Abstract
Recent studies have detected mutations in the EDA gene, previously identified as causing X-linked hypohidrotic ectodermal dysplasia (XLHED), in two families with X-linked non-syndromic hypodontia. Notably, all affected males in both families exhibited isolated oligodontia, while almost all female carriers showed a milder or normal phenotype. We hypothesized that the EDA gene could be responsible for sporadic non-syndromic oligodontia in affected males. In this study, we examined 15 unrelated males with non-syndromic oligodontia. Three novel EDA mutations (p.Ala259Glu, p.Arg289Cys, and p.Arg334His) were identified in four individuals (27%). A genetic defect in the EDA gene could result in non-syndromic oligodontia in affected males.
Keywords: EDA gene, oligodontia, mutation, non-syndromic
Introduction
Tooth agenesis (MIM# 106600), the congenital absence of one or more permanent teeth, is a common human anomaly (Pemberton et al., 2005). In most populations, the reported prevalence of permanent tooth agenesis, excluding third molars, varies from 2.2 to 10.1% (Polder et al., 2004). In the majority of cases, persons are missing only one tooth (Daugaard-Jensen et al., 1997). The prevalence becomes progressively smaller as the number of missing teeth increases. Agenesis of more than two teeth occurs in approximately 1% of the population (Polder et al., 2004). Selective tooth agenesis is divided into 2 types: hypodontia, the agenesis of fewer than 6 teeth, and oligodontia, the agenesis of 6 or more permanent teeth. In both cases, the third molars (wisdom teeth) are not included. Oligodontia (MIM# 604625) is a rare anomaly, affecting approximately 0.1 to 0.3 % of the population (Polder et al., 2004).
Although several potential and verified environmental factors in tooth agenesis have been identified, genetic defects play a major role in the etiology (Graber, 1978; Burzynski and Escobar, 1983; Vastardis, 2000). So far, researchers have identified genetic defects that cause tooth agenesis either as a sole anomaly (isolated or non-syndromic) or as a part of multiple congenital anomalies (syndromic). One gene associated with syndromic tooth agenesis is the EDA gene (MIM# 300451), which underlies X-linked hypohidrotic ectodermal dysplasia (XLHED; MIM# 305100). XLHED is a rare condition, in which some of the symptoms are sparse hair, defective sweat gland development, tooth agenesis, and reduced tooth size (microdontia) (Kere et al., 1996; Mikkola and Thesleff, 2003).
Non-syndromic tooth agenesis has wide phenotypic heterogeneity and is classified as either sporadic or familial, which can be inherited in an autosomal-dominant, autosomal-recessive, or X-linked mode (Burzynski and Escobar, 1983). Dominant mutations in MSX1 (MIM# 142983), PAX9 (MIM# 167416), and AXIN2 (MIM# 604025) have been found in families with non-syndromic tooth agenesis (Vastardis et al., 1996; Stockton et al., 2000, Lammi et al., 2004). However, mutations in these genes were detected in only a few affected individuals, suggesting that there may be other unidentified genetic defects responsible for this disease (Scarel et al., 2000; Frazier-Bowers et al., 2002). Recently, a missense mutation (p.Arg65Gly) in the EDA gene has been reported in a Chinese family with X-linked non-syndromic hypodontia (Taoet al., 2006). Following this report, another mutation (p.Gln358Glu) was detected in affected members of an Indian family with X-linked hypodontia (Tarpey et al., 2007). Unlike XLHED, affected persons in both families had no other clinical features but tooth agenesis. Specifically, affected males in both families exhibited oligodontia, with 6-24 teeth absent, while almost all female carriers were normal or had a milder phenotype (Tao et al., 2006; Tarpey et al., 2007). Therefore, we hypothesized that the EDA gene might be a candidate gene for non-syndromic oligodontia in affected males.
Materials & Methods
Study Individuals
Fifteen non-consanguineous males were chosen from among the patients referred to the Department of Prosthodontics at Peking University School and Hospital of Stomatology for diagnostic evaluation and treatment of tooth agenesis. A dentist determined the status of the dentition through oral and radiographic examinations for the patients and their available parents. The shapes and the sizes of the residual teeth were observed by the dentist.
