Abstract
In the Drosophila ovary, bone morphogenetic protein (BMP) ligands maintain germline stem cells (GSCs) in an undifferentiated state. The activation of the BMP pathway within GSCs results in the transcriptional repression of the differentiation factor bag of marbles (bam). The Nanos-Pumilio translational repressor complex and the miRNA pathway also help to promote GSC self-renewal. How the activities of different transcriptional and translational regulators are coordinated to keep the GSC in an undifferentiated state remains uncertain. Data presented here show that Mei-P26 cell-autonomously regulates GSC maintenance in addition to its previously described role of promoting germline cyst development. Within undifferentiated germ cells, Mei-P26 associates with miRNA pathway components and represses the translation of a shared target mRNA, suggesting that Mei-P26 can enhance miRNA-mediated silencing in specific contexts. In addition, disruption of mei-P26 compromises BMP signaling, resulting in the inappropriate expression of bam in germ cells immediately adjacent to the cap cell niche. Loss of mei-P26 results in premature translation of the BMP antagonist Brat in germline stem cells. These data suggest that Mei-P26 has distinct functions in the ovary and participates in regulating the fates of both GSCs and their differentiating daughters.
Keywords: Germline, Repression, Stem cells, Translation, Tumor, Drosophila
INTRODUCTION
Understanding the mechanisms that control the balance between stem cell self-renewal and differentiation is one of the fundamental goals of stem cell biology. This balance often depends on the coordinated regulation of complex transcriptional and post-transcriptional hierarchies. The germline stem cells of the Drosophila ovary and testis have proven to be powerful platforms for studying gene regulation during the transition from a stem cell state to a differentiated state (Fuller and Spradling, 2007).
The germline stem cells (GSCs) of the Drosophila ovary reside within a well-characterized cellular niche, which includes cap cells that produce the bone morphogenetic protein (BMP) ligands Decapentaplegic (Dpp) and Glass bottom boat (Gbb) (Wharton et al., 1991; Xie and Spradling, 1998; Song et al., 2004). Dpp and Gbb activate heterodimeric BMP receptors, which include the type I receptors Thickveins (Tkv) and Saxophone (Sax) and the type II receptor Punt, in GSCs (Xie and Spradling, 1998; Iovino et al., 2009), leading to the phosphorylation of Mothers against dpp (Mad). Phosphorylated Mad (pMad) partners with Medea and together these proteins translocate into the nucleus and directly repress the transcription of bag of marbles (bam), a gene that is both necessary and sufficient for germline differentiation (McKearin and Ohlstein, 1995; Ohlstein and McKearin, 1997; Hudson et al., 1998; Chen and McKearin, 2003b; Chen and McKearin, 2003a; Song et al., 2004). When a GSC divides, one of the resulting daughter cells is typically displaced away from the cap cell niche. This cell, called a cystoblast, expresses bam and begins to differentiate into a multicellular cyst.
In addition to transcriptional regulation by the BMP pathway, the maintenance of GSCs also depends on at least two intrinsically acting translational repressors: the Nanos-Pumilio complex and the microRNA (miRNA) pathway. Disruption of either nanos or pumilio leads to GSC loss (Forbes and Lehmann, 1998; Gilboa and Lehmann, 2004; Wang and Lin, 2004; Chen and McKearin, 2005; Szakmary et al., 2005). Clonal experiments demonstrate that GSCs depend on nanos function throughout adult life for their maintenance (Wang and Lin, 2004). Several efforts have been made to identify mRNA targets of Nanos and Pumilio in specific tissues (Gerber et al., 2006). Within embryos, Nanos and Pumilio partner with the TRIM-NHL domain protein Brain tumor (Brat) and repress the translation of hunchback mRNA (Irish et al., 1989; Barker et al., 1992; Murata and Wharton, 1995; Wreden et al., 1997; Zamore et al., 1997; Wharton et al., 1998; Sonoda and Wharton, 1999; Sonoda and Wharton, 2001). By contrast, within GSCs brat mRNA itself appears to be a target of Nanos and Pumilio (Harris et al., 2011). Once Nanos levels drop within the cystoblast, Brat expression increases and represses the translation of Mad and dMyc (Diminutive – FlyBase), thereby dampening the responsiveness of these germline cells to BMP signals from the niche. Misexpression of brat results in a GSC loss phenotype (Harris et al., 2011).
The miRNA pathway also promotes Drosophila GSC self-renewal. miRNAs processed by the RNase Dicer-1 block the expression of specific targets by destabilizing transcripts or preventing their translation (Guo et al., 2010; Smibert and Lai, 2008). In Drosophila, Dicer-1 functions together with the double-stranded RNA-binding protein Loquacious (Loqs) to process miRNAs into their mature form (Forstemann et al., 2005; Saito et al., 2005; Liu et al., 2007). Once processed, miRNAs serve as guides for members of the Argonaute family of proteins by binding, through imperfect base pairing, to elements within the 3′UTR of target RNAs (Czech and Hannon, 2011). In Drosophila, Ago1 pairs with the conserved protein GW182 (Gawky – FlyBase) to form the functional miRNA-induced silencing complex (miRISC) (Eulalio et al., 2008; Chekulaeva et al., 2009). Mutations in Dicer-1, loqs and Ago1 lead to GSC loss (Forstemann et al., 2005; Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007), whereas overexpression of Ago1 results in an expansion of the GSC population (Yang et al., 2007). These findings indicate that the miRNA pathway promotes GSC self-renewal and that its activity must be attenuated before differentiation can proceed.
In differentiating germline cysts, a second TRIM-NHL domain protein called Mei-P26 antagonizes the miRNA pathway (Neumuller et al., 2008). mei-P26 mutant ovaries display a germline tumor phenotype marked by the accumulation of undifferentiated multicellular cysts (Page et al., 2000; Neumuller et al., 2008). Mei-P26 physically associates with Ago1 in S2 cells and loss of mei-P26 results in increased levels of miRNAs within the ovary (Neumuller et al., 2008). The mechanism by which Mei-P26 disrupts the miRNA pathway remains unknown.
In contrast to the role that Mei-P26 plays in germline cysts, several TRIM-NHL domain proteins in other organisms promote miRNA-dependent translational silencing. For example, NHL-2, a close C. elegans ortholog of Mei-P26, enhances the silencing of several miRNA targets including hbl-1 and let-60 (Ras) (Hammell et al., 2009). Further biochemical experiments demonstrated that NHL-2 physically interacts with the DEAD-box protein CGH-1 and core components of the C. elegans miRISC (Hammell et al., 2009). In mice, the related TRIM32 protein also associates with Ago1 and increases the activity of specific miRISCs (Schwamborn et al., 2009). These findings suggest that different TRIM-NHL domain proteins have evolutionarily distinct functions or that there are other activities of Mei-P26 that have not yet been described.
Using a series of genetic and biochemical experiments, we find that Mei-P26 promotes stem cell self-renewal and has additional functions beyond negatively regulating the miRNA pathway. These unexpected results suggest that Mei-P26 carries out at least two distinct functions in the ovary.
MATERIALS AND METHODS
Drosophila strains
mei-P26mfs1 and mei-P26mfs2 were gifts from R. S. Hawley (Page et al., 2000) and w1118;UASp-mei-P26 lines were gifts from J. Knoblich (Neumuller et al., 2008). Ago114, P{FRT(w[hs])}G13/CyO was kindly provided by D. Chen (Yang et al., 2007). Hsp83-lacZ-orb3′UTR was a gift from P. Schedl (Tan et al., 2001). w1118;bamP-Bam::HA/CyO;bgcnP-bgcn::GFP/TM3 (Li et al., 2009) and the bamΔ86 and bgcn1 alleles (Ohlstein and McKearin, 1997; Lavoie et al., 1999) have been described previously. P{neoFRT}18A, P{FRT(w[hs])}G13 and P{neoFRT}42D, hs-FLP stocks were obtained from the Bloomington Drosophila Stock Center.
