Abstract
It has not been established whether transmission of mutans streptococci occurs between unrelated children older than 4 years of age. The aim of the study was to investigate the possible transmission of mutans streptococci genotypes from child to child in kindergarten. We studied 96 children (ages 5-6 yrs) in three San Francisco Bay Area public schools. Mutans streptococci colonies from each child were isolated from selective culture on Mitis Salivarius Sucrose Bacitracin agar. We used arbitrary primed polymerase chain reactions to determine the mutans streptococci genotypes. Two children (not siblings) in each of the three schools (6%) shared an identical amplitype of S. mutans, unique to each pair. The 19 S. sobrinus amplitypes were found in 12 children, and all were unique to each child. The presence of matching genotypes of S. mutans demonstrates horizontal transmission of this species between unrelated children aged 5-6 years.
Keywords: horizontal transmission, diversity, mutans streptococci, children
Introduction
Dental caries is an infectious and transmissible disease in which mutans streptococci play a major role. Early acquisition of mutans streptococci has been shown in many studies to be a major risk factor for early childhood caries and future caries experience (reviewed in Berkowitz, 2006). Vertical transmission from mother to child has been suggested as the main pathway for mutans streptococci acquisition. Several studies reported similar mutans streptococci genotypes common to mother, father, and child (Redmo Emanuelsson and Wang, 1998; van Loveren et al., 2000; Hames-Kocabas et al., 2008). Recent studies have suggested that horizontal transmission occurs readily within families (Saarela et al., 1993; Redmo Emanuelsson and Wang, 1998; Kozai et al., 1999; Emanuelsson and Thornqvist, 2000; van Loveren et al., 2000; Redmo Emanuelsson and Thornqvist, 2001; Nie et al., 2002; Ersin et al., 2004). To our knowledge, there are only three reports of transmission between unrelated nursery school children ranging in age from 2 mos to 4 yrs (Mattos-Graner et al., 2001; Tedjosasongko and Kozai, 2002; Liu et al., 2007).
The aim of the present study was to investigate mutans streptococci diversity and transmission among children aged 5-6 yrs. The hypothesis to be tested was that transmission of mutans streptococci genotypes occurs between non-genetically-related children aged 5 and 6 yrs.
Materials & Methods
Study Population and Study Design
Kindergarten children aged 5-6 yrs participating in a longitudinal investigation of social status, biological responses to adversity, and child health in San Francisco Bay Area schools were recruited for a dental health study. The study was approved by the Committee on Human Research at the University of California, San Francisco and the University of California, Berkeley. A cohort of 96 children attending three public schools in the San Francisco East Bay Area was recruited, with one dropping out prior to saliva sampling. Exclusion criteria were: (a) the presence of cardiovascular disorders, (b) the presence of mental disabilities, and (c) if the child or parents were non-English-speaking. Written informed consent was obtained from the parents. There were five pairs of siblings (all twins) that were recruited. All twins were in separate classrooms. No other children were genetically related. Children were together in class for 4 to 8 mos before the dental study commenced. A dental examination was conducted in each child, and caries status was recorded. A swab saliva sample was also collected for each child for mutans streptococci enumeration. Children with positive mutans streptococci colonization were included in the study.
Clinical Procedures
A dental examination was performed on each child by one examiner using a head-light, dental mirror, and explorer. The dmfs/DMFS (d/D = decayed, m/M = missing, f/F = filled tooth surfaces; lower-case letters indicate primary dentition, while upper-case letters indicate permanent dentition) and dmft/DMFT (decayed, missing, and filled tooth number) scores were recorded according to the WHO criteria. Oral bacteria were sampled with the use of a cotton-tipped applicator that was swabbed over the buccal mucosa, gingival margin, and tongue and tooth surfaces until saturated with saliva. This is a reliable sampling method for this age group, and we previously cross-calibrated it with sampling using expectorated saliva (unpublished observations). The saliva-soaked tip was placed in a sterile tube containing 2 mL of phosphate-buffered saline (PBS). The tube was then uniquely coded, chilled on ice, and transferred for microbiological assay within 24 hrs.
