Abstract
Proteolytic enzymes serve important functions during dental enamel formation, and mutations in the kallikrein 4 (KLK4) and enamelysin (MMP20) genes cause autosomal-recessive amelogenesis imperfecta (ARAI). So far, only 1 KLK4 and 3 MMP20 mutations have been reported in ARAI kindreds. To determine whether ARAI in a family with a hypomaturation-type enamel defect is caused by mutations in the genes encoding enamel proteolytic enzymes, we performed mutational analysis on candidate genes. Mutational and haplotype analyses revealed an ARAI-causing point mutation (c.910G>A, p.A304T) in exon 6 of MMP20 that results in a single amino acid substitution in the hemopexin domain. Western blot analysis showed decreased expression of the mutant protein, but zymogram analysis demonstrated that this mutant was a functional protein. The proband and an affected brother were homozygous for the mutation, and both unaffected parents were carriers. The enamel of newly erupted teeth had normal thickness, but was chalky white and became darker with age.
Keywords: MMP20, hemopexin, enamel, tooth, amelogenesis imperfecta
Introduction
Amelogenesis imperfecta (AI) is a heterogeneous group of inherited enamel malformations that can be caused by defects in many genes. The common classification system categorizes the spectrum of enamel phenotypes into hypoplastic, hypocalcified, hypomaturation, and hypomaturation-hypoplastic with taurodontism. AI can have autosomal-dominant, autosomal-recessive, or X-linked patterns of inheritance (Witkop and Sauk, 1976). Based upon clinical phenotypes and modes of inheritance, 14 AI subtypes are recognized (Witkop, 1988). In the United States, the combined incidence of all types of AI is about 1:14,000 (Witkop and Sauk, 1976). Genes encoding enamel matrix proteins have been implicated in the etiology of AI. Defects in amelogenin (AMELX) cause X-linked AI (Kim et al., 2004; Kida et al., 2007), defects in enamelin (ENAM) cause autosomal AI (Kim et al., 2005a; Ozdemir et al., 2005b), and mutations in KLK4 (Hart et al., 2004) and MMP20 (Kim et al., 2005b; Ozdemir et al., 2005a; Papagerakis et al., 2008) cause autosomal-recessive AI. It has been recently identified that the mutations in the FAM83H gene are associated with hypocalcified AI with autosomal-dominant inheritance (Kim et al., 2008; Lee et al., 2008; Hart et al., 2009). However, it is believed that many additional candidate genes have yet to be identified.
The function of enamelysin in developing teeth has been identified by expression pattern and phenotype analysis of null mice as well as by activity-based assays. Mmp20 null mice exhibit an enamel layer that is thinner than normal and chips away from the underlying dentin (Caterina et al., 2002). The amount of enamel mineral is reduced by half, and its hardness is decreased by about one-third (Bartlett et al., 2004). The dentitions of humans with homozygous mutant MMP20 alleles have been described. The MMP20 IVS6-2A>T mutation is thought to interfere with the excision of intron 6 during RNA splicing. Retention of intron 6 or skipping of exon 7 could introduce a translation termination codon before the final exon, so that the final mRNA products are probably degraded by the nonsense-mediated decay (NMD) system, and the dental phenotype reflects a null condition (absence of MMP20 expression). The resulting phenotype is a pigmented hypomaturation type of AI with autosomal-recessive inheritance (Kim et al., 2005b). The teeth are normal in size, with decreased contrast between dentin and enamel on radiographs, have an agar-brown discoloration, a mottled and rough surface, and a tendency to chip.
In a previous study of another ARAI family, an MMP20 missense mutation (p.H226Q) did not interfere with MMP20 expression, but completely abolished MMP-20 proteolytic activity (Ozdemir et al., 2005a). The enamel phenotype was diagnosed as an autosomal-recessive hypomaturation type of AI. In affected individuals, the teeth showed normal enamel thickness, but were yellowish to brownish, with decreased radiographic contrast between dentin and enamel. The enamel was often softer than normal, and showed extensive attrition and caries of posterior teeth. A nonsense mutation (p.W34X) in exon 1 of the MMP20 gene in an ARAI family is thought to cause degradation of mRNA by the NMD system (Papagerakis et al., 2008). The clinical phenotype of this mutation is remarkably similar to that reported in previous cases.
