameric and intrapenta-meric disulfide bonds were formed in this particle. Molecular modeling in (A) was rendered with Chimera (UCSF) (Chen et al., 2000). Structures in (B) were obtained from http://viperdb.scripps.edu (Shepherd et al., 2006).
Previous studies utilizing cryoelectron microscopy to resolve native BPV1 virions at a resolution of 9 A have depicted protein protrusions that emanate from the pentavalent capsomeres into each of 5 neighboring hexavalent capsomeres (Trus et al., 1997). These protrusions are morphologically similar to the C-terminal “arm-invading” model of polyomaviruses, which reinforce the invading arms via an interpentameric disulfide bridge(s) (Baker et al., 1989, 1991; Stehle et al., 1994, 1996; Sapp et al., 1998; Jao et al., 1999). While detailed information regarding the intra- and interpentameric molecular interactions between pentamers cannot be garnered from the current atomic structure of HPV16, assumptions can be made from this model. One such assumption is that if the T=1 particle expanded into a T=7 particle, the C-terminal h4 helix may reach into a neighboring pentamer, thus forming a critical interpentameric disulfide bridge between 2 Cys428 residues (Fig. 4A) (Chen et al., 2000). This is a rational assumption, since the C-terminal “invading arm” of SV40 is stabilized by an N-terminal disulfide bridge (Sapp et al., 1998; Jao et al., 1999; Schelhaas et al., 2007). However, analyses of recombinant VLPs suggest that a critical interpentameric disulfide bridge occurs between Cys428 and Cys175, and potentially Cys185 in HPV16, rather than between neighboring Cys428 residues (Sapp et al., 1998; Chen et al., 2000; Kondo et al., 2007). In addition, biochemical analyses assessing the disulfide bonding patterns of HPV virions utilized mass spectroscopy of VLP capsid proteins, coupled with cryoelectron microscopy image reconstructions of native virions, to determine which cysteine residue(s) is/are critical in the formation of interpentameric disulfide bonds (Modis et al., 2002). Briefly, HPV18 VLP L1 dimers were excised from a non-reducing polyacrylamide gel. L1 dimers were digested with trypsin and analyzed by liquid chromatography +/- tandem mass spectrometry (LC-MS/MS) (Modis et al., 2002). LC-MS/MS analysis of trypsin-digested HPV18 VLP L1 dimers revealed a tryptic product that corresponded to an 11-residue tryptic fragment that flanked the Cys428 equivalent of HPV18, Cys429 (Modis et al., 2002). The tryptic fragment contained a 476-Da adduct on Cys429, which suggested that a tryptic peptide corresponding to Cys175 was linked to the Cys429 tryptic fragment via a disulfide bond (Modis et al., 2002). These results are in contrast to the assumptions made from the atomic structure of the HPV16, T=1, 12-pentamer VLP, in that it seems that interpentameric interactions take place via Cys175 and Cys428 rather than the predicted Cys428 and Cys428. This result calls into question the accuracy to which VLPs assemble in vitro, or if the highly conserved cysteine residues Cys428, and Cys175 are arranged differently among various papillomavirus types. Because of the uncertain accuracy of VLP assembly in comparison with native virions, it is necessary that high-resolution studies of VLPs be coupled with the analysis of native virions.
Genetic and Biochemical Structural Analyses of Synthetic and Native Virions
Similar to the results obtained from current high-resolution structures of papillomavirus particles, which are unable to verify the exact disulfide bonding patterns necessary for intra- and interpentameric contacts, a variety of genetic and biochemical analyses has further complicated experimental structural analyses regarding the extent to which papillomavirus capsids are disulfide-bonded. Multiple reports have depicted PsV and native virions as having a much more extensively disulfide-bonded phenotype than VLPs, while matured PsV have a phenotype that mirrors native virions (Sapp et al., 1998; Fligge et al., 2001; Buck et al., 2005). Through the use of non-reducing SDS-PAGE analysis, it is apparent that L1 from HPV1 native virions obtained from a hand wart form extensively disulfide-bonded multimers which migrate above 170 kDa, whereas 50% of L1 from HPV16, HPV18, and HPV18 VLPs predominantly migrate as the 50-kDa monomeric protein (Sapp et al., 1998). Such multimerization of L1 from native virions seems to depend on components that make up the particle, in that the incorporation of nucleic acid alone into a VLP enhances the multimeric state of L1, based on similar non-reducing SDS-PAGE analyses of purified VLPs and PsV (Fligge et al., 2001). These results suggest that VLPs, which lack certain components of native virions, such as a complete 8-kb viral genome, are less-ordered structures. Incorporation of nucleic acid into papillomavirus particles appears to reinforce and/or facilitate the formation of additional disulfide bonds which are absent in synthetic papillomavirus particles.