Bacteria Enumeration, Mutans Streptococci Isolation, and Storage
The swab samples were vortexed for 30 sec, serially diluted in PBS, plated on Mitis Salivarius Sucrose Bacitracin agar (MSSB, Difco, Detroit, MI, USA), and incubated anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C for 72 hrs. Mutans streptococci were enumerated under a dissecting microscope based on their colony morphology. The swabs were dried in a 60°C incubator overnight and weighed to obtain the saliva volume for CFU/mL mutans streptococci calculation.
Five colonies, typical of mutans streptococcal colonial morphology on MSSB agar, were subcultured from each child. A single colony from each plate was then inoculated on MSSB for 48 hrs anaerobically, harvested, suspended in TSB broth with 20% glycerol, and stored at -80°C for later assays.
S. mutans and S. sobrinus Identification and DNA
Stored mutans streptococci isolates from each child were recovered on MSSB plates and inoculated into 2 mL of TPY (1.5% tryptone, 0.4% polypeptone, 0.4% yeast extract, 1% glucose, 0.5% KH2PO4, 0.25% K2HPO4, 0.2% Na2CO3, and 0.2% NaCl), then incubated anaerobically for 18 hrs. Then a 0.1-mL quantity of bacteria suspension was dropped onto an Indicating FTA® Classic Card (Whatman BioScience, Newton, MA, USA) to save bacterial DNA. The card was dried overnight at room temperature and stored for AP-PCR.
Differentiation of S. mutans and S. sobrinus was based on standard fermentation tests with sorbitol, mannitol, melibiose, and raffinose (Shklair and Keene, 1974).
DNA Extraction and Arbitrarily Primed Polymerase Chain Reaction (AP-PCR)
A 2-mm-diameter punch was removed from the dried Indicating FTA® Classic Card for each strain and was washed 3 times in 200 µL of FTA Purification Reagent (Whatman BioScience) and twice with 1X TE Buffer. Punches were then dried in a 56°C incubator for 10 min and used for AP-PCR assay as DNA templates within 24 hrs.
The AP-PCR reaction was performed as previously described (Saarela et al., 1996). Two primers were used for each isolate (primer OPA-5, sequence 5′-AGGGGTCTTG-3′, and primer OPA013, sequence 5′-CAGCACCCAC-3′; Gibco BRL, Rockville, CA, USA). A 12-µL quantity of the AP-PCR product was separated by electrophoresis in Mupid-2 Mini Gel Migration Through (Cosmo Bio Co. LTD, Tokyo, Japan) on a 1.0 % agarose gel stained with ethidium bromide for 25 min at 100 V, against 1 Kb Plus DNA Ladder (Invitrogen, Carlsbad, CA, USA). The electrophoresis gels were then photographed by an electrophoresis gel imaging system (DigiDoc-It® Imaging System, UVP, Upland, CA, USA). The electrophoresis patterns of each mutans streptococci isolate were then compared within children and between children on gel images by a side-by-side visual comparison by two operators. All AP-PCR patterns were compared when the AP-PCR reactions were conducted during the same run. Fingerprints were considered similar when all major bands were identical. AP-PCR with primer OPA 05 (Operon Technologies, Huntsville, AL, USA) was performed on all mutans streptococci isolates. The isolates with the same pattern were then tested by primer OPA 13 for further comparison. The number of different amplitypes in each child was determined following comparison of the AP-PCR patterns of isolates from each child with these two different primers. We used the above two primers to confirm the uniqueness of each genotype and to eliminate false-positives that could occur with just one primer.
Statistical Analysis
The bacteria counts were converted to log10 (bacteria count +1) for statistical analysis. The means and standard deviation were calculated for each of the caries scores and log values of bacteria counts. Relationships were not analyzed statistically between transmitted and non-transmitted groups, due to the small sample size of children with transmission.
Results
Study Population Demographics, Dental Caries Status, and Mutans Streptococci Levels
The children’s ages ranged from 5.2 to 6.5 yrs (38 girls and 58 boys; 41 white, 49 non-white, six other) from eight kindergarten classrooms in three public schools. The mean annual household income of the children’s parents ranged from $60,000-$80,000. Parental education levels ranged from grade school to professional degree with a median of college graduation. The children were in class about 3.5-4 hrs per day for 5 days a wk. Five pairs of twins were included in the study population, and all other children were genetically unrelated.