The purpose of our current study was to investigate the genetic etiology of a family with a hypomaturation-type enamel defect, to broaden our understanding of amelogenesis.
Materials & Methods
Identification of a Kindred and Enrollment of Study Participants
The study protocol and participant consents were independently reviewed and approved by the Institution Review Board at the Seoul National University Dental Hospital. Informed consent was obtained according to the Declaration of Helsinki.
Polymerase Chain-reaction (PCR) and Sequencing
Genomic DNA was isolated from peripheral whole blood with the use of the QuickGene DNA whole-blood kit S with QuickGene-Mini80 equipment (Fujifilm, Tokyo, Japan). The purity and concentration of the DNA were quantitated by spectrophotometry, as measured by the OD260/OD280 ratio. Mutational analyses, including exons and nearby intron sequences, were completed for KLK4 (Hart et al., 2004), MMP20 (Kim et al., 2005b), and AMELX (Kim et al., 2004). PCR amplifications were performed with the HL DNA polymerase (Bioneer, Daejeon, Korea). PCR amplification products were purified by the QIAquick PCR Purification Kit according to the manufacturer′s protocol (Qiagen Inc., Valencia, CA, USA). DNA sequencing was performed at the DNA sequencing center (Macrogen, Seoul, Korea).
Haplotype Analysis
Two microsatellite markers, D11S898 (Forward, GTTTCTTAG-CACCATTTGCTGAGACTG; Reverse, TGTATTTGTATCG-ATTAACCAACTT) and D11S2000 (Forward, AGTAGAGA-ACAAAACACTGTGGC; Reverse, GTTTCTTTGAAGATC-TGTGAAATGTGC), were used for haplotype analysis. PCR products for each family member, with fluorescent-labeled primers, were genotyped at the National Instrumentation Center for Environmental Management (NICEM), Seoul National University, Korea. Haplotypes were then generated based upon allele transmission.
Western Blotting and Zymogram Analysis
Human MMP20 cDNA, generated by an RT-PCR reaction with pfu enzyme (Elpis Bio, Taejeon, Korea) and PCR primers (sense, CCTAAGCTTCTACTGTGAGGGGATGAAGG; antisense, CCT-CTAGATTTCTATTTAGCAACCAATCC; amplicon size: 1489 bp), was cloned into the pTOP blunt V2 vector (Enzynomics, Daejeon, Korea) and subsequently subcloned into the pcDNA3.1 mammalian expression vector after double-digestion with HindIII and XbaI restriction endonucleases. We performed PCR mutagenesis to replace G with A (c.910G>A) using primers (sense, CAGCTCATCCTTTGACACTGTGACAATGCTG; antisense, CAGCATTGTCACAGTGTCAAAGGATGAGCTG). Sequences of normal and mutant pcDNA3.1 vectors were confirmed by direct plasmid sequencing. Normal and mutant MMP20 pcDNA3.1 vectors were transiently transfected into HEK293 cells with lipofectamine plus reagent (Invitrogen, Carlsbad, CA, USA). Culture media (Dulbecco’s Modified Eagle Media) without serum were harvested and concentrated by means of an Amicon ultra-4 centrifugal filter unit (Millipore, Bedford, MA, USA).
Concentrated media were run on a 10% SDS-polyacry1amide gel and subjected to Western blot and zymogram analysis as described previously (Ozdemir et al., 2005a). Primary antibody (Y051377, anti-human MMP-20) was purchased from Applied Biological Materials, Inc. (Richmond, ON, Canada).