Standard FLP/FRT-mediated recombination was used to generate mei-P26 and Ago1 mutant germline clones. Females were placed on wet yeast 2 days prior to clone induction. These females were subjected to 1-hour heat shocks at 37°C twice per day for 3 consecutive days.
Immunohistochemistry
Ovaries were processed (McKearin and Ohlstein, 1995) and imaged on a Zeiss LSM 510 confocal microscope, SP5 upright Leica confocal microscope or a Zeiss ApoTome. Primary antibodies included rabbit anti-GFP (1:500; Invitrogen), mouse anti-GFP (1:500; Abcam), goat anti-Vasa (1:200; Santa Cruz Biotechnology), rabbit anti-Ago1 (1:200; Abcam) and rat anti-HA (1:500; Roche). Mouse anti-Bam A7 (1:50), mouse anti-Hts 1B1 (1:20), mouse anti-Orb (1:10) and mouse anti-Sxl (1:10) were obtained from the Developmental Studies Hybridoma Bank. Rabbit anti-Mei-P26 antibody was kindly provided by J. Knoblich (Neumuller et al., 2008) and P. Lasko (Liu et al., 2009). Additional antibodies include rabbit anti-Nanos (1:1000; a gift from A. Nakamura, RIKEN Center for Developmental Biology, Kobe, Japan), guinea pig anti-GW182 [1:1000; a gift from E. Izaurralde (Eulalio et al., 2008)], rabbit anti-Brat [1:200; a gift from J. Knoblich (Betschinger et al., 2006)], rabbit anti-Bruno [1:5000; a gift from M. Lilly (Sugimura and Lilly, 2006)], rabbit polyclonal anti-Rbp9 (1:5000) (Kim-Ha et al., 1999), guinea pig anti-A2bp1 (1:5000) (Tastan et al., 2010), mouse anti-β-galactosidase (1:1000; Promega) and rabbit anti-pSmad (1:10; Cell Signaling Technology). Secondary antibodies were conjugated to Alexa 568, Alexa 488, FITC, Cy3 or Cy5 (1:500; Molecular Probes and Jackson ImmunoResearch).
Immunoprecipitation from ovaries
For immunoprecipitation (IP) experiments, 100 pairs of wild-type ovaries or 200 pairs of bamΔ86 or c587-gal4>UAS-dpp ovaries were extracted using lysis buffer (50 mM Hepes pH 7.5, 250 mM NaCl, 0.1% Nonidet P40, 0.2% Triton X-100). A protease inhibitor mixture (Roche) was added to the lysate. The extract was spun in a microcentrifuge at maximum speed (16,100 g) for 10 minutes at 4°C. The supernatant was added to 10 μl Protein G Sepharose 4 Fast Flow beads (GE Healthcare). After a 1-hour incubation at 4°C, the beads were pelleted at 8000 g and the supernatant collected.
Antibodies were then added to the supernatant. Rabbit anti-Mei-P26 (Liu et al., 2009) and rabbit anti-Ago1 (Abcam) were added at 1:200 and rabbit anti-Myc (Abcam) was added at 1:1000. Ovary lysate and antibody were mixed and incubated at 4°C for 2 hours. For Mei-P26 IP with peptide competition, the same peptide (NLKTVLSDDASNSSVLED) used for antibody production (Neumuller et al., 2008) was synthesized by Covance and added to the lysate at a final concentration of 100 ng/μl. Protein G Sepharose 4 Fast Flow beads (10 μl) were added and incubated with the lysate and the corresponding antibody overnight at 4°C. The beads were washed four times in lysis buffer and an equal volume of protein loading buffer was added, boiled for 5 minutes, loaded on an SDS-PAGE gel, and analyzed by western blotting. The loads in all experiments represent 10% of the lysate used for the IP.
Immunoblots were probed using anti-Myc (1:10,000; Abcam), anti-Ago1 (1:2000), anti-GW182 (1:10,000), anti-Brat (1:1000), anti-Actin (1:100; Developmental Studies Hybridoma Bank) and anti-Mei-P26 (1:1000). Secondary antibodies included goat anti-rat HRP, goat anti-rabbit HRP, goat anti-mouse and goat anti-guinea pig (1:3000; Bio-Rad).
Immunoprecipitation RT-PCR
IPs used in combination with RT-PCR were performed using RT-PCR lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Nonidet P40, 5 mM EDTA, 5 mM DTT). Protease inhibitor mixture (Roche) and RNase inhibitor (Roche) were added to the lysate. Protein IPs were performed as described above. 0.5 μl RNase inhibitor (40 U/μl) and 0.5 μl RNase-free DNase (1 U/μl) (Promega) were added to the final beads and incubated at 37°C for 10 minutes. The DNase was inactivated by incubating the beads at 75°C for 10 minutes. Specific primers were used to detect orb 3′UTR and Actin 5C 3′UTR (supplementary material Table S1).
The RT-PCR reactions were performed in two steps. Reverse transcription (RT) was carried out using 2 μl 5×reaction buffer, 2 μl template, 1 μl 10mM dNTPs, 1 μl reverse primer, 0.25 μl RNase inhibitor (40 U/μl), 0.25 μl reverse transcriptase (20 U/μl) (Roche). Total RNA (100 ng) isolated with Trizol (Invitrogen) from whole ovaries was used as the load template. For IP samples, 2 μl beads were used as template. The RT reaction was incubated at 50°C for 1 hour. PCR using Taq (Roche) was performed on 1 μl of RT product according to the manufacturer’s instructions. Different numbers of cycles (24-28) were performed and the resulting products were run on 1% agarose gels and visualized by ethidium bromide staining.
Quantitative (qPCR) was also used to quantify the RT product. A CHROMO4 continuous fluorescence detector (Bio-Rad CH001234) was used to perform qPCR and Opticon Monitor 4 software was used for analysis. qPCR was run at 95°C for 10 minutes, then 95°C for 15 seconds and 60°C for 1 minute for 40 cycles. Each reaction contained 1 μl DNA from the RT product, 0.4 μl 10 μM primer, 10 μl master mix (Power SYBR Green PCR Master Mix, Applied Biosystems) and 8.2 μl distilled water. Each IP sample was analyzed in triplicate. The primers used for qPCR are listed in supplementary material Table S1.
To generate a standard curve, 1 μl of RT product from the input sample or the appropriate cDNA (1 ng/μl) was diluted 1:1, 1:4, 1:16, 1:64, 1:256 and 1 μl from each dilution used as template in a qPCR reaction, performed in triplicate. The Opticon Monitor 4 program calculated the standard curve automatically. The relative amounts of mRNA in the IP pellet or control IP pellet were calculated based on this standard curve. We then determined the ratio of mRNA in the IP pellet versus control IP pellet.
Reporter constructs
Reporters were generated to compare the expression pattern of full-length orb 3′UTR (FL-orb3′UTR) with that of orb 3′UTR containing mutated miRNA binding sites (Δmir-orb3′UTR) (supplementary material Fig. S2). The vasa promoter (Sano et al., 2002) was amplified from genomic DNA and fused with venus sequence (Nagai et al., 2002) using splicing by overlapping extension PCR (SOE-PCR). The product was cloned into a pCasper vector (Drosophila Genomics Resource Center, Indiana University, Bloomington, IN, USA). Mutations in miRNA binding sites were generated using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) or by SOE-PCR. Primers used to introduce these mutations are listed in supplementary material Table S1. After making mutations, orb 3′UTR sequence was cloned into KpnI and NotI sites downstream of the vasa promoter and venus sequence in the pCasper vector. The reporter constructs were injected into flies (Rainbow Transgenic Flies) and multiple resulting lines carrying each transgene were dissected and immunostained.
