Abstract
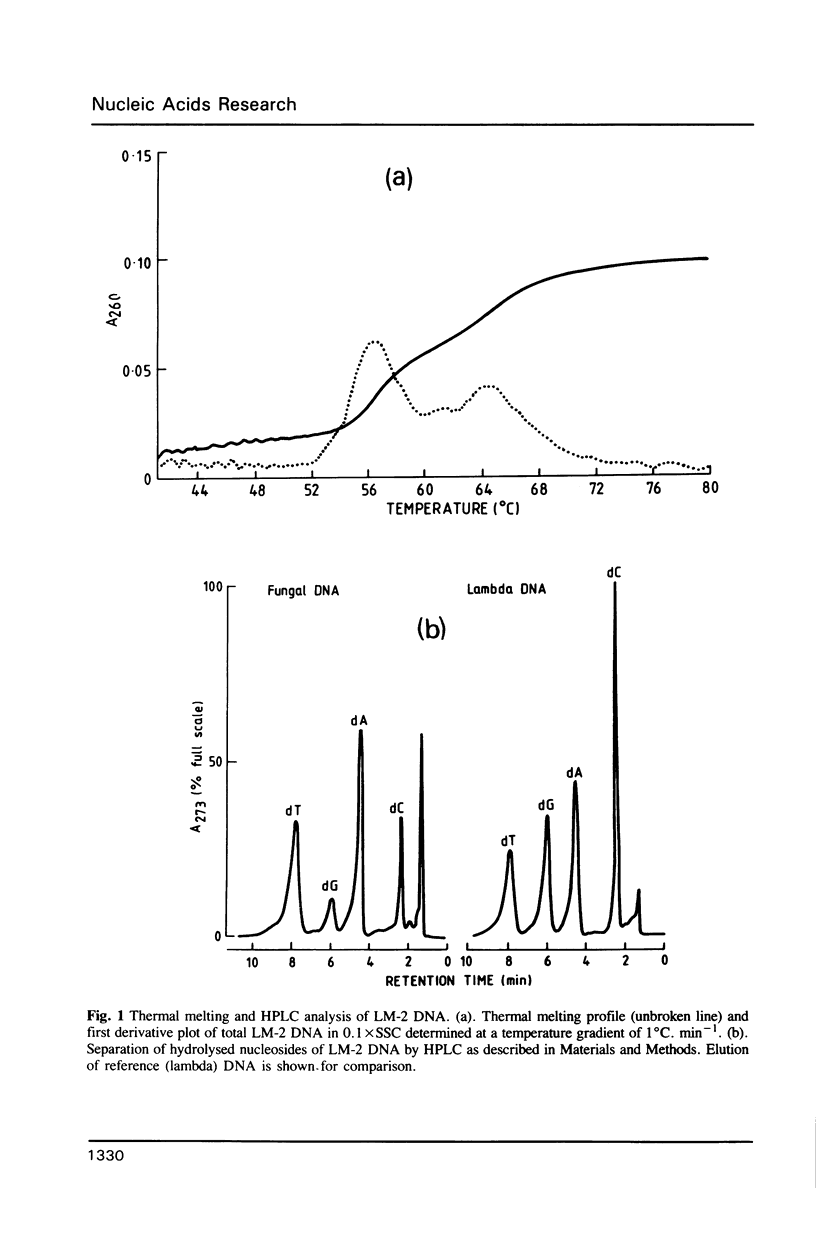
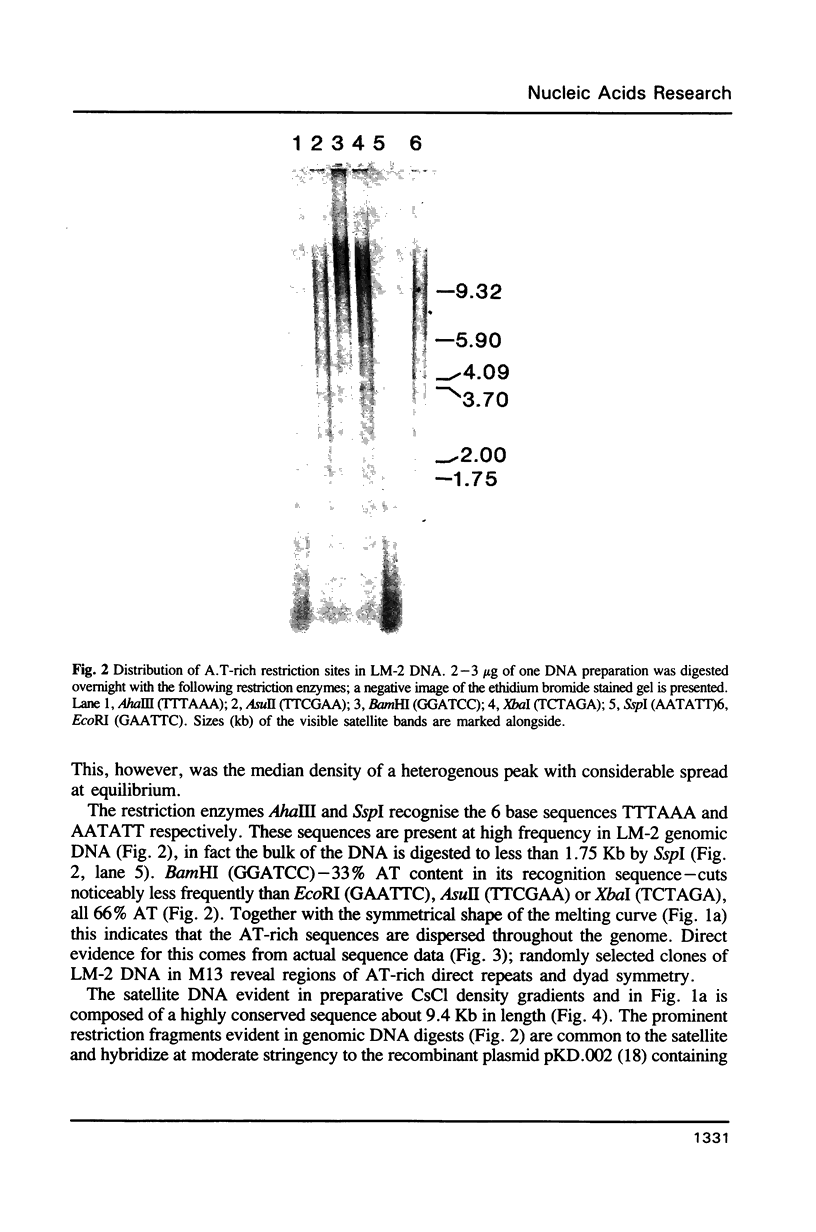
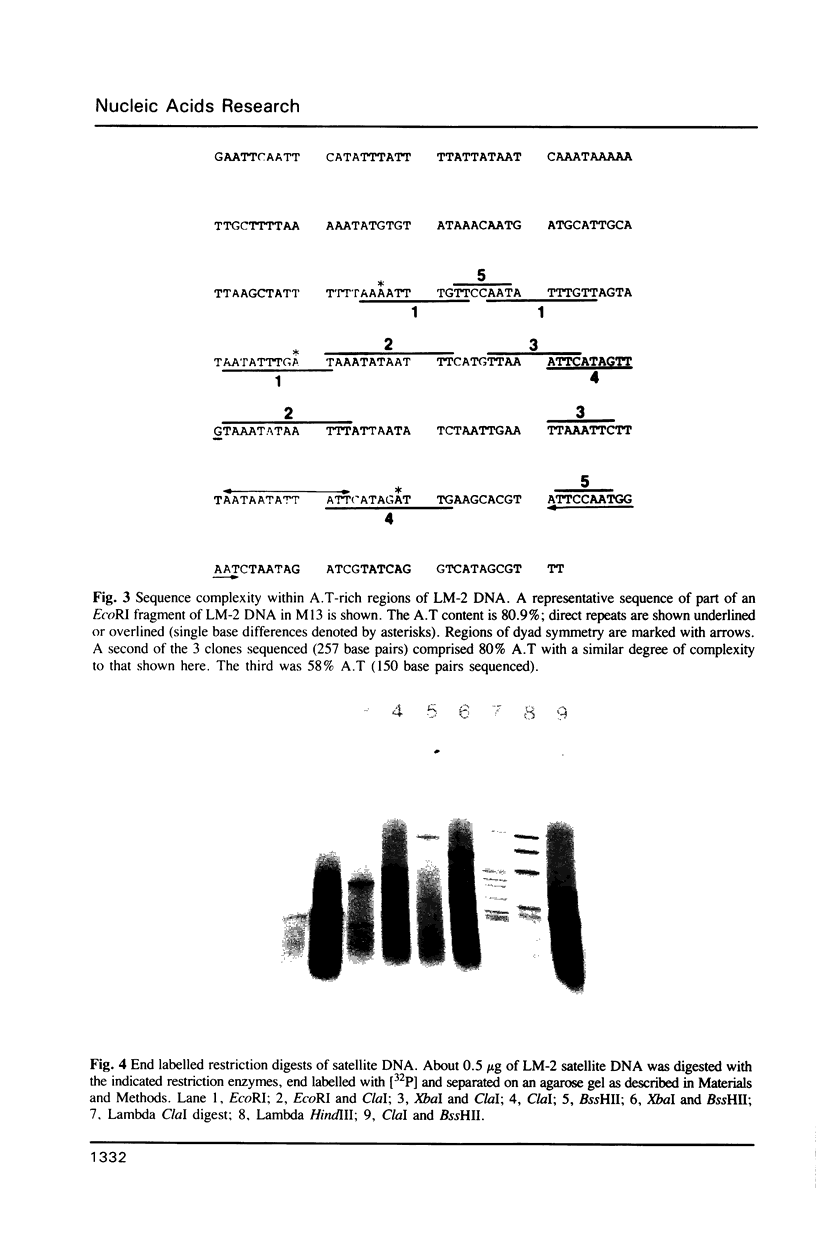
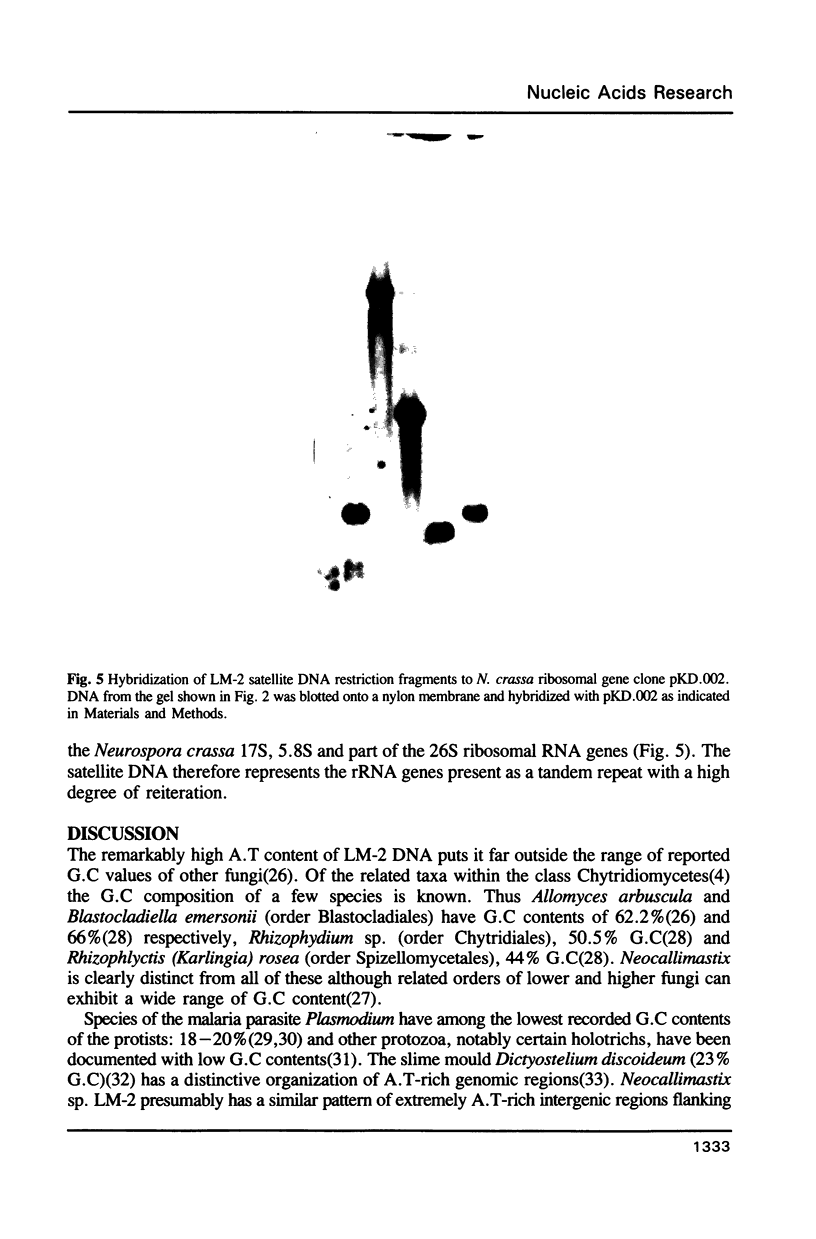
The genomic DNA of an anaerobic rumen phycomycete of the genus Neocallimastix has been purified and characterized. The non-repetitive fraction of the DNA has a G.C content of only 13%. The ribosomal RNA genes are highly reiterated, making up about 30% of the total DNA, and are evident as a more G.C-rich satellite with a repeating unit of about 9.4 kilobases (Kb). A.T-rich regions of DNA are highly dispersed and possess some sequence complexity. Chemical analysis of the DNA constituents reveals no evidence of modified bases. The genome of this anaerobic fungus has the highest A.T content of any organism so far described.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akin D. E., Gordon G. L., Hogan J. P. Rumen bacterial and fungal degradation of Digitaria pentzii grown with or without sulfur. Appl Environ Microbiol. 1983 Sep;46(3):738–748. doi: 10.1128/aem.46.3.738-748.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F., Tamame M., Villanueva J. R., Santos T. DNA methylation in the fungi. J Biol Chem. 1984 Jul 10;259(13):8033–8036. [PubMed] [Google Scholar]