The mean ± SD of dmfs/DMFS and ds/DS of the study population were 4.47 ± 7.89 and 1.56 ± 3.29, respectively. Forty-six (48%) children had dmfs/DMFS > 0 (range, 1-38), and 34 (35%) children had unfilled dental caries. The mean ± SD of dmfs/DMFS and ds/DS of 34 children who had unfilled dental caries were 9.24 ± 9.22 and 4.35 ± 4.28, respectively.
Forty-seven (49%) of the 95 children sampled had detectable mutans streptococci and were included in the study. The mean ± SD of log10 (mutans streptococci) CFU/mL was 3.66 ± 1.08 in the 47 mutans-streptococci-infected children. Thirty-five children (74%) with detectable mutans streptococci had dmfs/DMFS > 0, and 28 (60%) of them had unfilled caries. Among the 48 non-mutans-streptococci-infected children, 38 (79%) had a zero dmfs/DMFS score, and 43 (90%) had zero unfilled caries surfaces.
Mutans Streptococci Diversity
In total, 224 mutans streptococci isolates were collected and genotyped by AP-PCR. Typical AP-PCR results with OPA 05 and OPA 13 for one child are shown in Fig. 1. Based on fermentation and AP-PCR results, there were 69 S. mutans amplitypes and 19 S. sobrinus amplitypes identified. The mean ± SD number of mutans streptococci amplitypes per child was 1.85 ± 0.78 (range, 1-3) for the 47 mutans-streptococci-positive children. Among the 47 mutans-streptococci-infected children, 29 (62%) had ≥ 2 mutans streptococci amplitypes. Six children (13%) had both S. mutans and S. sobrinus. Thirty-five children (74%) had only S. mutans, while the other six (13%) had only S. sobrinus. Log(mutans streptococci) and mutans streptococci diversity are presented in Table 1.
Figure 1.
Typical AP-PCR results in one child. Each lane represents 1 of 5 different isolates. (A) Primer OPA 05. (B) Primer OPA 13. M, marker.
Table 1.
Salivary Mutans Streptococci (MS) Levels and Mutans Streptococci Diversity of Children with Mixed or Single S. mutans and S. sobrinus Infection
| Mutans Streptococci Infection | Log MS Mean ± SD | Number of MS Amplitypes per Child Mean ± SD |
|---|---|---|
| S. mutans only (n = 35) | 3.40 ± 1.20 | 1.74 ± 0.74 |
| S. sobrinus only (n = 6) | 4.21 ± 1.27 | 1.83 ± 0.98 |
| S. mutans + S. sobrinus (n = 6) | 4.10 ± 0.92 | 2.67 ± 0.52 |
| S. mutans 1.33 ± 0.52 | ||
| S. sobrinus 1.33 ± 0.52 |
Sharing of Mutans Streptococci in Non-related Children and Siblings
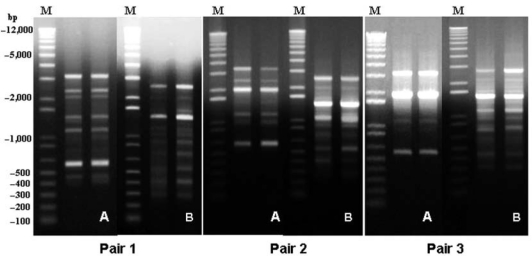
Among the 47 mutans-streptococci-infected children, one pair in each of the three schools shared an identical amplitype of S. mutans, unique to each pair (Fig. 2). Children 1 and 2 (Table 2) were from different classrooms in the same school, while the other two pairs (children 3 and 4 and children 5 and 6) were in the same classroom. None of the children who carried identical amplitypes were genetically related. No sharing of S. sobrinus amplitypes was found in this study.
Figure 2.
AP-PCR gel of three pairs of children sharing S. mutans amplitypes. One lane from each child is shown for each pair. (A) Primer OPA 05. (B) Primer OPA 13. M, marker.
Table 2.
