Abstract
In bacterial infection, Nucleotide-binding Oligomerization Domain (NOD) 1 and NOD2 induce innate immune responses by recognizing fragments of the bacterial component peptidoglycan (PGN). To determine the roles of these receptors in detection of periodontal pathogens, we stimulated human embryonic kidney cells expressing NOD1 or NOD2 with heat-killed Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Fusobacterium nucleatum or their soluble PGNs (sPGNs). All bacteria and their sPGNs could stimulate activation of NF-κB. However, there were differences in NOD1- and NOD2-stimulatory activities among the species of bacteria. P. gingivalis showed weaker NOD1- and NOD2-stimulatory activities than did other bacteria. These differences in activities were confirmed by production of interleukin-8 from oral epithelial cells stimulated with sPGNs. These findings indicate that both NOD1 and NOD2 might be involved in the recognition of periodontal pathogens, and that the weak NOD-stimulatory property of P. gingivalis might be helpful for survival in the periodontal pocket.
Keywords: NOD, peptidoglycan, periodontal pathogens
Introduction
Periodontitis is a chronic inflammatory disease caused by periodontopathic bacteria and is characterized by inflammation of supportive tissue surrounding teeth. Gram-negative bacteria such as Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Fusobacterium nucleatum have been reported to be associated with periodontitis (Slots et al., 1980; Socransky et al., 1998; Nishihara and Koseki, 2004).
Peptidoglycan (PGN) is a constituent of the cell membrane of bacteria and is one of the endotoxins that induce inflammatory responses such as the production of cytokines and inflammatory mediators (Stewart-Tull, 1980; Yoshimura et al., 1999). PGN is a heteropolymer built of glycan strands cross-linked through peptide chains. The peptide chains are made of two kinds of short peptides, stem peptides that are linked to the glycan chain, and a cross-bridge that links the stem peptides. The glycan of PGN consists of repeating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), but there is a diversity in the composition and sequence of both the stem peptides and the cross-bridge in PGN in bacterial species (Schleifer and Kandler, 1972). A notable difference is that the third amino acid of the stem peptide is meso-diaminopimelic acid (meso-DAP) in PGN of Gram-negative bacteria, in contrast to L-Lys in that of Gram-positive bacteria. The number of amino acids of stem peptides ranges from 2 to 5 (mostly tetrapeptides).
Recently, Nucleotide-binding Oligomerization Domain (NOD) 1 and NOD2 were identified as cytosolic sensors for fragments of PGN. Nod1 recognizes gamma-D-glutamyl-meso-DAP, which is preferentially found in Gram-negative bacteria (Chamaillard et al., 2003; Girardin et al., 2003a), whereas Nod2 recognizes muramyl dipeptide (MDP), which is found in PGN of virtually all bacteria (Girardin et al., 2003b). NOD1 and NOD2 have been reported to be involved in the recognition of infectious bacteria, the induction of immune responses, and the elimination of bacteria through activation of NF-κB (Viala et al., 2004; Kim et al., 2008).
NOD1 and NOD2 are expressed in oral and periodontal pocket epithelium (Sugawara et al., 2006). Because of increased expression of NOD1 and NOD2 at the inflammatory gingival site, NOD1 and NOD2 are thought to play a role in the development of periodontitis. However, it has been reported that some periodontal pathogens, such as P. gingivalis and F. nucleatum, have L.L-DAP and meso-Lanthionine in PGN, respectively, instead of meso-DAP, which is essential for recognition by NOD1 (Vasstrand et al., 1979; Barnard and Holt, 1985). Since NOD1- and NOD2-stimulatory activities of muropeptides are greatly affected by the kind of third amino acid, as well as by the number of amino acids of stem peptides (Girardin et al., 2003c; Magalhães et al., 2005), it is not clear whether a periodontal pathogen or its PGN is potent to stimulate NOD1 and NOD2. The aim of this study was to determine the roles of NOD1 and NOD2 in detection of periodontal pathogens.