DNA Sequence Analysis of the EDA Gene
This study was conducted with the approval of the Ethics Committee of Peking University Health Science Center. All participants and/or their legal representatives were fully aware of the scope of this study and signed an informed consent form prior to enrollment. Blood samples were obtained and genomic DNA was isolated by a standard high-salt method. Genomic DNA from 60 females (each female has two X chromosomes) was used as a control. Screening of the EDA gene was performed by direct sequencing of 8 PCR fragments that cover the entire cDNA of 8 exons and intron-exon junctions of more than 100 base pairs. Gel-purified PCR fragments were sequencedon an Applied Biosystems 377 automated sequencer in an ABI PRISM fluorescent dye terminator system (Perkin Elmer, Foster City, CA, USA).
Protein Structure Analysis
We used the crystal structure of EDA-A1, PDB coordinate 1RJ7 (Hymowitz et al., 2003), as a scaffold for the protein structure analysis. The structures were analyzed with the Insight II (2000) software package (Accelrys Inc., San Diego, CA, USA). Figures were created with PyMOL (The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA, USA; http://www.pymol.org).
Results
Clinical Details
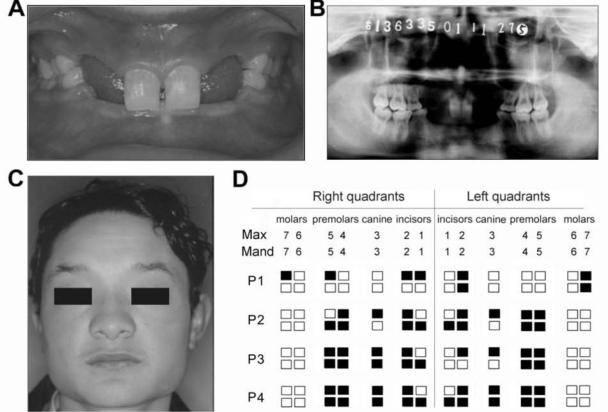
Although the missing teeth were distributed extensively in both dentitions, the shapes and the sizes of the residual teeth were observed to be normal (Fig. 1A). A panoramic radiograph was taken to confirm the diagnosis of oligodontia for the 15 study individuals (Fig. 1B). None of their parents had congenitally missing teeth, and their mothers did not have small or conical teeth. Phenotypic characteristics of scalp and body hair, skin, nails, tolerance to heat, and ability to sweat were examined through observation, palpation, and inquiry. All participants reported normal sweating and lachrymal secretions. They had no complaints about feeling of dry mouth, intolerance to heat, or susceptibility to respiratory tract infections. The participants had hair on the body and scalp, and their facial features, skin, and nails were observed to be normal (Fig. 1C).
Figure 1.
Clinical characteristics of persons with non-syndromic oligodontia with EDA mutations. (A) Clinical phenotype of participant 4 shows congenital tooth agenesis. (B) The panoramic radiograph of participant 4. (C) The facial photograph of participant 4. (D) Schematic presentation of congenitally missing teeth among the four non-syndromic oligodontia individuals with EDA mutations. The missing tooth is represented by a filled square. Max, maxillary; Mand, mandibular; P, participant.
DNA Sequence Analysis of the EDA Gene
In this study, 3 novel missense mutations (p.Ala259Glu, p.Arg289Cys, and p.Arg334His) were identified in four out of 15 non-syndromic oligodontia males (Table). The numbers and positions of the congenitally missing teeth of these four persons are shown in Fig. 1D. Although the manifestation of oligodontia was not uniform among these individuals, their maxillary and mandibular first molars were all present.
Table.
Details of EDA Pathogenic Mutations Detected in Persons with Non-syndromic Oligodontia.
| Patient No. | Exon | Mutation Type | Nucleotide* Change | Codon Position | Amino Acid Change | Origin |
|---|---|---|---|---|---|---|
| 1 | 6 | Missense | c.776C>A | 259 | p.Ala259Glu | de novo |
| 2 | 6 | Missense | c.776C>A | 259 | p.Ala259Glu | — |
| 3 | 7 | Missense | c.865C>T | 289 | p.Arg289Cys | Maternal |
| 4 | 8 | Missense | c.1001G>A | 334 | p.Arg334His | Maternal |
The A of the ATG translation initiation codon is numbered +1.