For the quantification of Venus signal in specific cell types, we analyzed confocal images using Amira software. Within one germarium, we obtained the average signal strength for both a stem cell and a 16-cell cyst. The ratio of the signal strength in these two regions was then calculated for at least 11 germaria for each sample.
RESULTS
Mei-P26 functions in female GSCs
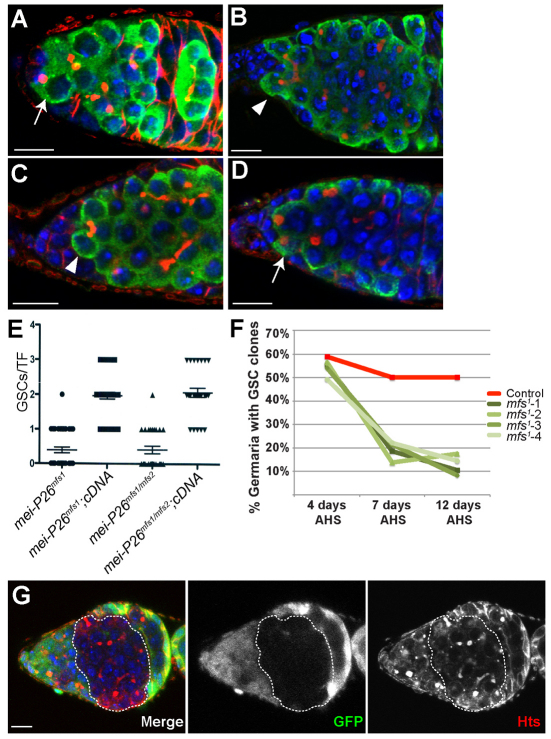
Our interest in the mechanisms that promote germline cyst development prompted us to further characterize the function of Mei-P26. As a first step, we stained control and mei-P26 mutant ovaries for the germ cell marker Vasa and the fusome marker Hts (monoclonal antibody 1B1) (Fig. 1A-C). Fusomes are specialized germline organelles and changes in fusome morphology that can be used to trace germline differentiation (de Cuevas and Spradling, 1998). Control ovaries exhibited the expected progression of germline development: GSCs immediately adjacent to cap cells contained small round fusomes, which became branched as the germ cells left the GSC niche and initiated cyst development. mei-P26 mutant ovaries contained germline cystic tumors as previously reported (Page et al., 2000; Neumuller et al., 2008). Surprisingly, however, mei-P26mfs1 homozygotes and mei-P26mfs1/mei-P26mfs2 transheterozygotes often contained multicellular cysts immediately adjacent to the cap cells (Fig. 1B,C). Moreover, single cells with round fusomes immediately adjacent to the cap cells were scarce within these mei-P26 mutant ovaries, suggesting that disruption of mei-P26 results in a GSC loss phenotype. Quantification of these observations revealed that the number of GSC-like cells with single round fusomes was reduced in mei-P26 mutants relative to controls, dropping from an average of two in wild-type samples to 0.5 in mei-P26 mutants (Fig. 1E). A chromosomal duplication encompassing the mei-P26 gene or a mei-P26 full-length cDNA transgene restored the fertility of mei-P26 mutants and rescued the GSC loss phenotype (Fig. 1D,E).
Fig. 1.
Disruption of mei-P26 results in GSC loss. (A-D) Germaria stained for Vasa (green), Hts (red) and DNA (blue). (A) Wild-type germaria contain two to three germline stem cells (GSCs) (arrow) marked by small round fusomes. mei-P26mfs1 homozygotes (B) and mei-P26mfs1/mei-P26mfs2 transheterozygotes (C) often have multicellular germline cysts, marked by branched fusomes (arrowheads), adjacent to the cap cells. (D) mei-P26mfs1 homozygotes carrying a duplication that contains the mei-P26 gene exhibit the normal number of GSCs (arrow). (E) Quantification of the mei-P26 mutant phenotype. Genotypes are displayed on the x-axis. The y-axis lists the number of GSCs per terminal filament (TF). Each point signifies a TF counted (mei-P26mfs1, n=41; mei-P26mfs1 cDNA, n=35; mei-P26mfs1/mfs2, n=27; mei-P26mfs1/mfs2 cDNA, n=27). A full-length wild-type mei-P26 cDNA transgene driven by a nanos-gal4::VP16 germline driver rescued the mei-P26 phenotype. Error bars indicate s.e.m. (F) Quantification of clonal analysis using the mei-P26mfs1 allele. Germline clones of a control chromosome (red line) and four independent recombinant mei-P26mfs1 mutant chromosomes (green lines) were induced using FRT/FLP-mediated mitotic recombination. The number of GSC clones was assayed over three time points (n≥100 germaria/sample/time point). Although nearly 50% of the germaria counted contained control clones, the number of clones observed for mei-P26 mutant chromosomes rapidly decreased over time, indicating that disruption of mei-P26 results in a stem cell loss phenotype. (G) A mei-P26mfs1 clonal germarium stained for GFP (green), Hts (red) and DNA (blue). A cystic tumor clone is outlined. Scale bars: 10 μm.
To test whether disruption of mei-P26 results in a GSC loss phenotype, we employed FRT/FLP-mediated recombination and compared the retention of mei-P26 mutant GSCs with that of control GSCs at multiple time points (Fig. 1F). Four independent mei-P26mfs1 recombinant chromosomes (mfs1-1 to mfs1-4) were tested. This analysis revealed that mei-P26 mutant GSC clones were rapidly lost from the cap cell niche over time. Four days after clone induction, nearly 50% of both the control and mei-P26 mutant germaria contained negatively marked homozygous GSC clones. The number of control GSC clones remained high during the course of the experiment. By contrast, the total number of mei-P26 mutant GSC clones rapidly decreased, so that 7 days after clone induction only 20% of the mei-P26 ovarioles contained homozygous negatively marked GSC clones (Fig. 1F,G). This number continued to decrease 12 days after clone induction. Once the mei-P26 mutant clones moved away from the cap cells, they formed tumorous multicellular cysts (Fig. 1G). We never observed transient mei-P26 mutant germline clones giving rise to normal egg chambers, suggesting that the mei-P26 phenotypes in GSCs and differentiating cysts are independent from one another. These data provide further evidence that disruption of mei-P26 leads to a GSC loss phenotype and that mei-P26 acts in a cell-autonomous manner to promote both GSC self-renewal and GSC daughter differentiation.