Recent studies suggest that nucleic acid alone, encapsidated within an L1/L2 capsid, is not sufficient to create a mature papillomavirus particle; thus, previous PsV protocols created papillomavirus particles which lacked the ordered structure of native virions (Buck et al., 2004, 2005; Gambhira et al., 2007). Since previous PsV protocols produced particles which lacked the disulfide bonds present in native virions, a maturation step was added to the PsV production protocol (Buck et al., 2005). This maturation step allowed for increased disulfide bond formation, presumably since the virions were exposed to an oxidizing environment for an extended period of time. In comparison with immature PsV, mature PsV do appear more resistant to proteolysis and chemical reduction, while also obtaining a more ordered structure, as seen via TEM; however, it is unknown if the disulfide bonds formed during PsV maturation are formed as they would in vivo (Buck et al., 2005).
Genetic analyses of VLPs and PsV have also provided clues into the structural differences among the various papillomavirus particles, mostly by substituting the 12 highly conserved L1 cysteine residues for irrelevant amino acids. In one such example, a series of recombinant HPV16 VLPs, each containing a serine residue substituted for each of 12 conserved L1 cysteine residues, was produced and analyzed for their structural stabilities via their resistance to chemical reduction and tryptic proteolysis, in addition to their gross morphologic appearance via TEM (Kondo et al., 2007). Analysis of the recombinant VLPs suggested that Cys428, Cys175, and Cys185 are pivotal residues, with Cys428Ser mutations leading to the complete dissociation of VLPs, Cys175Ser mutations leading to the production of large tube-like capsid structures, and Cys185Ser mutations leading to the production of small, approximately 40-nm capsids (Kondo et al., 2007). Analysis of these data suggests a prominent role for Cys428 in the formation of interpentameric disulfide bonds, and reinforces previous assumptions made from the crystal structure of the HPV16, T=1 12-pentamer VLP (Chen et al., 2000; Kondo et al., 2007). These results contrast, however, with previous work using recombinant HPV33 VLPs, which depicted equivalent roles for Cys427 and Cys176 (i.e., the equivalent of Cys428 and Cys175 in HPV16) in the formation of interpentameric disulfide bonds, which supports previous claims that pentamers from HPV18 VLPs form interpentameric disulfide bonds with each other (Sapp et al., 1998; Modis et al., 2002). These results call into question whether disulfide-bonding patterns differ between HPV16 VLPs, and HPV18 and HPV33 VLPs, or whether the various techniques utilized to produce synthetic papillomavirus particles lead to disulfide interactions that do not normally occur.
To complicate the structural analysis of papillomavirus particles further, recent studies have also suggested that HPV16 PsV containing the Cys428Ser mutation, which led to the complete dissociation of VLPs, retains residual infectivity. This suggests that PsV assembly can occur despite the loss of Cys428, which is thought to be critical in the formation of interpentameric disulfide bonds (Buck et al., 2005). In this same study, HPV16 PsV which contain the Cys175Ser mutation retain 70-90% infectivity, suggesting that this residue does not form interpentameric disulfide linkages with Cys428 in HPV16 PsV (Buck et al., 2005). How these interpentameric interactions form in native virions has not been examined.
Structural Alterations Affecting Function
Many studies have detailed the viral components that comprise the papillomavirus capsid (L1, L2, viral DNA, and histones), in addition to viral and cellular components (i.e., E2, E4, E5, E7, molecular chaperones, karyopherins, etc.) that may either directly or indirectly affect the structure of the capsid (Doorbar and Gallimore, 1987; Larsen et al., 1987; Heino et al., 2000; Fligge et al., 2001; Chromy et al., 2003, 2006; Gu et al., 2004; Bordeaux et al., 2006; Richards et al., 2006; Kondo et al., 2007; Bird et al., 2008; Buck et al., 2008). Many other unknown viral and/or cellular factors are undoubtedly necessary for the proper assembly of native virions regarding regulation of redox states within the cell, proper cellular localization and initial interaction of capsid proteins, the correct formation/regulation of disulfide bonds, and regulation of capsid protein expression. Advances in synthetic papillomavirus particle technology have yielded information regarding the structural roles of various components, and as such, these synthetic technologies have adapted over time as investigators attempt to create a more native-like viral particle.