Salivary Bacterial Levels, Caries Status, and Mutans Streptococci (MS) Diversity of Children with Shared S. mutans Amplitypes (In pair 1, child #2 was the twin.)
| Part 1 |
Part 2 |
Part 3 |
||||
|---|---|---|---|---|---|---|
| Child 1 | Child 2 | Child 3 | Child 4 | Child 5 | Child 6 | |
| LogMS | 4.81 | 2.57 | 2.48 | 2.95 | 2.83 | 3.85 |
| dmfs/DMFS | 3 | 0 | 9 | 15 | 10 | 6 |
| ds/DS | 0 | 0 | 1 | 1 | 10 | 6 |
| # of S. mutans amplitypes/child | 1 | 1 | 1 | 1 | 2 | 3 |
| # of S. sobrinus amplitypes/child | 0 | 0 | 0 | 0 | 0 | 0 |
Of the five pairs of twins in the study, only one child had detectable mutans streptococci with only one S. mutans present. The twin who carried the S. mutans amplitype shared it with a non-genetically-related child from another classroom in the same school (pair 1, Table 2).
Discussion
In the present study, we have demonstrated genotypic diversity of mutans streptococci. Further, analysis of the data suggests horizontal transmission between non-genetically-related children aged 5 and 6 yrs in the United States. Matching genotypes of S. mutans in one pair of children in each of three schools were identified.
Several methods have been used for the investigation of mutans streptococci transmission, including phenotypic characterizations, serotypic and genotypic characterizations, plasmid DNA profiling, restriction endonuclease analysis, ribotyping, and AP-PCR. Characterization of oral bacteria by AP-PCR or DNA fingerprinting has become well-accepted (Saarela et al., 1996; Li and Caufield, 1998; Gronroos and Alaluusua, 2000; Li et al., 2001; Toi et al., 2003; Ersin et al., 2004; Klein et al., 2004; Liu et al., 2004; Napimoga et al., 2004; Nascimento et al., 2004; Kamiya et al., 2005; Li et al., 2005; Cogulu et al., 2006; Liu et al., 2007; Hames-Kocabas et al., 2008). Moreover, AP-PCR has been shown to have good discriminating ability for mutans streptococci differentiation (Saarela et al., 1996). The choice of the primers (OPA 05 and OPA 13) used in the present study was based on previous studies where they were used either independently (Gronroos and Alaluusua, 2000; Toi et al., 2003; Ersin et al., 2004; Klein et al., 2004; Napimoga et al., 2004; Nascimento et al., 2004; Kamiya et al., 2005; Cogulu et al., 2006; Hames-Kocabas et al., 2008) or in combination (Saarela et al., 1996). The use of two primers for AP-PCR increases the sensitivity and specificity to identify different mutans streptococci amplitypes. The analysis of AP-PCR fingerprints by side-by-side visual comparison has been used in most studies (Gronroos and Alaluusua, 2000; Li et al., 2001; Toi et al., 2003; Ersin et al., 2004; Cogulu et al., 2006; Liu et al., 2007; Hames-Kocabas et al., 2008).
Initial acquisition and transmission of mutans streptococci have been widely studied. Mother-child transmission has been identified as the major route for early-infancy mutans streptococci acquisition. However, the reported maternal transmission rates range from 24% (Hames-Kocabas et al., 2008) to 100% in eight Turkish families (Ersin et al., 2004), clearly demonstrating that mothers are not the only source of mutans streptococci transmission.
Others have reported transmission between family members, such as spouse-spouse, father-child, and sibling-sibling (Saarela et al., 1993; Li and Caufield, 1998; Redmo Emanuelsson and Wang, 1998; Kozai et al., 1999; Emanuelsson and Thornqvist, 2000; van Loveren et al., 2000; Redmo Emanuelsson and Thornqvist, 2001; Nie et al., 2002; Tedjosa-songko and Kozai, 2002; Ersin et al., 2004; Hames-Kocabas et al., 2008). Although there have been three reports of transmission between unrelated young infants in nursery schools in China, Brazil, and Japan (Mattos-Graner et al., 2001; Tedjosasongko and Kozai, 2002; Liu et al., 2007), these children ranged in age from 2 mos to 4 yrs, when salivary transfer is more likely than in older children. To our knowledge, transmission from child-child outside of families for children older than nursery school has not been previously reported in the United States or elsewhere.