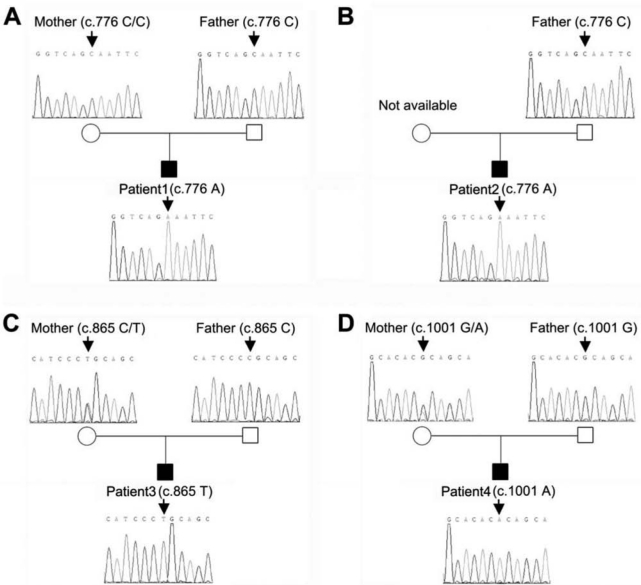
For participant 1, the nucleotide sequence showed a C to A transition at nucleotide 776 (c.776C>A) of the coding sequence in exon 6 of EDA, changing codon 259 from encoding Ala to Glu. Neither parent carried the mutation (Fig. 2A, Table), suggesting that the mutation was de novo. The same p.Ala259Glu (c.776C>A) mutation was also identified in participant 2 (Fig. 2B, Table). Although his maternal sample was not available, the possibility of participants 1 and 2 having inherited the mutation from a common ancestor was excluded, because participant 1’s mutation is most likely de novo.
Figure 2.
Sequence analyses of EDA gene among four sporadic persons with congenital non-syndromic oligodontia. (A) A de novo mutation c.776C>A was found in participant 1, but not in his parents. (B) Mutation c.776C>A was found in participant 2, but his mother was not available for this study. (C) Mutation c.865C>T was found in participant 3, who inherited the mutant allele from his mother. (D) Mutation c.1001G>A was found in participant 4, who also inherited the mutant allele from his mother. All mutated nucleotides are identified by arrows.
The p.Arg289Cys (c.865C>T) mutation was detected in participant 3. It is a C to T transition at nucleotide 865 of the coding sequence in exon 7 of EDA, changing codon 289 from encoding Arg to Cys. Analyses of his parents revealed that the mutant allele came from his mother (Fig. 2C, Table).
In participant 4, the p.Arg334His (c.1001G>A) mutation is due to a G to A transition at nucleotide 1001 of the coding sequence in exon 8 of EDA, changing codon 334 from encoding Arg to His. Sequence analyses of his parents revealed that the mutant allele also came from his mother (Fig. 2D, Table). All mutant alleles identified in this study were not detected in the 120 control X chromosomes (60 females).
Protein Structure Analysis
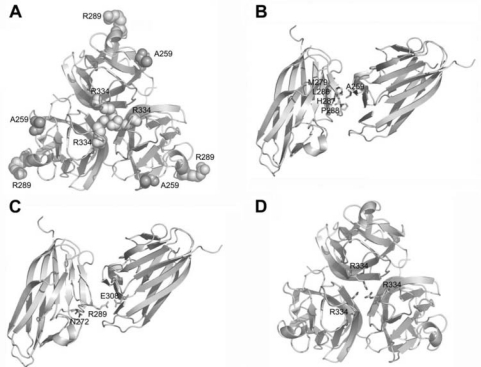
Structural analysis of the EDA protein revealed that, in the wild-type three-dimensional EDA structure, residue Ala259 is located at the outer surface of the homotrimer (Fig. 3A), interacting hydrophobically with non-polar residues Met279, Leu286, His287, and Pr0288 from the adjacent trimer (Fig. 3B). The substitution of Ala with Glu, which is a hydrophilic amino acid containing a hydrophilic amino acid with a side-chain longer than Ala, may eliminate the hydrophobic interaction.
Figure 3.
Mutant residues in the three-dimensional EDA-A1 trimer. (A) The location of the Ala259 (A259), Arg289 (R289), and Arg334 (R334) residues. The trimer of EDA-A1 is shown as a ribbon, with relevant side-chains represented by spheres. (B) Ala259 and 4 residues of the adjacent trimer (Met279, Leu286, His287, and Pr0288) interact hydrophobically. The relevant side-chains are represented by strips. (C) Arg289 forms hydrogen bonds with Asn272 (N272), and also forms hydrogen bonds and has electrostatic interactions with Glu308 (E308) of the adjacent homotrimer(s). Relevant side-chains are represented by strips. (D) Three Arg334 residues organize a hydrogen-bond network in an EDA-A1 trimer. The Arg 334 side-chain is represented by a strip.
Similarly, the residue Arg289 is also situated at the outer surface of the homotrimer (Fig. 3A), where it forms hydrogen bonds with Asn 272, and forms hydrogen bonds and electrostatic interactions with Glu308 of the adjacent homotrimer(s) to stabilize the multi-trimer asymmetric unit (Fig. 3C). The mutation p.Arg289Cys would abolish these interactions.