Incorporation of histone-associated plasmids, for example, has been shown to increase the infectivity of PsV 4- to 5-fold over PsV that contain histone-free plasmids (Fligge et al., 2001). This increase in infectivity suggests that the presence of histones alone may alter the overall structure of PsV, rendering them more infectious, hindering post-entry endonuclease digestion of viral genomes, or facilitating early transcriptional events within the host cell. Earlier discoveries that the incorporation of plasmid DNA into VLPs increases the multimeric state of L1 support the idea that histones can alter the final conformation of the papillomavirus particle (Sapp et al., 1998; Fligge et al., 2001). Cryoelectron microscopy images of native BPV1 virions further support a role for chromatin in the final structure of native virions, in that the nucleohistone core was shown to have a thin link to the inner walls of hexavalent capsomere protrusions, and between capsomeres (Trus et al., 1997). In either case, it is clear that the use of synthetic papillomavirus particles leads to many questions regarding the authenticity of the particles being tested.
Incorporation of L2 into VLPs also ostensibly alters the conformation of the virus particle. In vitro studies of VLP assembly under reducing conditions have depicted an important role for L2 in the initial assembly of L1/L2 aggregates (Kondo et al., 2007). L2 was also able to aggregate mutant HPV16 L1 proteins that lacked the critical interpentameric disulfide-bond-forming Cys428. L1/L2 aggregates, presumably, are the nucleation site for further capsid assembly, suggesting that L2 plays an important role in initial, regulated assembly of VLPs. Further linking the importance of L2 in native virion assembly is its ability to bind both L1 and DNA, suggesting that L2 is the mediator that facilitates assembly (Fligge et al., 2001; Fay et al., 2004; Bordeaux et al., 2006). Further, the analysis of in vitro assembly of L1/L2 VLPs has shown that L2 is incorporated into VLPs only when co-expressed with L1, or with L1 capsomeres (Kondo et al., 2007). L2 will not bind to intact VLPs, which suggests that L2 plays an early nucleation role in HPV assembly.
While the importance of L2 remains convincing in VLP assembly, and in binding assays, growth of HPV31 native virions containing 2 artificially inserted internal stop codons, which effectively shut down synthesis of L2 in organotypic culture, yields some infectious virus (Holmgren et al., 2005). Although the L2-deficient native virions were more than 100-fold less infectious than their wild-type counterparts, such important roles ascribed to L2 via VLP- and PsV-dependent experiments—such as initial capsid assembly, DNA encapsidation, capsid disassembly, and ferrying DNA into the nucleus—unearth additional questions as to the redundancy of native virions. Are there other viral and/or cellular proteins that exist within stratifying/differentiating epithelia that can assist L2 in the above activities, and if so, how do these proteins interact with L1-only virions to produce an infectious virus? These same experiments utilizing L2-deficient native virions depicted a role for L2 in DNA encapsidation and/or structural stability of the virions, since fewer DNase-resistant genomes were detected in the L2-deficient samples when compared with the wild-type native virions (Holmgren et al., 2005). Over the years, many publications have cited the appearance of viral and/or cellular factors that co-purify with native virions, such as E2, E4, E5, E7, and histones (Doorbar and Gallimore, 1987; Larsen et al., 1987; Heino et al., 2000; Fligge et al., 2001; Chromy et al., 2003, 2006; Gu et al., 2004; Bordeaux et al., 2006; Richards et al., 2006; Kondoet al., 2007; Bird et al., 2008; Buck et al., 2008).
A recent report has suggested that structural alterations of HPV16 PsV dramatically decrease infectivity of these particles (Ishii et al., 2007). Specifically, the addition of a variety of thiol-reactive reagents such as DTNB and NEM alkylate free C146, C225, and C229 sulfhydryls of L1 (Ishii et al., 2007). Alkylation of these sulfhydryls leads to marked decreases in infectivity, while C146A, C225A, and C229A substitutions did not appreciably alter infectivity (Ishii et al., 2007). The authors suggest that either local structural changes perturb entry of these particles, or that the free C146, C225, and C229 sulfhydryls interact with an as-of-yet unknown cellular protein involved in uncoating, since the loss of disulfide bonding via dithiothreotol or 2-mercaptoethanol-mediated reduction of disulfide bonds within PsV does not affect infectivity (Buck et al., 2006; Ishii et al., 2007). In either case, these results highlight the functional discrepancies that may arise given subtle conformational changes in the viral capsid.