Mei-P26 associates with components of the miRISC in GSCs
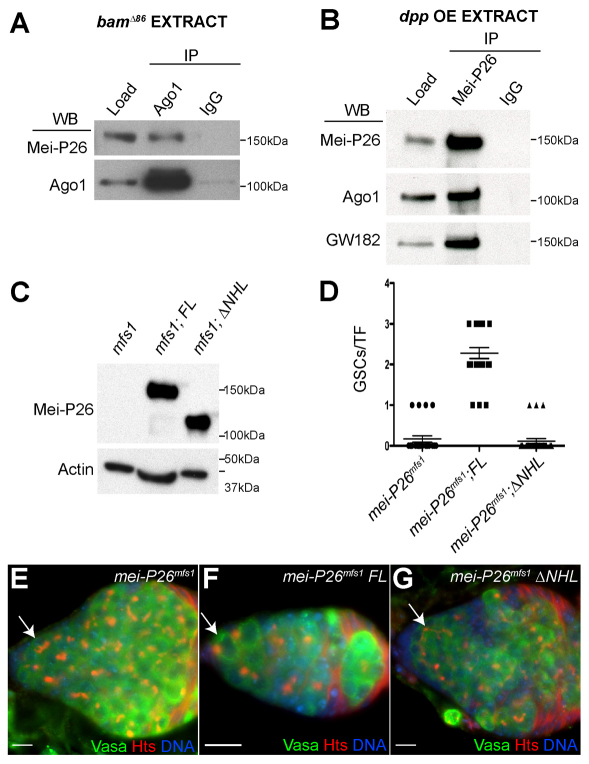
Genetic data indicate that components of the miRNA pathway (Jin and Xie, 2007; Park et al., 2007; Yang et al., 2007) and mei-P26 (this paper) are essential for GSC maintenance. A previous study had shown that Mei-P26 associates with Ago1 in S2 cells (Neumuller et al., 2008). Co-staining in wild-type ovaries showed that GSCs express Mei-P26 as well as two components of the miRISC: Ago1 and GW182 (supplementary material Fig. S1A,B). Mei-P26 co-immunoprecipitated with Ago1 from wild-type ovarian extracts (supplementary material Fig. S1C). This interaction did not depend on the presence of RNA and was disrupted by a Mei-P26 blocking peptide (supplementary material Fig. S1D-F).
To test whether the in vivo Mei-P26 and Ago1 interactions detected by immunoprecipitation (IP) occurred only within mature ovarian cells and not in GSCs, co-IP experiments were repeated using two different sources of undifferentiated stem cell-like cells: bam mutant ovaries and dpp-overexpressing (dpp OE) ovaries (Fig. 2A,B). Previous efforts showed that dpp driven by c587-gal4 (c587-gal4>UAS-dpp) results in the formation of GSC tumors, which have many of the characteristics of bona fide GSCs within the normal cap cell niche (Song et al., 2004). Using these genetic backgrounds, the resulting extracts contained material from GSC-like cells and not from differentiating cysts. Mei-P26 and Ago1 interacted in ovary extracts from both bam mutants and those overexpressing dpp (Fig. 2A,B). Moreover, Mei-P26 physically interacted with GW182 in these extracts (Fig. 2B), further supporting the notion that Mei-P26 associates with functional miRISC in undifferentiated germ cells. Interaction with Ago1 requires the Mei-P26 NHL domain (Neumuller et al., 2008). If the interaction between Mei-P26 and Ago1 was functionally significant in GSCs, a transgene lacking the NHL domain should not rescue the mei-P26 GSC loss phenotype. To test this, equivalent amounts of a full-length mei-P26 and a truncated version lacking the NHL domain (ΔNHL) were expressed in a mei-P26 mutant background using a nanos-gal4 driver (Fig. 2C). The full-length transgene rescued the mei-P26 GSC loss phenotype but the ΔNHL construct did not (Fig. 2D-G).
Fig. 2.
Mei-P26 associates with miRISC components in undifferentiated germ cells. (A) Co-immunoprecipitation (co-IP) from bamΔ86 mutant ovarian extracts using an anti-Ago1 antibody. The resulting pellets were analyzed by western blot using antibodies against Mei-P26 and Ago1. (B) co-IP from c587-gal4;UAS-dpp (dpp overexpressing, OE) ovarian extracts using an anti-Mei-P26 antibody. The resulting pellets were analyzed by western blot using antibodies against Mei-P26, Ago1 and GW182. (C) Western blot comparing levels of Mei-P26 protein in mei-P26mfs1, mei-P26mfs1;nanos-gal4::VP16>UAS-full-length mei-P26 (FL) and mei-P26mfs1;nanos-gal4::VP16>UAS-mei-P26ΔNHL (ΔNHL) ovaries. Actin served as a loading control. (D) Quantification of the mei-P26 mutant phenotype. Genotypes are displayed on the x-axis. The y-axis lists the number of GSCs per TF. Each point signifies a TF counted (mei-P26mfs1, n=24; mei-P26mfs1 cDNA, n=25; mei-P26mfs1 ΔNHL, n=26). A full-length wild-type mei-P26 cDNA transgene driven by a nanos-gal4::VP16 germline driver rescued the mei-P26 GSC loss phenotype but a transgene lacking the NHL domain did not. Error bars indicate s.e.m. (E-G) mei-P26mfs1 (E), FL (F) and ΔNHL (G) ovaries stained for Vasa (green), Hts (red) and DNA (blue). The arrows point to where GSCs would normally reside. Both mei-P26mfs1 and ΔNHL ovaries often had multicellular cysts at the anterior of the germarium, whereas FL ovaries appeared to be rescued. Scale bars: 10 μm.
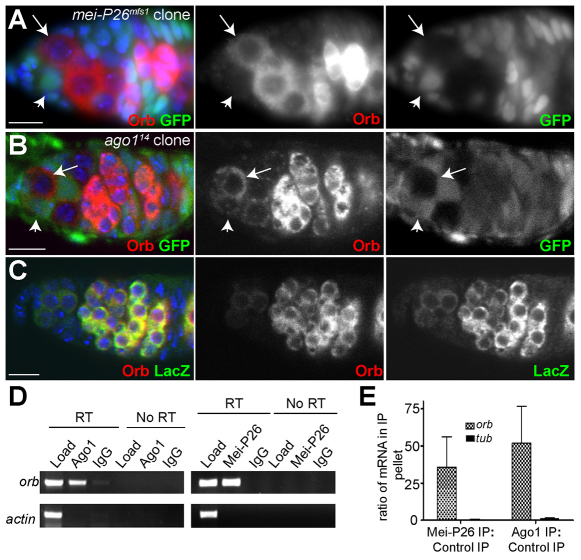
To test whether Mei-P26 and miRISC function together, we sought to identify an mRNA that served as a common target for Mei-P26 and miRISC. We screened for proteins showing increased expression in GSCs in the absence of either mei-P26 or Ago1 by staining mutant germline clones with a small number of publicly available antibodies directed against proteins with known functions during Drosophila oogenesis, including A2bp1, Bruno (Arrest – FlyBase), Rbp9 and Nanos. One potential target identified using this approach was oo18 RNA binding protein (orb), which encodes a CPEB-like protein that has multiple roles in germline development (Lantz et al., 1992; Christerson and McKearin, 1994; Lantz et al., 1994). Comparing mutant GSC clones with their GFP-positive heterozygous GSC neighbors showed that Orb protein levels increased in mei-P26 or Ago1 mutant clones, suggesting that Mei-P26 and Ago1 both negatively regulate the expression of this protein in GSCs (Fig. 3A,B). The 3′UTR of orb mRNA contains predicted binding sites for several miRNAs, including miR-190, miR-280 and miR-4/miR-8. Consistent with previous reports (Lantz et al., 1994), Orb protein expression closely matched the expression of an orb 3′UTR reporter driven by the broadly expressed Hsp83 promoter (Tan et al., 2001) (Fig. 3C). These results suggest that orb mRNA might be a shared target of Mei-P26 and miRISC.
Fig. 3.
Ago1 and Mei-P26 regulate orb translation in GSCs. (A) mei-P26mfs1 clonal germarium stained for GFP (green), Orb (red) and DNA (blue). The negatively marked mei-P26 mutant GSC (arrow) expresses higher levels of Orb protein than its heterozygous neighbor (arrowhead). (B) Ago114 clonal germarium stained for GFP (green), Orb (red) and DNA (blue). The negatively marked Ago1 mutant GSC (arrow) expresses higher levels of Orb protein than its heterozygous neighbor (arrowhead). (C) A germarium expressing an Hsp83-lacZ-orb3′UTR reporter stained for lacZ (green), Orb (red) and DNA (blue). (D) RT-PCR of orb and Actin 5C mRNA from Ago1 and Mei-P26 IPs. orb mRNA was consistently enriched in the Ago1 and Mei-P26 IP pellets but not in the IgG control pellets. (E) The ratio of mRNA pulled down in a Mei-P26 IP versus a control IP and in an Ago1 IP versus a control IP as measured by real-time PCR. Error bars indicate s.d. Scale bars: 10 μm.