- Barr D. J. How modern systematics relates to the rumen fungi. Biosystems. 1988;21(3-4):351–356. doi: 10.1016/0303-2647(88)90032-9. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Codon usage and genome composition. J Mol Evol. 1985;22(4):363–365. doi: 10.1007/BF02115693. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Eick D., Fritz H. J., Doerfler W. Quantitative determination of 5-methylcytosine in DNA by reverse-phase high-performance liquid chromatography. Anal Biochem. 1983 Nov;135(1):165–171. doi: 10.1016/0003-2697(83)90746-7. [DOI] [PubMed] [Google Scholar]
- Elton R. A. The relationship of DNA base composition and individual protein composition in micro-organisms. J Mol Evol. 1973 Nov 27;2(4):263–276. doi: 10.1007/BF01654095. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Firtel R. A., Bonner J. Characterization of the genome of the cellular slime mold Dictyostelium discoideum. J Mol Biol. 1972 May 28;66(3):339–361. doi: 10.1016/0022-2836(72)90419-6. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., McCune R. A., Gama-Sosa M. A., Ehrlich M., Kuo K. C. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984 Sep 28;301(1):199–219. doi: 10.1016/s0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24(1-2):39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Sequence organization in Dictyostelium: unique structure at the 5'-ends of protein coding genes. Nucleic Acids Res. 1983 Jan 25;11(2):541–552. doi: 10.1093/nar/11.