Abstract
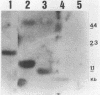
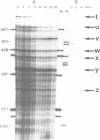
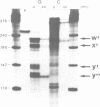
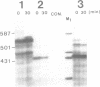
The degradation of the polycistronic tra-mRNA of the resistance plasmid R1-19 leads to the accumulation of a well defined series of stable mRNA species. The majority of the most stable mRNAs contains the message for the traA gene only. The differently sized stable mRNAs possess a common 3'terminus within the traL gene but vary at their 5' ends. The 3'terminus probably results from protection against exoribonucleases by a secondary structural feature. We propose that the 5' ends are generated by endoribonucleolytic cleavage. The stability of this part of the tra-mRNA exceeds 30 minutes and probably increases the rate of expression of the traA gene product propilin, the precursor of the sex pilus subunit. The expression of propilin and its processing into a protein of the molecular weight of mature pilin is demonstrated with the isolated gene. The sequence of the so far unknown genes traL and traE of R1-19 is presented.
Full text
PDF





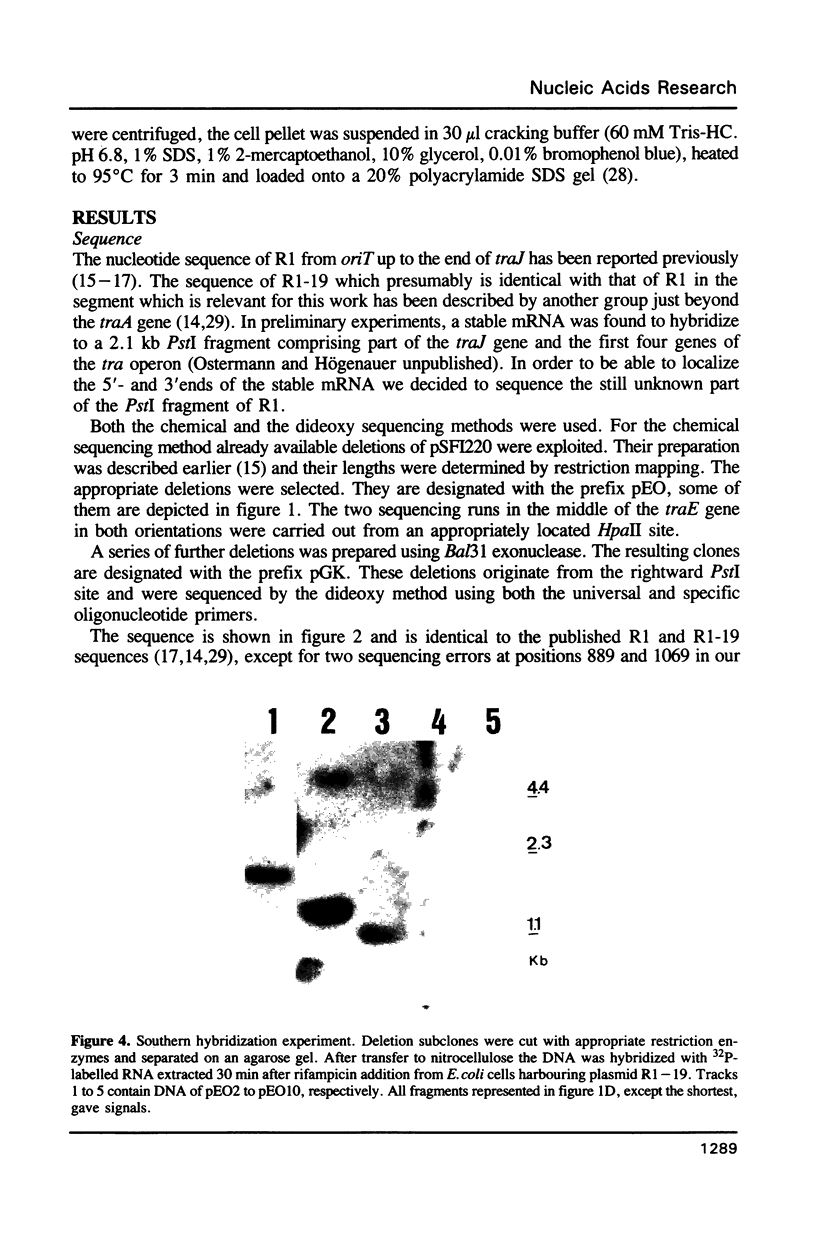
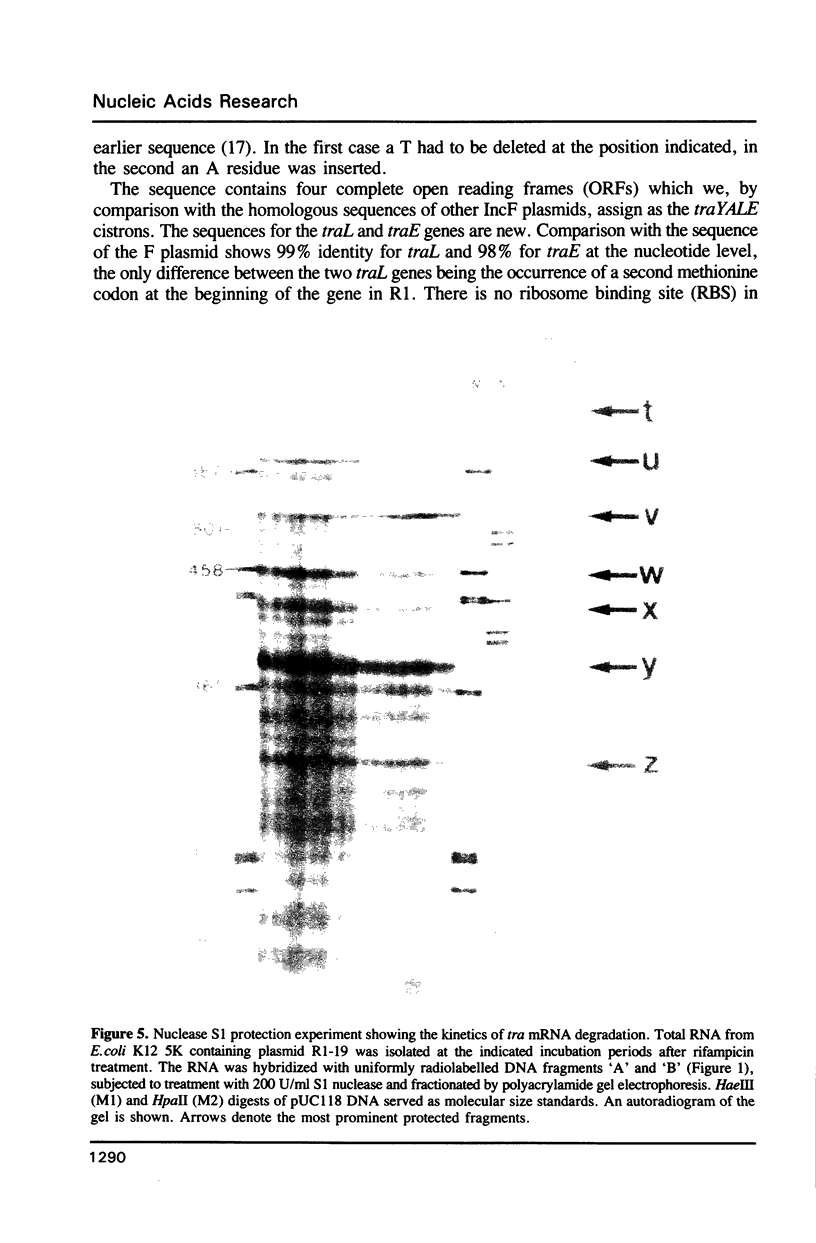
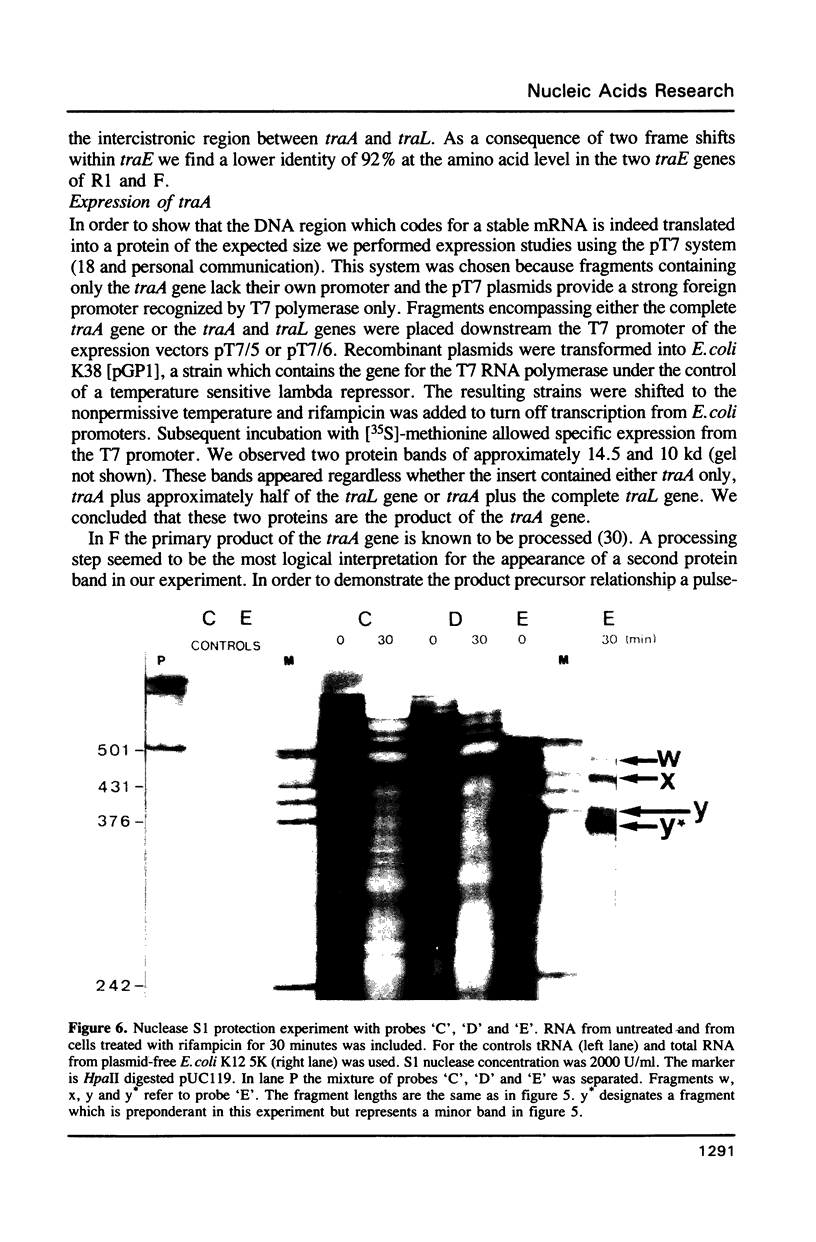



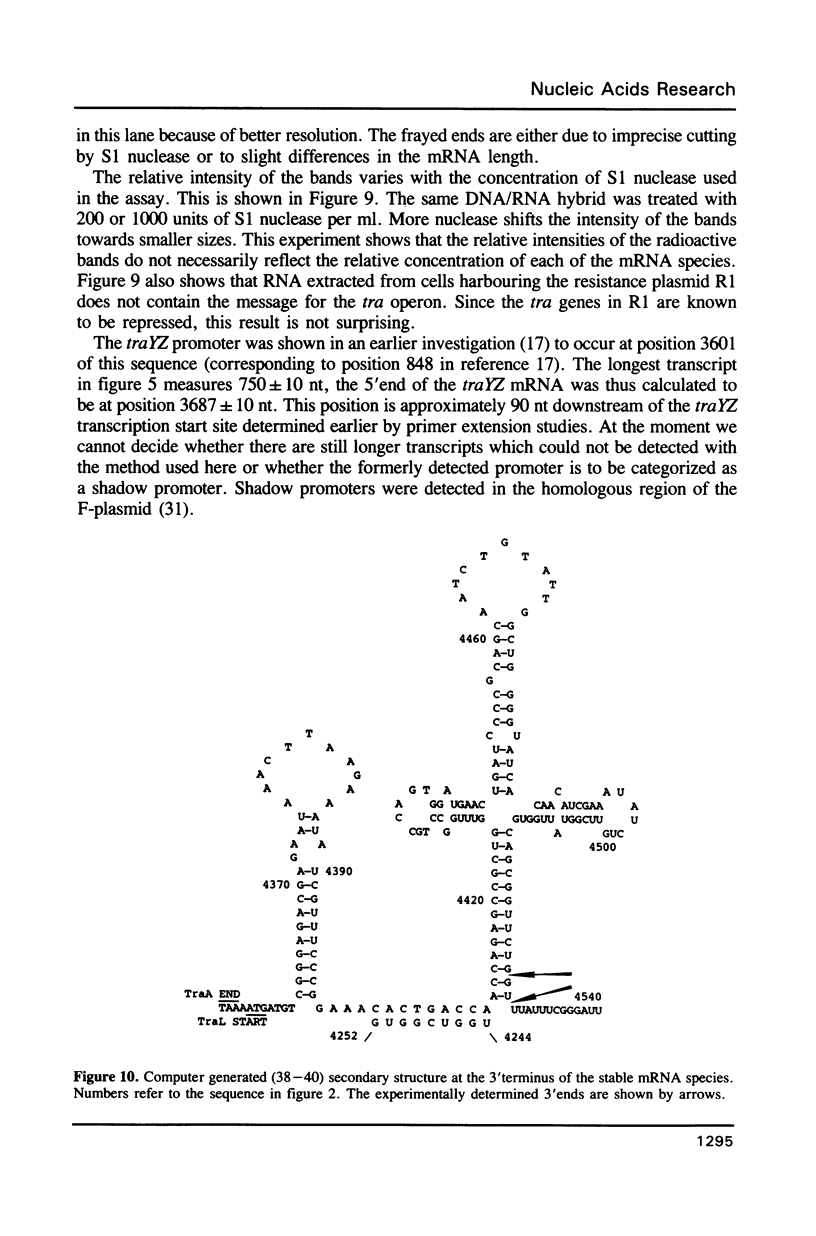



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Cole J. R., Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986 Apr 5;188(3):383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. E. coli RNases: making sense of alphabet soup. Cell. 1985 Apr;40(4):731–732. doi: 10.1016/0092-8674(85)90330-7. [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Frost L. S., Paranchych W. Nucleotide sequence of the tra YALE region from IncFV plasmid pED208. J Bacteriol. 1986 Nov;168(2):990–998. doi: 10.1128/jb.168.2.990-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Frost L. S., Paranchych W. Nucleotide sequences of the R1-19 plasmid transfer genes traM, finP, traJ, and traY and the traYZ promoter. J Bacteriol. 1986 May;166(2):368–374. doi: 10.1128/jb.166.2.368-374.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T., Thompson R. Shadow promoters in the F plasmid transfer operon. Mol Gen Genet. 