Mei-P26 and Ago1 associate with orb mRNA
If Ago1 and Mei-P26 directly repress Orb translation, both proteins would be predicted to associate with orb mRNA. To test for such interactions, we performed Ago1 and Mei-P26 IPs from ovarian extracts and assayed the resulting pellets for the presence of orb mRNA by RT-PCR. orb RNA was enriched in the Ago1 IP relative to the IgG control pellet (Fig. 3D). Similar results were obtained with a Mei-P26 IP (Fig. 3D). To provide a quantitative assessment of the amount of orb mRNA that associated with Ago1 and Mei-P26, we performed real-time PCR of the resulting IP pellets (Fig. 3E). These experiments consistently showed that Ago1 and Mei-P26 associate with orb mRNA.
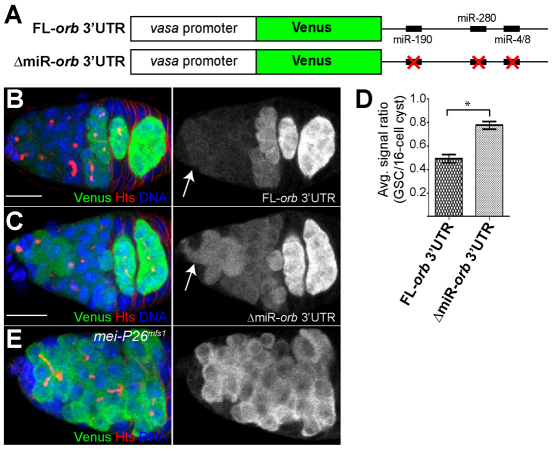
Next, we created two Venus (a GFP variant) orb 3′UTR reporter constructs under the control of the vasa promoter: one with an intact full-length orb 3′UTR and another with mutations in the seed regions of four predicted miRNA binding sites (Fig. 4A, supplementary material Fig. S2). The reporter carrying the intact orb 3′UTR (FL-orb3′UTR) was expressed in a pattern that was similar to both Orb protein and the aforementioned independent lacZ reporter construct (Fig. 4B, Fig. 3C). However, mutations in the miR-190, miR-280 and miR-4/8 sites (Δmir-orb3′UTR) resulted in increased reporter expression within GSCs (Fig. 4C). Quantification of Venus fluorescence in GSCs relative to 16-cell cysts showed a modest but reproducible increase in reporter expression in GSCs when the predicted miRNA binding sites were disrupted (Fig. 4D). In addition, the expression of the FL-orb3′UTR reporter increased in mei-P26 mutant germ cells (Fig. 4E), showing that this 3′UTR reporter responds to changes in Mei-P26 levels. In contrast to wild-type samples (Fig. 4B,C), mei-P26 mutant germline cells did not exhibit a second peak of reporter expression in 16-cell cysts within the posterior of the germarium (Fig. 4E) because they fail to reach these developmental stages (Tastan et al., 2010).
Fig. 4.
The 3′UTR of orb mRNA contains miRNA binding sites and responds to changes in Mei-P26 levels. (A) The reporter genes used to evaluate the importance of predicted miRNA binding sites within the 3′UTR of orb mRNA. (B,C) Full-length orb 3′UTR (FL-orb3′UTR) (B) and an orb 3′UTR reporter that contains mutated miRNA binding sites (ΔmiR-orb3′UTR) (C) stained for Venus (green), Hts (red) and DNA (blue). The FL-orb3′UTR reporter exhibits very low levels of expression in GSCs and high levels in 16-cell cysts, similar to the Hsp83-lacZ-orb3′UTR reporter and Orb protein (see Fig. 3C). By contrast, the ΔmiR-orb3′UTR reporter displays elevated levels of expression in GSCs. The arrows point to GSCs. (D) The average signal ratio between Venus expression in GSCs and that in 16-cell cysts for the indicated reporters (n=11; germaria from at least two different transgenic lines were evaluated for each reporter; *P<0.005). Error bars indicate s.d. (E) Cells adjacent to the cap cells express the FL-orb 3′UTR reporter in a mei-P26 mutant background. Scale bars: 10 μm.
Loss of mei-P26 mutant GSCs depends on Bam
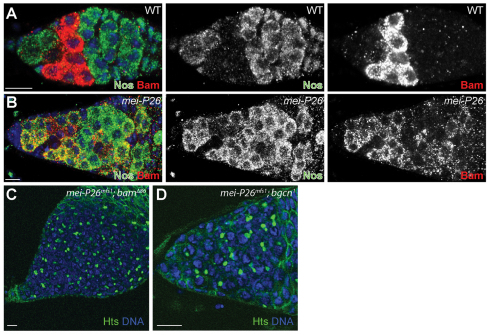
Although the data presented here suggest that Mei-P26 and miRISC can cooperate to modulate the translation of orb mRNA, overexpression of orb in GSCs does not result in GSC loss (data not shown). Therefore, we considered the possibility that Mei-P26 regulates the expression of other genes, dependent or independent of the miRNA pathway. One potential target was the differentiation factor bam. To test whether loss of mei-P26 results in inappropriate expression of Bam in GSCs immediately adjacent to cap cells, we co-stained control and mei-P26 mutant ovaries for Nanos and Bam. Control ovaries displayed mutually exclusive Nanos and Bam expression patterns: Nanos was expressed in GSCs and 16-cell cysts whereas Bam was expressed in cystoblasts and 2-, 4- or 8-cell cysts (Fig. 5A) as previously reported (Li et al., 2009). By contrast, mei-P26 mutants exhibited overlapping Nanos and Bam expression in germline cells immediately adjacent to the cap cells. This ectopic expression of Bam protein provided a possible explanation for the mei-P26 GSC loss phenotype.
Fig. 5.
mei-P26 mutants display ectopic Bam expression. (A,B) w1118 control (A) and mei-P26mfs1 homozygous (B) germaria stained for Nanos (Nos; green), Bam (red) and DNA (blue). In control samples, Nanos and Bam are expressed in mutually exclusive patterns. By contrast, mei-P26 mutants display overlapping Nanos and Bam expression. (C,D) mei-P26mfs1 bam (C) and mei-P26mfs1 bgcn (D) double mutants stained for Hts (green) and DNA (blue). Both double mutants form ovarian tumors that contain single cells with round fusomes. Scale bars: 10 μm.
Loss of bam, or its binding partner benign gonial cell neoplasm (bgcn) (Ohlstein et al., 2000), causes a complete block of germline differentiation, resulting in the formation of tumors that contain single cells with small round fusomes arrested in a ‘pre-cystoblast’ state. To test whether the premature differentiation of mei-P26 mutant GSCs depends on bam, we crossed the mei-P26mfs1 mutation into bam and bgcn mutant backgrounds. Both mei-P26;bam and mei-P26;bgcn homozygous double mutants displayed tumors containing single cells with round fusomes, similar to bam or bgcn single mutants (Fig. 5C,D). These results indicate that the inappropriate differentiation of mei-P26 mutant GSCs depends on both these genes.