Not only have structural alterations of HPV particles affected infectivity, but also they have altered the particle’s function as an immunogen (Kirnbauer et al., 1992; Breitburd et al., 1995; White et al., 1999; Christensen et al., 2001; Yang et al., 2005). Specifically, global conformational changes in VLPs can change the immunoreactivity profile against a panel of conformation-dependent neutralizing monoclonal antibodies (MAbs) (White et al., 1999; Christensen et al., 2001). For example, VLPs with a F50L substitution are not bound by the conformation-dependent MAbs, H16.V5 and H16.E70, while they are bound by MAbs that recognize surface linear epitopes (Christensen et al., 2001). Further, vaccination of mice with HPV16 VLPs containing the assembly-incompetent D223G, N327S, and F446S L1 substitutions failed to induce neutralizing antibodies, while HPV16 VLPs containing the assembly-competent D202H substitution led to the activation of bone-marrow-derived dendritic cells (BMDCs) and a potent L1-specific humoral response, but also failed to induce neutralizing antibodies. Analysis of these data implies that structural alteration of VLPs can change the cellular immune response to the immunogen (Yang et al., 2005). The elucidation of the correct structure of HPV is important for optimization of an HPV immunogen that will raise optimum neutralizing antibody titers, and an optimum cellular immune response against native HPV capsid structures.
Concluding Remarks
The efficiency by which synthetic papillomavirus particles are produced is invaluable in the efforts to uncover details regarding HPV structure, infectivity, transmission, and immunogenicity, and the protocols developed utilizing synthetic papillomavirus particles will be invaluable in future studies of native virions. Novel techniques continue to develop to construct synthetic papillomavirus particles with ever-increasing complexity, which seems to tighten the structural gap between synthetic and native virions. This being the case, detailed side-by-side comparisons of the structural aspects between these groups of particles are necessary for confident interpretation of the data.
Acknowledgments
We thank the Meyers’ lab for critical reading of the manuscript. M.J.C.
Footnotes
C.M. are supported by a PHS Grant from the NIAID R01AI57988.
References
- Anaya-Saavedra G, Ramirez-Amador V, Irigoyen-Camacho ME, Garcia-Cuellar CM, Guido-Jimenez M, Mendez-Martinez R, et al. (2008). High association of human papillomavirus infection with oral cancer: a case-control study. Arch Med Res 39:189-197 [DOI] [PubMed] [Google Scholar]
- Androphy EJ, Hubbert NL, Schiller JT, Lowy DR. (1987). Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J 6:989-992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. (1989). The capsid of small papova viruses contains 72 pentameric capsomeres: direct evidence from cryo-electron-microscopy of simian virus 40. Biophys J 55:243-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. (1991). Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 60:1445-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee NS, Genovese NJ, Noya F, Chien WM, Broker TR, Chow LT. (2006). Conditionally activated E7 proteins of high-risk and low-risk human papillomaviruses induce S phase in postmitotic, differentiated human keratinocytes. J Virol 80:6517-6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barksdale SK, Baker CC. (1993). Differentiation-specific expression from the bovine papillomavirus type 1 P2443 and late promoters. J Virol 67:5605-5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, McBride AA. (2000). Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134 [DOI] [PubMed] [Google Scholar]
- Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, et al. (1991). Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol 65:2254-2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, et al. (1996). Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol 259:249-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G, O’Donnell M, Moroianu J, Garcea RL. (2008). A possible role for cellular karyopherins in regulating polyomavirus and papillomavirus capsid assembly. J Virol 82:9848-9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeaux J, Forte S, Harding E, Darshan MS, Klucevsek K, Moroianu J. (2006). The l2 minor capsid protein of low-risk human papillomavirus type 11 interacts with host nuclear import receptors and viral DNA. J Virol 80:8259-8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis I, Roden RB, Gambhira R, Yang R, Tagaya M, Howley PM, et al. (2005). Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J Virol 79:6723-6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousarghin L, Touze A, Sizaret PY, Coursaget P. (2003). Human papillomavirus types 16, 31, and 58 use different endocytosis pathways to enter cells. J Virol 77:3846-3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitburd F, Kirnbauer R, Hubbert NL, Nonnenmacher B, Trin-Dinh-Desmarquet C, Orth G, et al. (1995). Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J Virol 69:3959-3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Kitchin D, Qadadri B, Neptune N, Batteiger T, Ermel A. (2006). The human papillomavirus type 11 E1–E4 protein is a transglutaminase 3 substrate and induces abnormalities of the cornified cell envelope. Virology 345:290-298 [DOI] [PubMed] [Google Scholar]