In the current study, we identified three pairs of 5- to 6-year-old non-genetically-related children, in three different schools, who shared S. mutans amplitypes (each unique to the pair of children). These results indicate child-child transmission of mutans streptococci in kindergartners in the United States. The suggested horizontal transmission rate in this population was relatively low, namely, 6.3% overall, and 13% among the mutans-streptococci-infected children. This means that one in six children was apparently infected by horizontal transmission. However, the children in the study population were enrolled in the schools for only 4-8 mos prior to saliva sampling. Finding one pair in each of the three different schools (and towns) indicates that horizontal transmission was not a chance occurrence.
Our study is the first to indicate horizontal transmission of S. mutans in kindergartners aged 5-6 yrs. The low transmission rate may be explained by short contact time and less intimate contact, such as sharing food or utensils, as compared with the nursery-school children. Besides the intimacy of contact, there are other factors that could potentially relate to transmission. In the present study, the children who carried identical amplitypes all had moderate to high levels of mutans streptococci. All of them had only S. mutans infection, and only one pair of children had multiple mutans streptococci amplitypes. All children but one had dental caries experience. However, the limited data make it difficult for definite conclusions to be drawn about the reasons for the indicated horizontal mutans streptococci transmission.
No data or samples were collected from the caregivers of these children, so we cannot exclude the possibility of mutans streptococci infection from a mutual caregiver. However, it is unlikely that this would occur for all three pairs in three different schools.
The strongly indicated horizontal mutans streptococci transmission between children aged 5-6 yrs in the present study illustrates the necessity of further prospective studies to explore the frequency, nature, and risk factors associated with horizontal cariogenic bacterial transmission between and among children of different age groups. Measures need to be found to break the chain of infection from child-child as part of caries-preventive therapies.
Footnotes
This investigation was supported by NIH Grant RO1 MH62320-03S1.
References
- Berkowitz RJ. (2006). Mutans streptococci: acquisition and transmission. Pediatr Dent 28:106-109 [PubMed] [Google Scholar]
- Cogulu D, Sabah E, Uzel A, Ozkinay F. (2006). Genotyping of Streptococcus mutans by using arbitrarily primed polymerase chain reaction in children with Down syndrome. Arch Oral Biol 51:177-182 [DOI] [PubMed] [Google Scholar]
- Emanuelsson IR, Thornqvist E. (2000). Genotypes of mutans streptococci tend to persist in their host for several years. Caries Res 34:133-139 [DOI] [PubMed] [Google Scholar]
- Ersin NK, Kocabas EH, Alpoz AR, Uzel A. (2004). Transmission of Streptococcus mutans in a group of Turkish families. Oral Microbiol Immunol 19:408-410 [DOI] [PubMed] [Google Scholar]
- Gronroos L, Alaluusua S. (2000). Site-specific oral colonization of mutans streptococci detected by arbitrarily primed PCR fingerprinting. Caries Res 34:474-480 [DOI] [PubMed] [Google Scholar]
- Hames-Kocabas EE, Ucar F, Kocatas Ersin N, Uzel A, Alpoz AR. (2008). Colonization and vertical transmission of Streptococcus mutans in Turkish children. Microbiol Res 163:168-172 [DOI] [PubMed] [Google Scholar]
- Kamiya RU, Napimoga MH, Rosa RT, Höfling JF, Gonçalves RB. (2005). Mutacin production in Streptococcus mutans genotypes isolated from caries-affected and caries-free individuals. Oral Microbiol Immunol 20:20-24 [DOI] [PubMed] [Google Scholar]