Results
Mutation Analyses
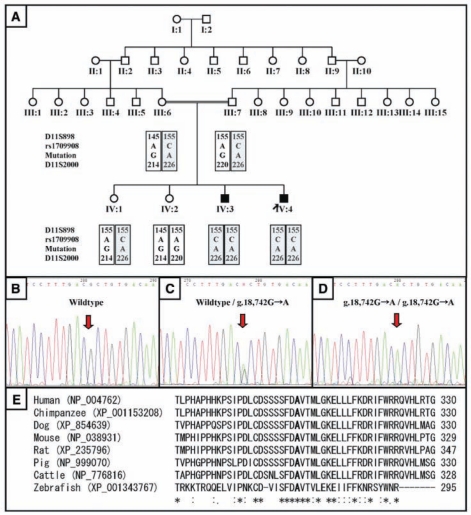
Mutational analyses were performed for candidate genes (MMP20, KLK4, and AMELX) for hypomaturation-type AI. No mutations were found in the KLK4 and AMELX genes. The identified mutation was g.18,742G>A in exon 6 (Figs. 1B-1D). This alteration changes the DNA codon for alanine (GCT) at amino acid position 304 to threonine (ACT), thus inducing the amino acid substitution p.A304T. This A304 is highly conserved among MMP20 sequences from human (NP_004762), chimpanzee (XP_001153208), dog (XP_854639), mouse (NP_038931), rat (XP_235796), pig (NP_999070), cattle (NP_776816), and zebrafish (XP_001343767) (Fig. 1E). The proband (IV:4) and his affected brother (IV:3) were both homozygous for this mutation. Both parents (III:6 and III:7) and one unaffected sister (IV:1) were carriers of the mutation. The other unaffected sister (IV:2) had the wild-type (normal) sequence in both alleles. Sequence analysis of 100 healthy normal control individuals did not reveal this sequence alteration, indicating that this mutation is not a common variation.
Figure 1.
Pedigrees, mutational analyses of the AI kindred, and alignment of amino acid sequences. (A) Pedigree and haplotype of the kindred with the (g.18,742G>A) MMP20 mutation. (B) DNA sequencing chromatogram of the normal individual IV:2. (C) DNA sequencing chromatogram of the carrier individual IV:1. (D) DNA sequencing chromatogram of the proband IV:4. (E) MMP20 amino acid sequence alignment of various species. Note the high conservation of the affected alanine (A) among the species.
Haplotype Analysis
It is confirmed that affected individuals have the same segment identical by descent. Haplotype analysis showed that the proband (IV:4) and his affected brother (IV:3) had inherited the same parental haplotypes and were homozygous for all sequence variations (Fig. 1A).
Western Blotting and Zymogram Analysis
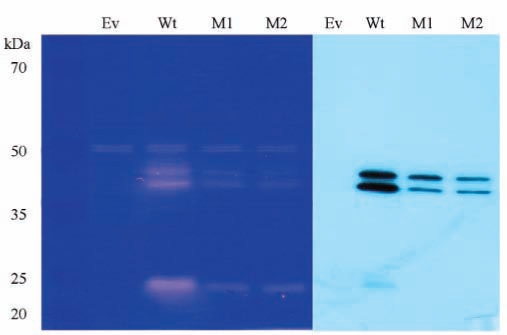
Western blotting analysis showed the decreased expression of mutant MMP20 compared with the wild-type protein. However, zymogram analysis revealed that the mutant MMP20 was functionally active (Fig. 4).
Figure 4.
Zymography and Western blot analysis. The zymogram revealed that mutant MMP20 was catalytically active; however, Western blot analysis showed that the amount of mutant MMP20 in the culture media was much less than that of normal MMP20. Lane Ev indicates empty expression vector only; Wt indicates normal MMP20; M1 and M2 indicate mutant MMP20.
Clinical Findings
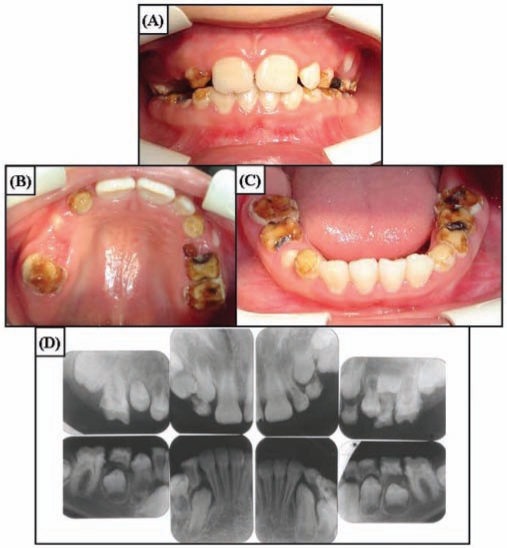
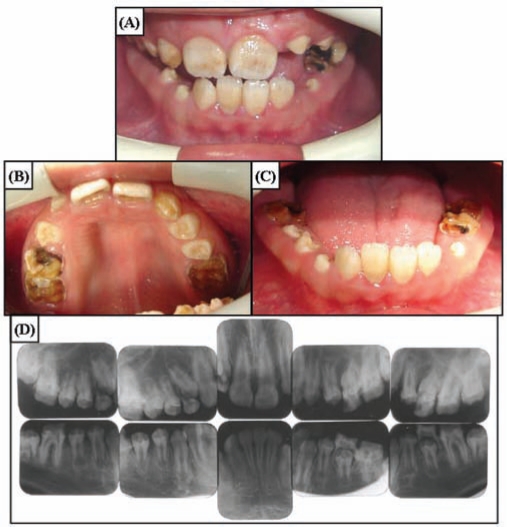
The newly erupted teeth of the 10-year-old proband showed chalky white hypomaturation enamel with minimal discoloration (Fig. 2). The enamel thickness of the newly erupted and unerupted teeth was within normal limits. The posterior teeth showed significant attrition that was secondary to the tendencies for the enamel to chip, to wear rapidly, and to develop dental caries. Radiographically, the teeth showed almost no contrast between dentin and enamel. The anterior teeth of the 12-year-old brother of the proband had darker pigmentation, which was reportedly acquired following tooth eruption (Fig. 3). Heterozygote carriers exhibited no apparent enamel defects.