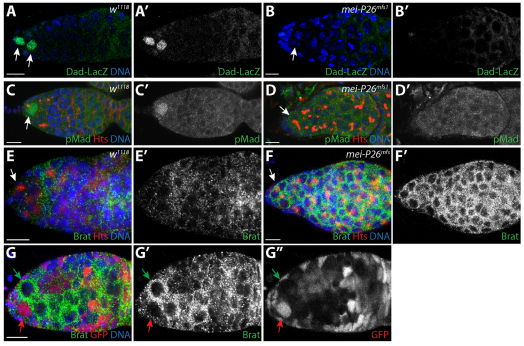
Previous studies have shown that Dpp signaling represses the transcription of bam in GSCs (Chen and McKearin, 2003a; Chen and McKearin, 2003b; Song et al., 2004). We considered the possibility that loss of mei-P26 compromises BMP signaling within GSCs. To test this, we first examined the expression of a Dad-lacZ enhancer trap in a mei-P26 mutant background. Dad is a downstream transcriptional target of the Dpp pathway and this enhancer trap has previously been used as a reporter for pathway activation in the ovary and in other tissues (Kai and Spradling, 2003; Casanueva and Ferguson, 2004; Song et al., 2004). In control ovaries, detectable levels of Dad-lacZ expression were confined to GSCs and newly formed cystoblasts (Fig. 6A,A′), as previously reported (Song et al., 2004). By contrast, we could not detect Dad-lacZ expression in the germline of mei-P26 mutants, even in those germline cells closest to the cap cells (Fig. 6B,B′). We also compared pMad levels in control and mei-P26 mutant ovaries. pMad was detected in control GSCs but not in the germline cells of mei-P26 mutant ovaries (Fig. 6C-D′). Together, the lack of Dad-lacZ and pMad expression indicates that the germline cells of mei-P26 mutants display compromised Dpp signaling, which in turn results in the inappropriate derepression of bam expression. Our clonal analysis suggested that mei-P26 acts in a cell-autonomous manner. However, to exclude the possibility that loss of mei-P26 results in reduced dpp expression within the niche, we performed RT-PCR to compare dpp mRNA levels in bam versus mei-P26 bam double-mutant ovaries (supplementary material Fig. S3). These experiments were performed in a bam mutant background to control for the size and overall morphology of the ovaries being compared. Both samples exhibited similar levels of dpp expression, suggesting that loss of mei-P26 compromised Dpp signaling within GSCs downstream of ligand production.
Fig. 6.
mei-P26 mutants display defects in BMP signal transduction. (A-B′) w1118 control (A,A′) and mei-P26mfs1 homozygotes (B,B′) stained for Dad-lacZ (green), Hts (red) and DNA (blue). (A′,B′) Dad-lacZ staining alone. Arrows indicate germ cells adjacent to cap cells. (C-D′) w1118 control (C,C′) and mei-P26mfs1 homozygous (D,D′) germaria stained for pMad (green), Hts (red) and DNA (blue). (C′,D′) pMad staining alone. Lack of Dad-lacZ and pMad expression in germline cells immediately adjacent to cap cells in mutant samples suggests that BMP signaling is compromised in the absence of mei-P26. Arrows indicate germ cells adjacent to cap cells. (E-F′) w1118 control (E) and mei-P26mfs1 (F) germaria stained for Brat (green), Hts (red) and DNA (blue). (E′,F′) Brat expression shown alone. Germline cells throughout mei-P26 mutant germaria express higher levels of Brat than the control samples. Arrows indicate germ cells adjacent to cap cells. (G-G″) mei-P26mfs1 clonal germarium stained for Brat (green), GFP (red) and DNA (blue) (G). The homozygous mei-P26 mutant GSC clone (green arrow) expresses more Brat protein than the heterozygous control clone (red arrow). (G′,G″) Brat and GFP staining alone. Scale bars: 10 μm.
Recent results showed that Nanos silences the expression of the translational repressor Brat in GSCs (Harris et al., 2011). Once expressed, Brat promotes germline differentiation by targeting components of the BMP signaling pathway for silencing. We hypothesized that Mei-P26 might cooperate with Nanos to repress the expression of Brat in GSCs and that inappropriate expression of Brat might be responsible for the GSC loss phenotype observed in mei-P26 mutants. Brat expression in control and mei-P26 mutant ovaries was assayed (Fig. 6E,F). As previously described (Harris et al., 2011), Brat expression appeared low in control GSCs but increased once the germ cells moved away from the cap cell niche and began to differentiate (Fig. 6E,E′). By contrast, mei-P26 mutants displayed robust Brat expression in germ cells immediately adjacent to the cap cells and throughout the germarium (Fig. 6F,F′). Clonal analysis was used to directly compare Brat protein expression levels in control and mei-P26 mutant cells. Clones homozygous for the mei-P26mfs1 mutation clearly exhibited higher levels of Brat protein expression when compared with heterozygous controls (Fig. 6G, green versus red arrow). Interestingly, Ago1 mutant clones did not exhibit increased Brat expression relative to adjacent control cells (supplementary material Fig. S4). These results suggest that Mei-P26 might negatively regulate the expression of Brat in GSCs in an miRISC-independent manner. However, the ability of Mei-P26 to regulate Brat expression still depends on the NHL domain (supplementary material Fig. S5), consistent with its failure to rescue the stem cell loss phenotype (Fig. 2D,G), suggesting that Mei-P26 interacts with additional proteins to promote BMP signaling in GSCs.
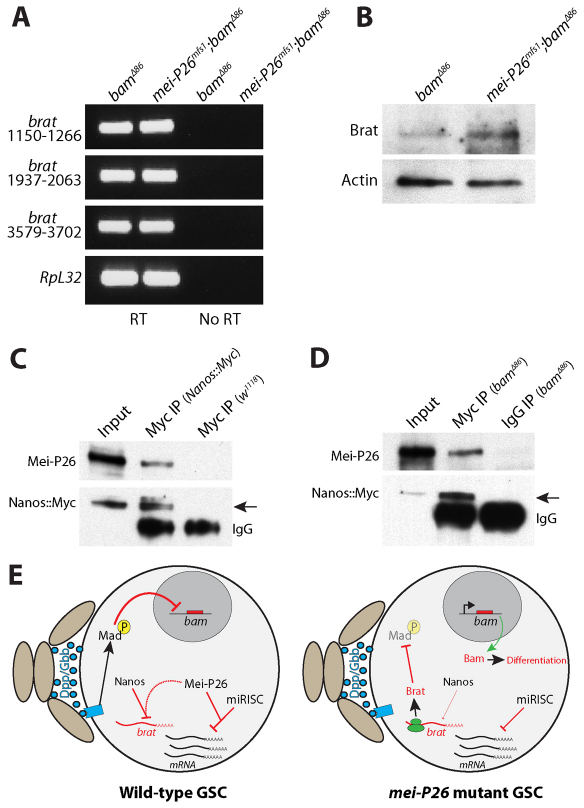
We performed RT-PCR and western blot analyses to begin to characterize the regulation of brat expression in mei-P26 mutants (Fig. 7). These assays were performed in a bamΔ86 mutant background to compare brat mRNA and protein levels in extracts enriched for undifferentiated germ cells. RT-PCR analysis using three different primer sets suggested that brat mRNA levels are similar in bam and mei-P26 bam double mutants (Fig. 7A). By contrast, western blot analysis showed a modest increase in Brat protein levels in mei-P26 bam double mutants when compared with bam mutant controls (Fig. 7B). This finding was consistent with the increase in Brat staining observed in whole-mount preparations (Fig. 6E-G) and suggests that Mei-P26 might regulate Brat expression at the level of translation.
Fig. 7.