Materials & Methods
Bacteria
The bacteria used in this study were P. gingivalis strains ATCC 33277, W83, TDC60, TDC117, TDC275, SU63, and GAI7802, A. actinomycetemcomitans Y4, F. nucleatum ATCC 10953, Escherichia coli MC4100, and Aerococcus viridans ATCC 10400. The cells were washed with sterilized distilled water and lyophilized.
Peptidoglycan Purification
PGN purification was performed according to a previous report (de Jonge et al., 1992). Purified Gram-negative and Gram-positive insoluble PGNs were re-suspended in PBS (2 mg/mL) and treated with mutanolysin (80 U/mL, Sigma, St. Louis, MO, USA) for 24 hrs at 37°C for solubilization. Mutanolysin cleaves the bond between MurNAc and GlcNAc. After digestion, each sample was boiled for 10 min and centrifuged, and the supernatant was used as soluble PGN (sPGN) stock solution.
Measurement of NF-κB Activation
Human embryo kidney (HEK) 293T cells were kindly provided by D. Golenbock (University of Massachusetts Medical School). The cells were seeded into 96-well plates and incubated in DMEM (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS). Then the cells were transfected for 24 hrs with pUNO/vector, pUNO/hNod1 or pUNO/hNod2 plasmid (Invivogen, San Diego, CA, USA), plus NF-κB-firefly luciferase and renilla luciferase reporter plasmid, with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The luciferase activities were measured at 12 hrs after stimulation with bacterial samples. The activity of firefly luciferase was normalized to that of renilla luciferase. Each value was divided by the value for unstimulated cells that were transfected with pUNO/vector and represented as an NF-κB fold activation. A-iE-DAP (AnaSpec, San Jose, CA, USA) and MDP (Bachem AG, Bubendorf, Switzerland) were used as ligands for NOD1 and NOD2, respectively. Drosophila S2★ cells stably transfected with diptericin-luciferase reporter plasmids were maintained in Schneider’s drosophila medium (Gibco) supplemented with 10% FBS and 800 µg/mL of G418 (Kaneko et al., 2005). After differentiation into macrophage-like cells by incubation with 1 µM 20-hydroxyecdysone for 24 hrs, the cells were stimulated with bacterial samples for 4 hrs, and luciferase activities were measured.
Measurement of Interleukin-8 Production
Human oral epithelial cell line HSC-2 was provided by the RIKEN CELL BANK (Tsukuba, Japan), and the cells were maintained in MEM (Gibco) supplemented with 10% FBS. After treatment with 1000 units/mL of interferon (IFN)-γ (Sigma) for 3 days, the cells were stimulated with sPGN samples in a serum-starved condition (no FBS) to increase the uptake of bacterial samples into the cells. The concentration of FBS in the medium was increased to 10% by the addition of FBS at 2 hrs after stimulation, and the cells were further incubated for 22 hrs. The interleukin-8 (IL-8) levels in culture medium were measured by an enzyme-linked immunosorbent assay (DuoSet Kit, R&D Systems, Minneapolis, MN, USA).
Statistical Analysis
The data were analyzed with one-factor ANOVA and Fisher’s PLSD test with StatView software (HULINKS, Tokyo, Japan). A probability level of P < 0.05 was considered to be significant.
Results
We performed overexpression assays in HEK293T cells to explore functions of NODs, because these cells are highly transfectable and lack most of the endogenous Toll-like receptors (TLRs) that mediate bacterial recognition and activation of NF-κB. MDP could not stimulate NF-κB activation in HEK/vector cells, but A-iE-DAP weakly stimulated the cells due to the expression of endogenous NOD1 (Fig. 1A and data not shown). However, strong NF-κB activation was observed in HEK/NOD1 and HEK/NOD2 cells after stimulation with A-iE-DAP and MDP, respectively. These results indicated that expressed NOD1 and NOD2 functioned in the HEK293T cells, and that this overexpression system is useful for monitoring cell response specific for the activation of NOD1 or NOD2.