Figure 2.
Clinical photographs and radiographs of proband (IV:4) at age 10 yrs. (A) Frontal view. Overjet and overbite are within normal limits. Anterior teeth have dull, chalky hypomatured enamel. (B) Maxillary occlusal view. Enamel breakdown can be seen in the permanent first molars and deciduous teeth. (C) Mandibular occlusal view. Anterior teeth have slight staining on the labial cervical portion. (D) Full-mouth intra-oral radiographs. Enamel has reduced radiopacity, but normal thickness can be seen in the unerupted teeth.
Figure 3.
Clinical photographs and radiographs of the affected brother of the proband (IV:3) at age 12 yrs. (A) Frontal view. Overjet and overbite are minimal, showing the edge-to-edge bite. Anterior teeth have dark brown pigmented areas. (B) Maxillary occlusal view. Enamel breakdown can be seen in the permanent first molars and deciduous teeth. Newly erupting premolars do not have pigmentation. (C) Mandibular occlusal view. Anterior teeth have dark staining. (D) Full-mouth intra-oral radiographs. Enamel has reduced radiopacity, but normal thickness can be seen in the newly erupted premolars.
Discussion
The clinical phenotype of hypomaturation AI in this study displays similarities to those reported previously (Hart et al., 2004; Kim et al., 2005b; Ozdemir et al., 2005a; Papagerakis et al., 2008). Enamel of normal thickness, but showing a reduced level of mineralization, was common in all reports of KLK4 and MMP20 mutations. The coloration of the anterior teeth was different between our two affected participants. The newly erupted teeth of the proband had dull, chalky white hypomaturation enamel with minimum discoloration in the cervical area of the lower anterior teeth. Anterior teeth of the affected brother had dark brown-pigmented areas; however, erupting premolars had features of hypomaturation enamel without pigmentation. From this finding, at least in this family, we attribute discoloration of the affected enamel to extrinsic rather than intrinsic staining.
This is the fourth report of an MMP20 mutation that causes autosomal-recessive AI. All 4 known mutations are point mutations, and each was found in both MMP20 alleles (a likely consequence of consanguinity) in affected individuals. The IVS6-2A>T mutation blocks expression at the mRNA level, precluding the synthesis and secretion of the MMP20 protein (Kim et al., 2005b). The p.H226Q substitution destroys the MMP20 active site. The protein was secreted, but was not catalytically active (Ozdemir et al., 2005a). The p.W34X nonsense mutation results in no functional MMP20 during tooth formation (Papagerakis et al., 2008). In contrast, the p.A304T substitution reported here is in the hemopexin domain. In this case, the MMP20 protein would potentially be expressed at normal levels, with an intact catalytic domain but a defective hemopexin domain. Another possibility is that the p.A304T substitution precluded proper folding of the hemopexin domain, leading to destruction of the entire protein by the endoplasmic-reticulum-associated degradation system (Romisch, 2005). To detect a mutational effect on MMP20 expression, we performed transient transfection and analyzed the amount and function of the mutant MMP20 by Western blotting and zymogram analyses. Data showed significantly decreased expression of mutant MMP20; however, we demonstrated that the mutant MMP20 was catalytically active.