- Bryan JT, Brown DR. (2000). Association of the human papillomavirus type 11 E1()E4 protein with cornified cell envelopes derived from infected genital epithelium. Virology 277:262-269 [DOI] [PubMed] [Google Scholar]
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. (2004). Efficient intracellular assembly of papillomaviral vectors. J Virol 78(2):751-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. (2005). Maturation of papillomavirus capsids. J Virol 79:2839-2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Thompson CD, Roberts JN, Muller M, Lowy DR, Schiller JT. (2006). Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog 2(7):e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, et al. (2008). Arrangement of L2 within the papillomavirus capsid. J Virol 82:5190-5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. (2000). Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell 5:557-567 [DOI] [PubMed] [Google Scholar]
- Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, et al. (2001). Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324-334 [DOI] [PubMed] [Google Scholar]
- Chromy LR, Pipas JM, Garcea RL. (2003). Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc Natl Acad Sci USA 100:10477-10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy LR, Oltman A, Estes PA, Garcea RL. (2006). Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J Virol 80:5086-5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conger KL, Liu JS, Kuo SR, Chow LT, Wang TS. (1999). Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J Biol Chem 274:2696-2705 [DOI] [PubMed] [Google Scholar]
- Cripe TP, Haugen TH, Turk JP, Tabatabai F, Schmid PG, 3rd, Durst M, et al. (1987). Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J 6:3745-3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp TD, Christensen ND. (2004). Kinetics of in vitro adsorption and entry of papillomavirus virions. Virology 319:152-161 [DOI] [PubMed] [Google Scholar]
- Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. (2004). Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279:21749-21758 [DOI] [PubMed] [Google Scholar]
- D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. (2007). Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 356:1944-1956 [DOI] [PubMed] [Google Scholar]
- Day PM, Lowy DR, Schiller JT. (2003). Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1-11 [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. (2004). Classification of papillomaviruses. Virology 324:17-27 [DOI] [PubMed] [Google Scholar]
- Doorbar J. (2005). The papillomavirus life cycle. J Clin Virol 32(Suppl 1):S7-S15 [DOI] [PubMed] [Google Scholar]
- Doorbar J, Gallimore PH. (1987). Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J Virol 61:2793-2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Campbell D, Grand RJ, Gallimore PH. (1986). Identification of the human papilloma virus-1a E4 gene products. EMBO J 5:355-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Foo C, Coleman N, Medcalf L, Hartley O, Prospero T, et al. (1997). Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52 [DOI] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. (1989). The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937 [DOI] [PubMed] [Google Scholar]
- Egawa K. (2003). Do human papillomaviruses target epidermal stem cells? Dermatology 207:251-254 [DOI] [PubMed] [Google Scholar]
- Embers ME, Budgeon LR, Culp TD, Reed CA, Pickel MD, Christensen ND. (2004). Differential antibody responses to a distinct region of human papillomavirus minor capsid proteins. Vaccine 22:670-680 [DOI] [PubMed] [Google Scholar]
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. (2008). Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100:261-269 [DOI] [PubMed] [Google Scholar]
- Favre M. (1975). Structural polypeptides of rabbit, bovine, and human papillomaviruses. J Virol 15:1239-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre M, Orth G, Croissant O, Yaniv M. (1975). Human papillomavirus DNA: physical map. Proc Natl Acad Sci USA 72:4810-4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Yutzy WH, 4th, Roden RB, Moroianu J. (2004). The positively charged termini of L2 minor capsid protein required for bovine papillomavirus infection function separately in nuclear import and DNA binding. J Virol 78:13447-13454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann F, Klumpp DJ, Laimins LA. (2003). Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J Virol 77:2819-2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnen RL, Erickson KD, Chen XS, Garcea RL. (2003). Interactions between papillomavirus L1 and L2 capsid proteins. J Virol 77:4818-4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fligge C, Schafer F, Selinka HC, Sapp C, Sapp M. (2001). DNA-induced structural changes in the papillomavirus capsid. J Virol 75:7727-7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Lambert PF. (1997). Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol 71:7167-7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquemont DW, Ward BE, Andersen WA, Crum CP. (1989). Prediction of ‘high-risk’ cervical papillomavirus infection by biopsy morphology. Am J Clin Pathol 92:577-582 [DOI] [PubMed] [Google Scholar]
- Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, et al. (2007). A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol 81:13927-13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison ML. (2004). Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol 31:744-754 [DOI] [PubMed] [Google Scholar]
- Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. (2001). Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565-1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss B, Bernard HU. (1990). The E6/E7 promoter of human papillomavirus type 16 is activated in the absence of E2 proteins by a sequence-aberrant Sp1 distal element. J Virol 64:5577-5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. (1996). Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol 70:2339-2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Li M, Zhao WM, Fang NX, Bu S, Frazer IH, et al. (2004). tRNASer(CGA) differentially regulates expression of wild-type and codon-modified papillomavirus L1 genes. Nucleic Acids Res 32:4448-4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagensee ME, Yaegashi N, Galloway DA. (1993). Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol 67:315-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagensee ME, Olson NH, Baker TS, Galloway DA. (1994). Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J Virol 68:4503-4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper DM, Franco EL, Wheeler C, Ferris DG, Jenkins D, Schuind A, et al. (2004). Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 364:1757-1765 [DOI] [PubMed] [Google Scholar]
- Heino P, Zhou J, Lambert PF. (2000). Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304-314 [DOI] [PubMed] [Google Scholar]
- Herrero R, Castellsague X, Pawlita M, Lissowska J, Kee F, Balaram P, et al. (2003). Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J Natl Cancer Inst 95:1772-1783 [DOI] [PubMed] [Google Scholar]
- Hindmarsh PL, Laimins LA. (2007). Mechanisms regulating expression of the HPV 31 L1 and L2 capsid proteins and pseudovirion entry. Virol J 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Broker TR, Chow LT. (1987). Enhancers and trans-acting E2 transcriptional factors of papillomaviruses. J Virol 61:2599-2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. (2005). The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol 79:3938-3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa T, Nakajima T, Uhara H, Gotoh M, Shimosato Y, Tsutsumi K, et al. (1990). Detection of human papillomavirus DNA in laryngeal squamous cell carcinomas by polymerase chain reaction. Laryngoscope 100:647-650 [DOI] [PubMed] [Google Scholar]
- Hummel M, Hudson JB, Laimins LA. (1992). Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol 66:6070-6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (2007). Human papillomaviruses. In: IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Geneva, Switzerland: WHO Press, pp. 45-177 [Google Scholar]
- Ishii Y, Kondo K, Matsumoto T, Tanaka K, Shinkai-Ouchi F, Hagiwara K, et al. (2007). Thiol-reactive reagents inhibits [sic] intracellular trafficking of human papillomavirus type 16 pseudovirions by binding to cysteine residues of major capsid protein L1. Virol J 4:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CC, Weidman MK, Perez AR, Gharakhanian E. (1999). Cys9, Cys104 and Cys207 of simian virus 40 Vp1 are essential for inter-pentamer disulfide-linkage and stabilization in cell-free lysates. J Gen Virol 80(Pt 9):2481-2489 [DOI] [PubMed] [Google Scholar]
- Kanaya T, Kyo S, Laimins LA. (1997). The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology 237:159-169 [DOI] [PubMed] [Google Scholar]
- Kaur P, Li A. (2000). Adhesive properties of human basal epidermal cells: an analysis of keratinocyte stem cells, transit amplifying cells, and postmitotic differentiating cells. J Invest Dermatol 114:413-420 [DOI] [PubMed] [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. (1992). Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA 89:12180-12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, et al. (1993). Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol 67:6929-6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight GL, Turnell AS, Roberts S. (2006). Role for Wee1 in inhibition of G2-to-M transition through the cooperation of distinct human papillomavirus type 1 E4 proteins. J Virol 80:7416-7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Ishii Y, Ochi H, Matsumoto T, Yoshikawa H, Kanda T. (2007). Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology 358:266-272 [DOI] [PubMed] [Google Scholar]
- Koths K, Dressler D. (1980). The rolling circle.capsid complex as an intermediate in phi X DNA replication and viral assembly. J Biol Chem 255:4328-4338 [PubMed] [Google Scholar]