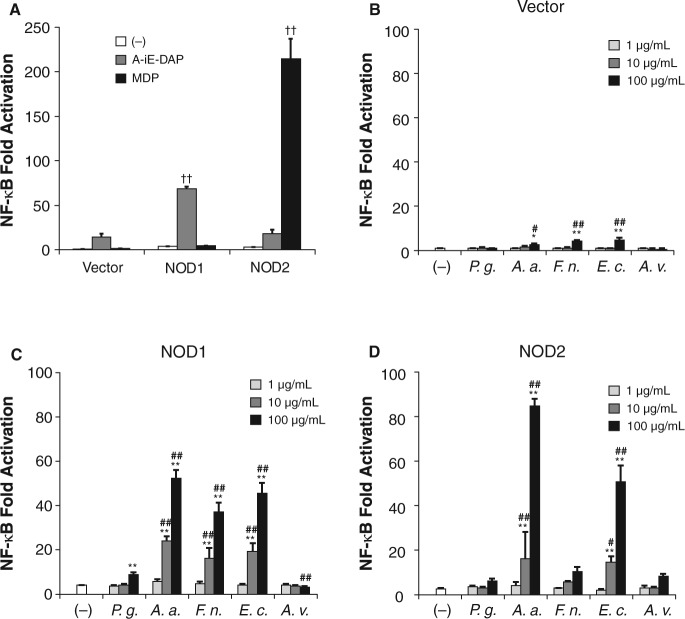
Figure 1.
Activation of NOD1 and NOD2 by stimulation with periodontopathic bacteria samples. HEK293T cells were transfected for 24 hrs with pUNO/vector, pUNO/hNod1 or pUNO/hNod2 plasmid, together with NF-κB-firefly luciferase and renilla luciferase reporter plasmid. After 24 hrs, the cells were stimulated with 1 µg/mL of A-iE-DAP, or 1 µg/mL of MDP or heat-killed bacteria. Luciferase activities were measured 12 hrs after stimulation and expressed as means ± SD of triplicate experiments. The results are representative of 3 different experiments. P.g., P. gingivalis ATCC 33277; A.a., A. actinomycetemcomitans; F.n., F. nucleatum; E.c., E. coli; A.v., A. viridans.††P < 0.01 vs. control vector; *P < 0.05, **P < 0.01 vs. (-); #P < 0.05, ##P < 0.01 vs. P. g.
Next, the cells were stimulated with heat-killed periodontopathic bacteria. A. actinomycetemcomitans, F. nucleatum, and E. coli weakly, but significantly, induced activation of NF-κB in HEK/vector cells, while P. gingivalis and A. viridans failed to stimulate the cells (Fig. 1B). A. actinomycetemcomitans, F. nucleatum, and E. coli strongly stimulated HEK/NOD1 cells in a dose-dependent manner, and significant activations were observed at concentrations of 10-100 µg/mL (Fig. 1C). P. gingivalis also stimulated HEK/NOD1 cells, but its activity was much weaker than the activities of the other 3 Gram-negative bacteria. Gram-positive A. viridans could not activate NOD1. In the case of HEK/NOD2 cells, A. actinomycetemcomitans and E. coli activated NOD2 in a dose-dependent manner, and those activities were statistically significant at concentrations of 10-100 µg/mL. In contrast, P. gingivalis, F. nucleatum, and A. viridans could not activate NOD2 significantly, even at the maximum concentration of 100 µg/mL (Fig. 1D).