1986 Mar;202(3):509–511. doi: 10.1007/BF00333285. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Finlay B. B., Opgenorth A., Paranchych W., Lee J. S. Characterization and sequence analysis of pilin from F-like plasmids. J Bacteriol. 1985 Dec;164(3):1238–1247. doi: 10.1128/jb.164.3.1238-1247.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Paranchych W., Willetts N. S. DNA sequence of the F traALE region that includes the gene for F pilin. J Bacteriol. 1984 Oct;160(1):395–401. doi: 10.1128/jb.160.1.395-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAREN A., LEVINTHAL C. A fine-structure genetic and chemical study of the enzyme alkaline phosphatase of E. coli. I. Purification and characterization of alkaline phosphatase. Biochim Biophys Acta. 1960 Mar 11;38:470–483. doi: 10.1016/0006-3002(60)91282-8. [DOI] [PubMed] [Google Scholar]
- Golub E. I. 'One minute' transformation of competent E. coli by plasmid DNA. Nucleic Acids Res. 1988 Feb 25;16(4):1641–1641. doi: 10.1093/nar/16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Kennedy N., Beutin L., Achtman M., Skurray R., Rahmsdorf U., Herrlich P. Conjugation proteins encoded by the F sex factor. Nature. 1977 Dec 15;270(5638):580–585. doi: 10.1038/270580a0. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V. E., Bauer E., Högenauer G. The traM gene of the resistance plasmid R1: comparison with the corresponding sequence of the Escherichia coli F factor. Gene. 1985;36(1-2):79–86. doi: 10.1016/0378-1119(85)90071-x. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Högenauer G. The sequences of the traJ gene and the 5' end of the traY gene of the resistance plasmid R1. Mol Gen Genet. 1986 Apr;203(1):137–142. doi: 10.1007/BF00330394. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Meynell E., Datta N. Mutant drug resistant factors of high transmissibility. Nature. 1967 May 27;214(5091):885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann E., Kricek F., Högenauer G. Cloning the origin of transfer region of the resistance plasmid R1. EMBO J. 1984 Aug;3(8):1731–1735. doi: 10.1002/j.1460-2075.1984.tb02039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]