Mei-P26 binds to Nanos and regulates Brat protein levels. (A) Ethidium bromide-stained agarose gel showing products from RT-PCR reactions with (RT) and without (No RT) reverse transcriptase. Three different primer pairs specific for different regions of brat mRNA were used to amplify total RNA isolated from bam mutant and mei-P26 bam double-mutant samples. RpL32 was amplified as a loading control. In the RT samples, bam and mei-P26 bam mutants exhibited similar levels of brat mRNA. (B) Western blot analysis of ovarian extracts from bam and mei-P26 bam mutants probed with Brat and Actin antibodies. The mei-P26 bam double-mutant extracts exhibited higher levels of Brat expression than the control extract. (C) Western blot of an anti-Myc co-IP from ovary extract expressing a Myc-tagged nanos transgene. The specificity of the anti-Myc resin was tested using extract from ovaries that did not express the Myc-tagged nanos transgene. (D) Western blot of an anti-Myc co-IP from bamΔ86 mutant extract expressing a Myc-tagged nanos transgene. A rabbit IgG IP of bamΔ86 mutant extract was used as a negative control. co-IP of Mei-P26 with Myc-Nanos was seen in both control ovary and bam mutant ovary extracts (arrows). (E) Model describing the translational regulatory cascades within wild-type and mei-P26 mutant GSCs. Niche cells (light brown ovals) produce Dpp and Gbb, which activate a signal transduction cascade in wild-type GSCs that results in the transcriptional repression of bam. Within wild-type GSCs, Mei-P26 cooperates with both Nanos and miRISC to repress the translation of specific mRNAs. In the absence of Mei-P26, Brat is inappropriately translated resulting in the repression of Mad. Loss of pMAD results in the expression of bam, which causes these mei-P26 mutant GSCs to partially differentiate.
Recent results showed that the well-characterized translational repressor Nanos regulates Brat expression in GSCs (Harris et al., 2011). We speculated that Mei-P26 acts with Nanos to repress Brat translation. Two different IP experiments were performed to test whether Mei-P26 physically interacts with Nanos in germline cells: one using extracts from whole ovaries (Fig. 7C) and a second using extracts from bam mutant ovaries (Fig. 7D). Mei-P26 co-immunoprecipitated with Nanos from both extracts, suggesting that Mei-P26 coordinates with Nanos to repress expression of Brat in early germ cells.
DISCUSSION
Here we provide evidence that Mei-P26 promotes GSC self-renewal in addition to its previously described role in negatively regulating the miRNA pathway during germline cyst development. Disruption of mei-P26 results in a bam-dependent GSC loss phenotype and further characterization reveals that Mei-P26 fosters BMP signal transduction within GSCs by repressing Brat protein expression. In addition, Mei-P26 also appears to participate in the miRNA-mediated silencing of orb mRNA in GSCs. These results indicate that Mei-P26 carries out multiple functions within the Drosophila ovary and might be at the center of a molecular hierarchy that controls the fates of GSCs and their differentiating daughters.
Mei-P26 regulates GSC maintenance
Three observations suggest that mei-P26 functions within GSCs. First, the average number of GSCs per terminal filament decreases from an average of two to well below one in mei-P26 mutant ovaries. Second, mei-P26 mutant germline clones are rapidly lost from the GSC niche. Third, syncytial cysts and Bam-expressing cells are often observed immediately adjacent to the cap cells in mei-P26 mutant ovaries.
Research over the last ten years has shown that BMP ligands emanating from cap cells at the anterior of the germarium initiate a signal transduction cascade in GSCs that results in the transcriptional repression of bam. Stem cell daughters one cell diameter away from the cap cell niche express bam, suggesting that a steep gradient of Dpp availability or responsiveness exists between GSCs and cystoblasts. Recent work has shed light on how various mechanisms antagonize BMP signaling in cystoblasts. For example, the ubiquitin ligase Smurf (Lack – FlyBase) promotes germline differentiation and partners with the serine/threonine kinase Fused to reduce levels of the Dpp receptor Tkv in cystoblasts (Casanueva and Ferguson, 2004; Xia et al., 2010). The TRIM-NHL domain protein Brat also functions in cystoblasts, serving to translationally repress Mad expression (Harris et al., 2011). Notably, inappropriate expression of Brat within GSCs results in a stem cell loss phenotype. Brat itself is translationally repressed in GSCs by the Pumilio-Nanos complex. Mutant phenotypes and co-IP experiments presented here support a model in which Mei-P26 partners with Nanos to repress Brat expression in GSCs (Fig. 7E). This negative regulation of Brat expression protects the BMP signal transduction pathway in GSCs from inappropriate deactivation.
Mei-P26 modulates translational repression in both miRNA-dependent and -independent manners
Mei-P26 appears to enhance miRNA-dependent translational silencing within GSCs based on several lines of experimental evidence. First, co-IP experiments using ovarian extracts from c587-gal4>UAS-dpp and bam mutants suggest that Mei-P26 physically associates with Ago1 and GW182 in undifferentiated germ cells. Second, disruption of mei-P26 results in a GSC loss phenotype, similar to the effects of disrupting components of the miRNA pathway tested to date. Third, Mei-P26 and Ago1 can physically associate with the same target mRNA. Finally, disruption of either Ago1 or mei-P26 results in increased expression of this target in GSCs. The evidence that Mei-P26 promotes miRNA action in certain contexts is consistent with the established activities of its close homologs NHL-2 and TRIM32 (Hammell et al., 2009; Schwamborn et al., 2009).
Multiple functions for Mei-P26
We propose that Mei-P26 regulates GSC self-renewal and early germ cell differentiation through distinct mechanisms (Fig. 7E). In GSCs, Mei-P26 promotes self-renewal by repressing the expression of Brat and potentially other negative regulators of BMP signal transduction. Within stem cells, Mei-P26 also functions together with miRISC to attenuate the translation of specific mRNAs. miRISC does not appear to target brat mRNA based on clonal data (supplementary material Fig. S4). However, we cannot rule out the possibility that the enhancement of miRNA-mediated silencing of some mRNAs by Mei-P26 contributes to stem cell self-renewal. Interestingly, recent findings suggest that Pumilio can function together with the miRNA pathway in certain contexts (Kedde et al., 2010). In BJ primary fibroblasts, Pumilio 1, miR-221 and miR-222 regulate the expression of p27 in a 3′UTR-dependent manner. In response to growth factors, Pumilio 1 becomes phosphorylated, which in turn increases its RNA binding activity. Pumilio 1 binding to p27 mRNA results in a conformational change in the 3′UTR that allows miR-221 and miR-222 to bind more efficiently, resulting in greater repression of p27 (Kedde et al., 2010). Perhaps, together, Drosophila Pumilio, Nanos, Ago1 and Mei-P26 also silence specific messages in specific contexts. Identifying more direct in vivo targets for these proteins within GSCs will be crucial for testing this idea.
In cystoblasts, Mei-P26 promotes germline cyst development by antagonizing the miRNA pathway (Neumuller et al., 2008). Here we show that Mei-P26 can also promote miRNA translational repression in another cell, the GSC. We provide evidence that Mei-P26 physically associates with miRISC and co-regulates translation of at least one mRNA, orb, through specific elements within its 3′UTR. In cystoblasts and early developing cysts, the induction of Bam expression might cause Mei-P26 to switch from an miRISC-associated silencer to an miRNA antagonist. How Bam activates this switch is currently under investigation. The finding that Mei-P26 functions in both GSCs and differentiating cysts hints at a mechanism whereby different translational repression programs coordinate changes in cell fate.