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P. DNA base sequence complementarity and the definition of fungal taxa. Microbiol Sci. 1984 May;1(2):44–48. [PubMed] [Google Scholar]
- Mandel M., Igambi L., Bergendahl J., Dodson M. L., Jr, Scheltgen E. Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J Bacteriol. 1970 Feb;101(2):333–338. doi: 10.1128/jb.101.2.333-338.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T. F., Dame J. B., Miller L. H., Barnwell J. Evolutionary relatedness of Plasmodium species as determined by the structure of DNA. Science. 1984 Aug 24;225(4664):808–811. doi: 10.1126/science.6382604. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Gautier F. Interactions of heteroaromatic compounds with nucleic acids. A - T-specific non-intercalating DNA ligands. Eur J Biochem. 1975 Jun;54(2):385–394. doi: 10.1111/j.1432-1033.1975.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Yamao F., Muto A., Jukes T. H., Ozeki H., Umesono K. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. W., Gordon G. L. Sugar and polysaccharide fermentation by rumen anaerobic fungi from Australia, Britain and New Zealand. Biosystems. 1988;21(3-4):377–383. doi: 10.1016/0303-2647(88)90036-6. [DOI] [PubMed] [Google Scholar]
- Pollack Y., Katzen A. L., Spira D. T., Golenser J. The genome of Plasmodium falciparum. I: DNA base composition. Nucleic Acids Res. 1982 Jan 22;10(2):539–546. doi: 10.1093/nar/10.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. J., Wagner S., Rodland K. D., Feinbaum R. L., Russell J. P., Bret-Harte M. S., Free S. J., Metzenberg R. L. Organization of the ribosomal ribonucleic acid genes in various wild-type strains and wild-collected strains of Neurospora. Mol Gen Genet. 1984;196(2):275–282. doi: 10.1007/BF00328060. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MANDEL M., LEVISOHN S., SMITH-SONNEBORN J. E., MARMUR J. Deoxyribonucleic acid base composition and taxonomy of some protozoa. Nature. 1962 Nov 24;196:795–796. doi: 10.1038/196795a0. [DOI] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Stevens J. N. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer C. E., Ames B. N. Sunlight ultraviolet and bacterial DNA base ratios. Science. 1970 Nov 20;170(3960):822–825. doi: 10.1126/science.170.3960.822. [DOI] [PubMed] [Google Scholar]