Three Arg334 residues, one from each monomer at the monomer-monomer interface (Fig. 3A), form hydrogen bonds in an interacting network (Fig. 3D). The mutation p.Arg334His would interrupt this interaction network. Therefore, structural analyses revealed that these mutations would produce conformational changes, potentially altering the stability of the EDA homotrimers.
Discussion
In the present study, identification of EDA mutations in 27% (four out of 15) of unrelated affected individuals indicates that EDA is a candidate gene involved in non-syndromic oligodontia. Although several mutations in the EDA gene leading to XLHED have been reported, all mutations causing non-syndromic oligodontia identified in this study, as well as the two mutations reported in previous studies, are novel missense mutations (Tao et al., 2006; Tarpey et al., 2007).
In this study, the mean number of missing teeth (excluding the third molars) in the four individuals with mutations was 14 (range, 8 to 18 teeth), which is less than the mean number of missing teeth in those with XLHED (22; range, 14 to 28; Lexner et al., 2007). In persons with XLHED, the incisors in the maxilla usually had a tapered and conical morphology (Lexner et al., 2007), but these tooth malformations were not observed in our study participants. The above evidence suggests that there is a noticeable correlation between the genotype and the phenotype.
Considering that all four individuals did not have central or lateral incisors of both dentitions, consistent with the phenotype observed in two previously reported families (Tao et al., 2006; Tarpey et al., 2007), we recommend that mutation analysis of the EDA gene be performed in persons with isolated oligodontia, especially when central and lateral incisors are absent in both dentitions. In addition, it was noticeable that several of our participants appeared to have congenitally missing maxillary and mandibular canines, which is very rare in isolated oligodontia with other genetic defects (Kim et al., 2006).
The EDA gene encodes the protein ectodysplasin-A (EDA), a member of the tumor necrosis factor (TNF) superfamily. EDA is a type II transmembrane protein with a C-terminal TNF homology domain consisting of 10 predicted anti-parallel β-sheets linked by variable loops (Ezer et al., 1999). The TNF homology domain forms a homotrimer (Ezer et al., 1999), which is required for interaction with the receptor at the monomer–monomer interface (Hymowitz et al., 2000). Mutations in the TNF domain affect the interaction of EDA with its receptors, ectodysplasin-A receptor (EDAR) andectodysplasin-A2 receptor (EDA2R; Schneider et al., 2001). The missense mutations observed in this study are all located within the TNF-like domain (residues 245–391) of EDA. We speculate that these mutations may also affect the interaction of EDA with its receptors, which correlates with the non-syndromic phenotype.
Among the previously reported XLHED-causing mutations, p.Gly255Cys, p.Gly291Arg, and p.Cys332Tyr are physically closest to the mutations p.Ala259Glu, p.Arg289Cys, and p.Arg334His, identified in this study. P.Gly255Cys and p.Gly291Arg are both mutations replacing the smallest amino acid Gly, with large residues that could not be accommodated structurally, probably globally destabilizing the ligand (Hymowitz et al., 2003). The p.Cys332Tyr mutation is located adjacent to the receptor specificity switch E308 and was predicted, based on the structure, to participate in the formation of active sites (Hymowitz et al., 2003). These XLHED-causing mutations may severely affect EDA function by impairing interactions with the receptors (Schneider et al., 2001). In a comparison of the mutations causing the isolated hypodontia phenotype with the mutations causing the full XLHED phenotypes, the mutations p.Ala259Glu and p.Arg289Cys, identified in the present study, were both located at the protein surface, away from the active site Glu308. We propose here that changes in these residues, including the p.Arg334His substitution, would influence the stability of only the homotrimers and partially affect EDA function. This proposal is supported by the observation that all XLHED-causing missense mutations in the TNF domain resulted in a protein that could not interact with EDAR and EDA2R, while p.Ser374Arg retained weak, but apparently specific, binding activity to EDAR, but not to EDA2R (Schneider et al., 2001). The family with the p.Ser374Arg mutation had two affected males and one affected maternal grandfather with isolated hypodontia, similar to the phenotype of isolated hypodontiain this study. Therefore, we speculate that the phenotypedifference between isolated oligodontia and XLHED might be related to the different effects of the mutations on EDA structure and function. While XLHED-causing mutations result in the loss of interaction of EDA with EDAR, mutations causing isolated oligodontia likely cause only a partial loss of EDA function, which specifically targets the development of teeth, as we saw in our study participants.