Human enamelysin is a matrix metalloproteinase having 483 amino acids, including the signal peptide, propeptide, catalytic, linker, and hemopexin domains. The only post-translational modification is a disulfide bridge connecting the first and last amino acids of the hemopexin domain (Yamada et al., 2003). In the pig, active MMP20 migrates as a doublet at 46 and 41 kDa on casein zymograms (Fukae et al., 1998). Both bands are comprised of the catalytic, linker, and hemopexin domains, but two bands are observed because “nicking’ (cleavage that does not release a peptide because of the disulfide bridge connection) of the hemopexin domain causes a change in its mobility on SDS-PAGE. This cleavage site is the peptide bond connecting T335 and I336 (Yamada et al., 2003). The structural divisions of human MMP20 are: signal peptide M1 to A22; propeptide A23 to N107; catalytic domain Y108 to G271; linker P272 to L295; and hemopexin domain C296 to C483 (Yamada et al., 2003).
Enamelysin cleaves off its own hemopexin domain during in vitro digestion, confounding attempts to characterize how the hemopexin domain might influence cleavage site selection. The previous report used recombinant porcine MMP20, consisting primarily of the catalytic domain, to digest recombinant porcine amelogenin (rP172); it generated amelogenin cleavage products that exactly matched those previously identified by mass spectrometry analyses of porcine enamel extracts (Ryu et al., 1999). The major amelogenin cleavage products observed in developing pig enamel are the 20-kDa amelogenin (M1 to S148), the tyrosine-rich amelogenin peptide (TRAP; M1 to W45), extended TRAP (M1 to H62 or A63), and the 13- (L46 to S148) and 11- (L64 to S148) kDa amelogenins. The MMP20 catalytic domain generated these products, but also cleaved amelogenin after P105, P107, and P136. These cleavages are unlikely to occur frequently in vivo, since they would prevent accumulation of the 13- and 11-kDa amelogenins. Recent research has shown that the enamelysin catalytic domain cleaves recombinant ameloblastin at the same sites that occur in vivo, but additional cleavage sites are catalyzed that have either not yet been identified from in vivo extracts or are catalyzed only by the MMP20 catalytic domain lacking the hemopexin domain (Iwata et al., 2007). Current research therefore supports that enamelysin, even without its hemopexin domain, cleaves enamel proteins at the same sites as those that occur in vivo, but it is still plausible that the hemopexin domain may restrict cleavage site selection, and that such restriction may be critical for proper processing of the enamel matrix. The hemopexin domains of other matrix metalloproteinases help define substrate specificity (Gomis-Ruth, 2004).
A deeper analysis of the primary, secondary, and tertiary sequences of hemopexin domains from all MMPs and other hemopexin proteins revealed a unique hemopexin domain structure, which is composed of 4 blades, each composed of 4 β-strands. The affected alanine in this study is highly conserved among most of the MMP sequences, being located in the R2 position of the β1 strand, at the rim of blade I of the hemopexin domain. Moreover, the presence of this small hydrophobic residue is a repeated feature in the same position at the beginning of every blade of the domain and, in each of them, is the second residue out of 5 that configure the funnel-shaped tunnel formed in the hemopexin domain (Gomis-Ruth, 2004). This tunnel is responsible for cation binding, mainly Ca2+, and is very important for the structural integrity of the domain, as well as for ligand binding, as described for fibronectin, heparin, and the MMP2 hemopexin domain (Libson et al., 1995; Wallon and Overall, 1997).
Hemopexin domains in other MMPs have been attributed to several important functions. The hemopexin domain of Gelatinase A (MMP2) participates in the activation of the MMP-2 zymogen by binding tissue inhibitor of metalloproteinases 2 (TIMP2). The TIMP2/MMP2 complex docks with membrane-type MMP on the cell membrane, which excises the MMP2 propeptide (Overall et al., 1999). The hemopexin domain of Gelatinase B (MMP9) binds low-density lipoprotein-receptor-related protein 1 (LRP1), a cargo receptor that mediates removal of the protease from the matrix by endocytosis (Van den Steen et al., 2006). Hemopexin domains appear to serve different functions for different MMPs. Some bind to tissue inhibitors of metalloproteinases in an association that facilitates conversion of the zymogen to the active enzyme, determine substrate specificity, or assist in binding trafficking molecules that control the bioavailability of the enzyme.