- Specht C. A., DiRusso C. C., Novotny C. P., Ullrich R. C. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982 Jan 1;119(1):158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- Speyer J. F. Mutagenic DNA polymerase. Biochem Biophys Res Commun. 1965 Oct 8;21(1):6–8. doi: 10.1016/0006-291x(65)90417-1. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman R., Rayner E. P. Physical characterization of deoxyribonucleic acids in Dictyostelium discoideum. Arch Biochem Biophys. 1971 May;144(1):127–137. doi: 10.1016/0003-9861(71)90462-0. [DOI] [PubMed] [Google Scholar]
- Wagner I., Capesius I. Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochim Biophys Acta. 1981 Jun 26;654(1):52–56. doi: 10.1016/0005-2787(81)90135-0. [DOI] [PubMed] [Google Scholar]
- Weber J. L. Analysis of sequences from the extremely A + T-rich genome of Plasmodium falciparum. Gene. 1987;52(1):103–109. doi: 10.1016/0378-1119(87)90399-4. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Bleyman M. A. Genetic code limit organisms--do they exist? J Mol Evol. 1972;1(3):223–229. doi: 10.1007/BF01660241. [DOI] [PubMed] [Google Scholar]
- Yarlett N., Orpin C. G., Munn E. A., Yarlett N. C., Greenwood C. A. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem J. 1986 Jun 15;236(3):729–739. doi: 10.1042/bj2360729. [DOI] [PMC free article] [PubMed] [Google Scholar]