Acknowledgments
We thank all the families for their participation.
Footnotes
This work was supported by the Chinese Ministry of Health 985 Project (985-2-035-39), the Natural Science Foundation of Beijing (7063099), and The International Collaborative Genetics Research Training Program (NIH D43 TW06176).
References
- Burzynski NJ, Escobar VH. (1983). Classification and genetics of numeric anomalies of dentition. Birth Defects Orig Artic Ser 19:95-106 [PubMed] [Google Scholar]
- Daugaard-Jensen J, Nodal M, Kjaer I. (1997). Pattern of agenesis in the primary dentition: a radiographic study of 193 cases. Int J Paediatr Dent 7:3-7 [DOI] [PubMed] [Google Scholar]
- Ezer S, Bayes M, Elomaa O, Schlessinger D, Kere J. (1999). Ectodysplasin is a collagenous trimeric type II membrane protein with a tumor necrosis factor-like domain and co-localizes with cytoskeletal structures at lateral and apical surfaces of cells. Hum Mol Genet 8:2079-2086 [DOI] [PubMed] [Google Scholar]
- Frazier-Bowers SA, Scott MR, Cavender A, Mensah J, D’Souza RN. (2002). Mutational analysis of families affected with molar oligodontia. Connect Tissue Res 43:296-300 [DOI] [PubMed] [Google Scholar]
- Graber LW. (1978). Congenital absence of teeth: a review with emphasis on inheritance patterns. J Am Dent Assoc 96:266-275 [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, O’Connell MP, Ultsch MH, Hurst A, Totpal K, Ashkenazi A, et al. (2000). A unique zinc-binding site revealed by a high-resolution x-ray structure of homotrimeric Ap02L/TRAIL. Biochemistry 39:633-640 [DOI] [PubMed] [Google Scholar]
- Hymowitz SG, Compaan DM, Yan M, Wallweber HJ, Dixit VM, Starovasnik MA, et al. (2003). The crystal structures of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity. Structure 11:1513-1520 [DOI] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, Ferguson B, et al. (1996). X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet 13:409-416 [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Lin BP, Hu JC. (2006). Novel MSX1 frameshift causes autosomal-dominant oligodontia J Dent Res 85:267-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Järvinen H, Lahermo P, Thesleff I, et al. (2004). Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74:1043-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexner MO, Bardow A, Hertz JM, Nielsen LA, Kreiborg S. (2007). Anomalies of tooth formation in hypohidrotic ectodermal dysplasia. Int J Paediatr Dent 17:10-18 [DOI] [PubMed] [Google Scholar]
- Mikkola ML, Thesleff I. (2003). Ectodysplasin signaling in development. Cytokine Growth Factor Rev 14:211-224 [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Das P, Patel PI. (2005). Hypodontia: genetics and future perspectives. Braz J Oral Sci 4:695-706 [Google Scholar]
- Polder BJ, Van’t Hof MA, Van der Linden FP, Kuijpers-Jagtman AM. (2004). A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent Oral Epidemiol 32:217-226 [DOI] [PubMed] [Google Scholar]
- Scarel RM, Trevilatto PC, di Hipolito O, Jr, Camargo LE, Line SR. (2000). Absence of mutations in the homeodomain of the MSX1 gene in patients with hypodontia. Am J Med Genet 92:346-349 [DOI] [PubMed] [Google Scholar]
- Schneider P, Street SL, Gaide O, Hertig S, Tardivel A, Tschopp J, et al. (2001). Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J Biol Chem 276:18819-18827 [DOI] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI. (2000). Mutation of PAX9 is associated with oligodontia. Nat Genet 24: 18-19 [DOI] [PubMed] [Google Scholar]
- Tao R, Jin B, Guo SZ, Qing W, Feng GY, Brooks DG, et al. (2006). A novel missense mutation of the EDA gene in a Mongolian family with congenital hypodontia. J Hum Genet 51:498-502 [DOI] [PubMed] [Google Scholar]
- Tarpey P, Pemberton TJ, Stockton DW, Das P, Ninis V, Edkins S, et al. (2007). A novel gln358glu mutation in ectodysplasin A associatedwith X-linked dominant incisor hypodontia. Am J Med Genet A 143:390-394 [DOI] [PubMed] [Google Scholar]
- Vastardis H. (2000). The genetics of human tooth agenesis: new discoveries for understanding dental anomalies. Am J Orthod Dentofacial Orthop 117:650-656 [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. (1996). A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet 13:417-421 [DOI] [PubMed] [Google Scholar]