Footnotes
This work was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A060010), a grant from the Korea Science and Engineering Foundation (KOSEF) through the Biotechnology R&D Program (No. M10646010003-08N4601-00310), a grant from the KOSEF through the Bone Metabolism Research Center (No. R11-2008-023-02003-0), and by NIDCR/NIH grant DE12769.
References
- Bartlett JD, Beniash E, Lee DH, Smith CE. (2004). Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. J Dent Res 83:909-913 [DOI] [PubMed] [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, et al. (2002). Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 277:49598-49604 [DOI] [PubMed] [Google Scholar]
- Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, et al. (1998). Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J Dent Res 77:1580-1588 [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F. (2004). Hemopexin domains. In: Handbook of metalloproteins. Vol. 3 Messerschmidt A, Bode W, Cygler M, editors. Chichester: John Wiley & Sons, Ltd, pp. 631-646 [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, et al. (2004). Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet 41:545-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Becerik S, Cogulu D, Emingil G, Ozdemir-Ozenen D, Han ST, et al. (2009). Novel FAM83H mutations in Turkish families with autosomal dominant hypocalcified amelogenesis imperfecta. Clin Genet 75:401-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, et al. (2007). Processing of ameloblastin by MMP-20. J Dent Res 86:153-157 [DOI] [PubMed] [Google Scholar]
- Kida M, Sakiyama Y, Matsuda A, Takabayashi S, Ochi H, Sekiguchi H, et al. (2007). A novel missense mutation (p.P52R) in amelogenin gene causing X-linked amelogenesis imperfecta. J Dent Res 86:69-72 [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hu YY, Lin BP-L, Boyd C, Wright JT, et al. (2004). Amelogenin p.M1T and p.W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J Dent Res 83:378-383 [DOI] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, et al. (2005a). ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res 84:278-282 [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, et al. (2005b). MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet 42:271-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, et al. (2008). FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet 82:489-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hu JC, Bartlett JD, Lee KE, Lin BP, Simmer JP, et al. (2008). Mutational spectrum of FAM83H: the C-terminal portion is required for tooth enamel calcification. Hum Mutat 29:E95-E99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libson AM, Gittis AG, Collier IE, Marmer BL, Goldberg GI, Lattman EE. (1995). Crystal structure of the haemopexin-like C-terminal domain of gelatinase A. Nat Struct Biol 2:938-942 [DOI] [PubMed] [Google Scholar]
- Overall CM, King AE, Bigg HF, McQuibban A, Atherstone J, Sam DK, et al. (1999). Identification of the TIMP-2 binding site on the gelatinase A hemopexin C-domain by site-directed mutagenesis and the yeast two-hybrid system. Ann NY Acad Sci 878:747-753 [DOI] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, et al. (2005a). MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res 84:1031-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. (2005b). Phenotype of ENAM mutations is dosage-dependent. J Dent Res 84:1036-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagerakis P, Lin HK, Lee KY, Hu Y, Simmer JP, Bartlett JD, et al. (2008). Premature stop codon in MMP20 causing amelogenesis imperfecta. J Dent Res 87:56-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romisch K. (2005). Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol 21:435-456 [DOI] [PubMed] [Google Scholar]
- Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. (1999). Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res 78:743-750 [DOI] [PubMed] [Google Scholar]
- Van den Steen PE, Van Aelst I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, et al. (2006). The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J Biol Chem 281:18626-18637 [DOI] [PubMed] [Google Scholar]
- Wallon UM, Overall CM. (1997). The hemopexin-like domain (C domain) of human gelatinase A (matrix metalloproteinase-2) requires Ca2+ for fibronectin and heparin binding. Binding properties of recombinant gelatinase A C domain to extracellular matrix and basement membrane components. J Biol Chem 272:7473-7481 [DOI] [PubMed] [Google Scholar]
- Witkop CJ., Jr (1988). Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol 17:547-553 [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Jr, Sauk JJ., Jr (1976). Heritable defects of enamel. In: Oral facial genetics. Stewart RE, Prescott GH, editors. St. Louis: C.V. Mosby Co, pp. 151-226 [Google Scholar]
- Yamada Y, Yamakoshi Y, Gerlach RF, Hu JC-C, Matsumoto K, Fukae M, et al. (2003). Purification and characterization of enamelysin from secretory stage pig enamel. Arch Comp Biol Tooth Enam 8:21-25 [Google Scholar]