- Klein MI, Flório FM, Pereira AC, Höfling JF, Gonçalves RB. (2004). Longitudinal study of transmission, diversity, and stability of Streptococcus mutans and Streptococcus sobrinus genotypes in Brazilian nursery children. J Clin Microbiol 42:4620-4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozai K, Nakayama R, Tedjosasongko U, Kuwahara S, Suzuki J, Okada M, et al. (1999). Intrafamilial distribution of mutans streptococci in Japanese families and possibility of father-to-child transmission. Microbiol Immunol 43:99-106 [DOI] [PubMed] [Google Scholar]
- Li S, Liu T, Xiao X, Yang J, Yang D, Zhuang H, et al. (2005). Detection of mutA genes in transmitted strains and nontransmitted strains of mutans streptococci. Caries Res 39:417-421 [DOI] [PubMed] [Google Scholar]
- Li Y, Caufield PW. (1998). Arbitrarily primed polymerase chain reaction fingerprinting for the genotypic identification of mutans streptococci from humans. Oral Microbiol Immunol 13:17-22 [DOI] [PubMed] [Google Scholar]
- Li Y, Caufield PW, Emanuelsson IR, Thornqvist E. (2001). Differentiation of Streptococcus mutans and Streptococcus sobrinus via genotypic and phenotypic profiles from three different populations. Oral Microbiol Immunol 16:16-23 [DOI] [PubMed] [Google Scholar]
- Liu J, Bian Z, Fan M, He H, Nie M, Fan B, et al. (2004). Typing of mutans streptococci by arbitrarily primed PCR in patients undergoing orthodontic treatment. Caries Res 38:523-529 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zou J, Shang R, Zhou XD. (2007). Genotypic diversity of Streptococcus mutans in 3- to 4-year-old Chinese nursery children suggests horizontal transmission. Arch Oral Biol 52:876-881 [DOI] [PubMed] [Google Scholar]
- Mattos-Graner RO, Li Y, Caufield PW, Duncan M, Smith DJ. (2001). Genotypic diversity of mutans streptococci in Brazilian nursery children suggests horizontal transmission. J Clin Microbiol 39:2313-2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napimoga MH, Kamiya RU, Rosa RT, Rosa EA, Höfling JF, Mattos-Graner RO, et al. (2004). Genotypic diversity and virulence traits of Streptococcus mutans in caries-free and caries-active individuals. J Med Microbiol 53(Pt 7):697-703 [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Höfling JF, Gonçalves RB. (2004). Streptococcus mutans genotypes isolated from root and coronal caries. Caries Res 38:454-463 [DOI] [PubMed] [Google Scholar]
- Nie M, Fan M, Bian Z. (2002). Transmission of mutans streptococci in adults within a Chinese population. Caries Res 36:161-166 [DOI] [PubMed] [Google Scholar]
- Redmo Emanuelsson IM, Wang XM. (1998). Demonstration of identical strains of mutans streptococci within Chinese families by genotyping. Eur J Oral Sci 106:788-794 [DOI] [PubMed] [Google Scholar]
- Redmo Emanuelsson IM, Thornqvist E. (2001). Distribution of mutans streptococci in families: a longitudinal study. Acta Odontol Scand 59:93-98 [DOI] [PubMed] [Google Scholar]
- Saarela M, Von Troil-Lindén B, Torkko H, Stucki AM, Alaluusua S, Jousimies-Somer H, et al. (1993). Transmission of oral bacterial species between spouses. Oral Microbiol Immunol 8:349-354 [DOI] [PubMed] [Google Scholar]
- Saarela M, Hannula J, Matto J, Asikainen S, Alaluusua S. (1996). Typing of mutans streptococci by arbitrarily primed polymerase chain reaction. Arch Oral Biol 41:821-826 [DOI] [PubMed] [Google Scholar]
- Shklair IL, Keene HJ. (1974). A biochemical scheme for the separation of the five varieties of Streptococcus mutans. Arch Oral Biol 19:1079-1081 [DOI] [PubMed] [Google Scholar]
- Tedjosasongko U, Kozai K. (2002). Initial acquisition and transmission of mutans streptococci in children at day nursery. ASDC J Dent Child 69:284-288 [PubMed] [Google Scholar]
- Toi CS, Bonecker M, Cleaton-Jones PE. (2003). Mutans streptococci strains prevalence before and after cavity preparation during Atraumatic Restorative Treatment. Oral Microbiol Immunol 18:160-164 [DOI] [PubMed] [Google Scholar]
- van Loveren C, Buijs JF, ten Cate JM. (2000). Similarity of bacteriocin activity profiles of mutans streptococci within the family when the children acquire the strains after the age of 5. Caries Res 34:481-485 [DOI] [PubMed] [Google Scholar]