- Kyo S, Tam A, Laimins LA. (1995). Transcriptional activity of human papillomavirus type 31b enhancer is regulated through synergistic interaction of AP1 with two novel cellular factors. Virology 211:184-197 [DOI] [PubMed] [Google Scholar]
- Laniosz V, Holthusen KA, Meneses PI. (2008). Bovine papillomavirus type 1: from clathrin to caveolin. J Virol 82:6288-6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PM, Storgaard L, Fey SJ. (1987). Proteins present in bovine papillomavirus particles. J Virol 61:3596-3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman CW, Botchan MR. (1998). Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA 95:4338-4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. (2004). Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev 68:362-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach H, Volkin DB, Troutman RD, Wang B, Luo Z, Jansen KU, et al. (2006). Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J Pharm Sci 95:2195-2206 [DOI] [PubMed] [Google Scholar]
- Madsen BS, Jensen HL, van den Brule AJ, Wohlfahrt J, Frisch M. (2008). Risk factors for invasive squamous cell carcinoma of the vulva and vagina—population-based case-control study in Denmark. Int J Cancer 122:2827-2834 [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. (2004). Evidence for the coexistence of two genital HPV types within the same host cell in vitro. Virology 321:173-180 [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. (2005). Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med 119:171-186 [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Wilson S, Mullikin B, Suzich J, Meyers C. (2003). Human papillomavirus type 45 propagation, infection, and neutralization. Virology 312:1-7 [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. (2004). Propagation, infection, and neutralization of authentic HPV16 virus. Virology 322:213-219 [DOI] [PubMed] [Google Scholar]
- McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. (2006). Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol 80:9530-9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin H, Friesland S, Lewensohn R, Dalianis T, Munck-Wikland E. (2000). Human papillomavirus (HPV) DNA in tonsillar cancer: clinical correlates, risk of relapse, and survival. Int J Cancer 89:300-304 [PubMed] [Google Scholar]
- Meyers C, Frattini MG, Hudson JB, Laimins LA. (1992). Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973 [DOI] [PubMed] [Google Scholar]
- Modis Y, Trus BL, Harrison SC. (2002). Atomic model of the papillomavirus capsid. EMBO J 21:4754-4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr IJ, Clark R, Sun S, Androphy EJ, MacPherson P, Botchan MR. (1990). Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699 [DOI] [PubMed] [Google Scholar]
- Mork J, Lie AK, Glattre E, Hallmans G, Jellum E, Koskela P, et al. (2001). Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med 344:1125-1131 [DOI] [PubMed] [Google Scholar]
- Morris JD, Crook T, Bandara LR, Davies R, LaThangue NB, Vousden KH. (1993). Human papillomavirus type 16 E7 regulates E2F and contributes to mitogenic signalling. Oncogene 8:893-898 [PubMed] [Google Scholar]
- Morrison EA, Ho GY, Vermund SH, Goldberg GL, Kadish AS, Kelley KF, et al. (1991). Human papillomavirus infection and other risk factors for cervical neoplasia: a case-control study. Int J Cancer 49:6-13 [DOI] [PubMed] [Google Scholar]
- Neary K, DiMaio D. (1989). Open reading frames E6 and E7 of bovine papillomavirus type 1 are both required for full transformation of mouse C127 cells. J Virol 63:259-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G, Pitteroff S, Herbst H, Shepherd P, Finn T, Anagnostopoulos I, et al. (1990). Detection of human papillomavirus type 16 DNA in carcinomas of the palatine tonsil. J Clin Pathol 43:918-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostwald C, Muller P, Barten M, Rutsatz K, Sonnenburg M, Milde-Langosch K, et al. (1994). Human papillomavirus DNA in oral squamous cell carcinomas and normal mucosa. J Oral Pathol Med 23:220-225 [DOI] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. (1997). Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol 71:5161-5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. (1998). Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol 72:2715-2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA, Meyers C. (1999). Two novel promoters in the upstream regulatory region of human papillomavirus type 31b are negatively regulated by epithelial differentiation. J Virol 73:3505-3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YY, Schermer A, Yu J, Sun TT. (1993). Suprabasal change and subsequent formation of disulfide-stabilized homo- and hetero-dimers of keratins during esophageal epithelial differentiation. J Cell Sci 104(Pt 3):727-740 [DOI] [PubMed] [Google Scholar]
- Parker JN, Zhao W, Askins KJ, Broker TR, Chow LT. (1997). Mutational analyses of differentiation-dependent human papillomavirus type 18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ 8:751-762 [PubMed] [Google Scholar]