Further work will be needed to determine the specific biochemical function of Mei-P26 when it associates with either the Nanos complex or miRISC. Like other TRIM-NHL domain proteins, Mei-P26 contains a RING domain that may have E3 ubiquitin ligase activity. Based on results presented here, we propose that Mei-P26 and perhaps other TRIM-NHL domain proteins act as effectors for multiple translational repressor complexes. In this model, Mei-P26 is targeted to specific mRNAs through sequence-directed RNA-binding proteins. Specific protein substrates of Mei-P26 in the germline have not yet been determined but identifying these targets will provide key insights into how Mei-P26 and other related TRIM-NHL domain proteins regulate translational repression. Furthermore, the Mei-P26 complex is likely to target additional mRNAs for silencing in both GSCs and developing cysts. Identifying more of these mRNAs will further elucidate the complex translational regulatory hierarchies that control the balance between stem cell self-renewal and differentiation.
Supplementary Material
Acknowledgments
We thank J. Knoblich, R. S. Hawley, Q. Liu, D. Chen, P. Schedl, P. Lasko, E. Izaurralde, R. Wharton, M. Lilly and A. Nakamura for reagents; members of the M.B. lab for comments on the manuscript; N. Shalaby, P. R. Hiesinger, B. Ohlstein and J. Wilhelm for helpful discussions; and M. Fuller for sharing data prior to publication.
Footnotes
Funding
This work was supported by the E. E. and Greer Garson Fogelson Endowment for Scholars Program (UTSW Medical Center) and by the National Institutes of Health [GM045820]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.077412/-/DC1
References
- Barker D. D., Wang C., Moore J., Dickinson L. K., Lehmann R. (1992). Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 6, 2312–2326 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 [DOI] [PubMed] [Google Scholar]
- Casanueva M. O., Ferguson E. L. (2004). Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development 131, 1881–1890 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W., Parker R. (2009). Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA 15, 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., McKearin D. M. (2003a). A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159–1170 [DOI] [PubMed] [Google Scholar]
- Chen D., McKearin D. M. (2003b). Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 13, 1786–1791 [DOI] [PubMed] [Google Scholar]
- Chen D., McKearin D. M. (2005). Gene circuitry controlling a stem cell niche. Curr. Biol. 15, 179–184 [DOI] [PubMed] [Google Scholar]
- Christerson L. B., McKearin D. M. (1994). orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Genes Dev. 8, 614–628 [DOI] [PubMed] [Google Scholar]
- Czech B., Hannon G. J. (2011). Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M., Spradling A. C. (1998). Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125, 2781–2789 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. (2008). GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15, 346–353 [DOI] [PubMed] [Google Scholar]
- Forbes A., Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690 [DOI] [PubMed] [Google Scholar]
- Forstemann K., Tomari Y., Du T., Vagin V. V., Denli A. M., Bratu D. P., Klattenhoff C., Theurkauf W. E., Zamore P. D. (2005). Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3, e236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M. T., Spradling A. C. (2007). Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402–404 [DOI] [PubMed] [Google Scholar]
- Gerber A. P., Luschnig S., Krasnow M. A., Brown P. O., Herschlag D. (2006). Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103, 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L., Lehmann R. (2004). Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 14, 981–986 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V. (2009). nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136, 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. E., Pargett M., Sutcliffe C., Umulis D., Ashe H. L. (2011). Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell 20, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J. B., Podos S. D., Keith K., Simpson S. L., Ferguson E. L. (1998). The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development 125, 1407–1420 [DOI] [PubMed] [Google Scholar]
- Iovino N., Pane A., Gaul U. (2009). miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 17, 123–133 [DOI] [PubMed] [Google Scholar]
- Irish V., Lehmann R., Akam M. (1989). The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338, 646–648 [DOI] [PubMed] [Google Scholar]
- Jin Z., Xie T. (2007). Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 17, 539–544 [DOI] [PubMed] [Google Scholar]
- Kai T., Spradling A. (2003). An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc. Natl. Acad. Sci. USA 100, 4633–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J. A., Elkon R., Agami R. (2010). A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12, 1014–1020 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kim J., Kim Y. J. (1999). Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol. Cell. Biol. 19, 2505–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz V., Ambrosio L., Schedl P. (1992). The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development 115, 75–88 [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang J. S., Horabin J. I., Bopp D., Schedl P. (1994). The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8, 598–613 [DOI] [PubMed] [Google Scholar]
- Lavoie C. A., Ohlstein B., McKearin D. M. (1999). Localization and function of Bam protein require the benign gonial cell neoplasm gene product. Dev. Biol. 212, 405–413 [DOI] [PubMed] [Google Scholar]
- Li Y., Minor N. T., Park J. K., McKearin D. M., Maines J. Z. (2009). Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. USA 106, 9304–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Han H., Lasko P. (2009). Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 23, 2742–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Park J. K., Jiang F., Liu Y., McKearin D., Liu Q. (2007). Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 13, 2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D., Ohlstein B. (1995). A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121, 2937–2947 [DOI] [PubMed] [Google Scholar]
- Murata Y., Wharton R. P. (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- Neumuller R. A., Betschinger J., Fischer A., Bushati N., Poernbacher I., Mechtler K., Cohen S. M., Knoblich J. A. (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., McKearin D. (1997). Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124, 3651–3662 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Lavoie C. A., Vef O., Gateff E., McKearin D. M. (2000). The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 155, 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., McKim K. S., Deneen B., Van Hook T. L., Hawley R. S. (2000). Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 155, 1757–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. K., Liu X., Strauss T. J., McKearin D. M., Liu Q. (2007). The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17, 533–538 [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M. C. (2005). Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 3, e235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Nakamura A., Kobayashi S. (2002). Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 112, 129–139 [DOI] [PubMed] [Google Scholar]
- Schwamborn J. C., Berezikov E., Knoblich J. A. (2009). The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Lai E. C. (2008). Lessons from microRNA mutants in worms, flies and mice. Cell Cycle 7, 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wong M. D., Kawase E., Xi R., Ding B. C., McCarthy J. J., Xie T. (2004). Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 131, 1353–1364 [DOI] [PubMed] [Google Scholar]
- Sonoda J., Wharton R. P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Wharton R. P. (2001). Drosophila Brain Tumor is a translational repressor. Genes Dev. 15, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura I., Lilly M. A. (2006). Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell 10, 127–135 [DOI] [PubMed] [Google Scholar]
- Szakmary A., Cox D. N., Wang Z., Lin H. (2005). Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 15, 171–178 [DOI] [PubMed] [Google Scholar]
- Tan L., Chang J. S., Costa A., Schedl P. (2001). An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development 128, 1159–1169 [DOI] [PubMed] [Google Scholar]
- Tastan O. Y., Maines J. Z., Li Y., McKearin D. M., Buszczak M. (2010). Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development 137, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin H. (2004). Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303, 2016–2019 [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Thomsen G. H., Gelbart W. M. (1991). Drosophila 60A gene, another transforming growth factor beta family member, is closely related to human bone morphogenetic proteins. Proc. Natl. Acad. Sci. USA 88, 9214–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R. P., Sonoda J., Lee T., Patterson M., Murata Y. (1998). The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1, 863–872 [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti A. C., Schisa J. A., Lieberfarb M. E., Strickland S. (1997). Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 124, 3015–3023 [DOI] [PubMed] [Google Scholar]
- Xia L., Jia S., Huang S., Wang H., Zhu Y., Mu Y., Kan L., Zheng W., Wu D., Li X., et al. (2010). The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell 143, 978–990 [DOI] [PubMed] [Google Scholar]
- Xie T., Spradling A. C. (1998). decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251–260 [DOI] [PubMed] [Google Scholar]
- Yang L., Chen D., Duan R., Xia L., Wang J., Qurashi A., Jin P. (2007). Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 134, 4265–4272 [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Williamson J. R., Lehmann R. (1997). The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3, 1421–1433 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.