- Patterson NA, Smith JL, Ozbun MA. (2005). Human papillomavirus type 31b infection of human keratinocytes does not require heparan sulfate. J Virol 79:6838-6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps WC, Yee CL, Münger K, Howley PM. (1988). The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547 [DOI] [PubMed] [Google Scholar]
- Pyeon D, Lambert PF, Ahlquist P. (2005). Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc Natl Acad Sci USA 102:9311-9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes W, O’Hara BA, Williams G, Atwood WJ. (2006). Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol 80:9402-9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RM, Lowy DR, Schiller JT, Day PM. (2006). Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci USA 103:1522-1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe M, Beer-Romero P, Glass S, Eckstein J, Berdo I, Theodoras A, et al. (1995). Reconstitution of p53-ubiquitinylation reactions from purified components: the role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP). Proc Natl Acad Sci USA 92:3264-3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanczuk H, Thierry F, Howley PM. (1990). Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol 64:2849-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RC, Bonnez W, Reichman RC, Garcea RL. (1993). Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol 67:1936-1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RC, Reichman RC, Bonnez W. (1994). Human papillomavirus (HPV) type 11 recombinant virus-like particles induce the formation of neutralizing antibodies and detect HPV-specific antibodies in human sera. J Gen Virol 75(Pt 8):2075-2079 [DOI] [PubMed] [Google Scholar]
- Sapp M, Fligge C, Petzak I, Harris JR, Streeck RE. (1998). Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J Virol 72:6186-6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmström J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, et al. (2007). Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131: 516-529 [DOI] [PubMed] [Google Scholar]
- Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, et al. (2006). VIPERdb: a relational database for structural virology. Nucleic Acids Res 34:386-389 (database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindoh M, Chiba I, Yasuda M, Saito T, Funaoka K, Kohgo T, et al. (1995). Detection of human papillomavirus DNA sequences in oral squamous cell carcinomas and their relation to p53 and proliferating cell nuclear antigen expression. Cancer 76:1513-1521 [DOI] [PubMed] [Google Scholar]
- Smith JL, Campos SK, Ozbun MA. (2007). Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J Virol 81:9922-9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, Harrison SC. (1994). Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160-163 [DOI] [PubMed] [Google Scholar]
- Stehle T, Gamblin SJ, Yan Y, Harrison SC. (1996). The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165-182 [DOI] [PubMed] [Google Scholar]
- Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. (1999). Human papillomavirus DNA replication compartments in a transient DNA replication system. J Virol 73:1001-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, Booy FP. (1997). Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol 4:413-420 [DOI] [PubMed] [Google Scholar]
- Venuti A, Manni V, Morello R, De Marco F, Marzetti F, Marcante ML. (2000). Physical state and expression of human papillomavirus in laryngeal carcinoma and surrounding normal mucosa. J Med Virol 60:396-402 [DOI] [PubMed] [Google Scholar]
- Vlazny DA, Kwong A, Frenkel N. (1982). Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA 79:1423-1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpers C, Schirmacher P, Streeck RE, Sapp M. (1994). Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology 200:504-512 [DOI] [PubMed] [Google Scholar]
- White WI, Wilson SD, Palmer-Hill FJ, Woods RM, Ghim SJ, Hewitt LA, et al. (1999). Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J Virol 73:4882-4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wheeler CM, Chen X, Uematsu S, Takeda K, Akira S, et al. (2005). Papillomavirus capsid mutation to escape dendritic cell-dependent innate immunity in cervical cancer. J Virol 79:6741-6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon CS, Kim KD, Park SN, Cheong SW. (2001). alpha(6) Integrin is the main receptor of human papillomavirus type 16 VLP. Biochem Biophys Res Commun 283:668-673 [DOI] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. (2004). Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360 [DOI] [PubMed] [Google Scholar]
- Zhang W, Carmichael J, Ferguson J, Inglis S, Ashrafian H, Stanley M. (1998). Expression of human papillomavirus type 16 L1 protein in Escherichia coli: denaturation, renaturation, and self-assembly of virus-like particles in vitro. Virology 243:423-431 [DOI] [PubMed] [Google Scholar]
- Zheng ZM, Baker CC. (2006). Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 11:2286-2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. (1996). Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta 1288:F55-F78 [DOI] [PubMed] [Google Scholar]