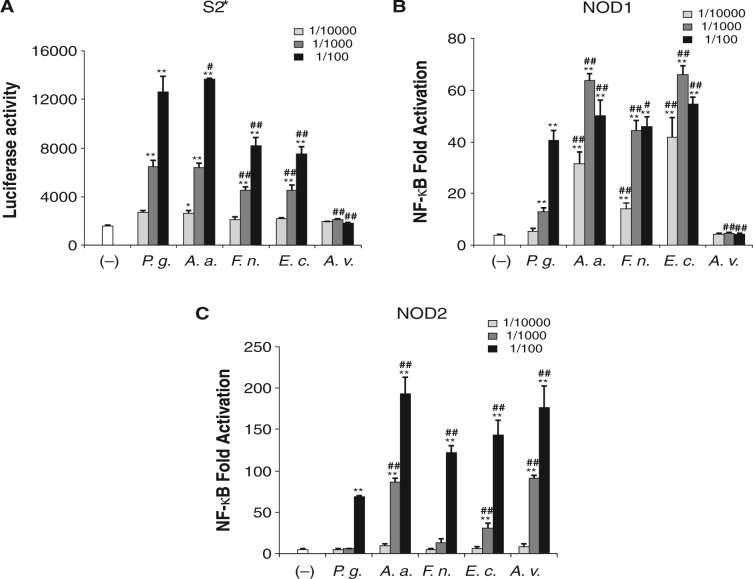
We then stimulated HEK/NOD1 and HEK/NOD2 cells with sPGNs. Before the experiment, we confirmed the biological activities of purified sPGNs by measuring the expression of antimicrobial peptide gene diptericin in Drosophila S2★ cells by a luciferase reporter assay. Diptericin is induced only by Gram-negative bacterial PGN stimulation in S2★ cells (Kaneko et al., 2004). All stock solutions of sPGNs obtained from Gram-negative bacteria showed almost the same activity against S2★ cells at the dilution of 1/1000 (Fig. 2A). A. viridans sPGN could not induce the expression of diptericin. As was found in experiments with heat-killed bacteria, HEK/NOD1 cells were strongly activated by stimulation with sPGNs from A. actinomycetemcomitans, F. nucleatum, and E. coli (Fig. 2B). The sPGN from P. gingivalis could activate NOD1 significantly, but its activity level was 10- to 100-times lower than those of sPGNs from other Gram-negative bacteria. The sPGN from A. viridans could not activate NOD1. In contrast, all sPGNs activated NOD2 (Fig. 2C). A. actinomycetemcomitans and A. viridans showed the strongest NOD2-stimulatory activities, followed by F. nucleatum and E. coli, and P. gingivalis showed the weakest activity. The sPGNs from F. nucleatum and P. gingivalis could activate NOD2 only at the dilution of 1/100.
Figure 2.
Activations of NOD1 and NOD2 by sPGNs from periodontopathic bacteria. (A) Diptericin expression in Drosophila S2* cells by stimulation with purified sPGNs from periodontopathic bacteria. S2* cells stably transfected with diptericin-luciferase reporter plasmid were seeded into 96-well plates and differentiated into macrophage-like cells by incubation with 1 µM 20-hydroxyecdysone for 24 hrs. Then the cells were stimulated with sPGN samples, and the luciferase activities were measured 4 hrs after stimulation and expressed as means ± SD of triplicate experiments. The results are representative of 3 different experiments. HEK293T cells were transfected for 24 hrs with pUNO/hNod1 (B) or pUNO/hNod2 plasmid (C) together with NF-κB-firefly luciferase and renilla luciferase reporter plasmid. After 24 hrs, the cells were stimulated with sPGN samples. Then luciferase activities were measured 12 hrs after stimulation and expressed as means ± SD of triplicate experiments. The results are representative of 3 different experiments. P.g., P. gingivalis ATCC 33277; A.a., A. actinomycetemcomitans; F.n., F. nucleatum; E.c., E. coli; A.v., A. viridans. *P < 0.05, **P < 0.01 vs. (-); #P < 0.05, ##P < 0.01 vs. P.g.
Both NOD1- and NOD2-stimulatory activities of P. gingivalis were weaker than those of other periodontal pathogens. Therefore, we investigated whether these weak activities were species-specific for P. gingivalis. HEK/NOD1 and HEK/NOD2 cells were stimulated with heat-killed P. gingivalis strains W83, TDC60, TDC117, TDC275, SU63, and GAI7802, in addition to ATCC 33277. Although all P. gingivalis strains exhibited weaker NOD1-stimulatory activity than the activity of E. coli, strains W83 and GAI7802 significantly activated NOD1 at the concentration of 10 µg/mL (Fig. 3A). Strains W83 and GAI7802 could activate NOD2 as strongly as E. coli could, but other strains did not show significant activity until the concentration was increased to 100 µg/mL (Fig. 3B).
Figure 3.
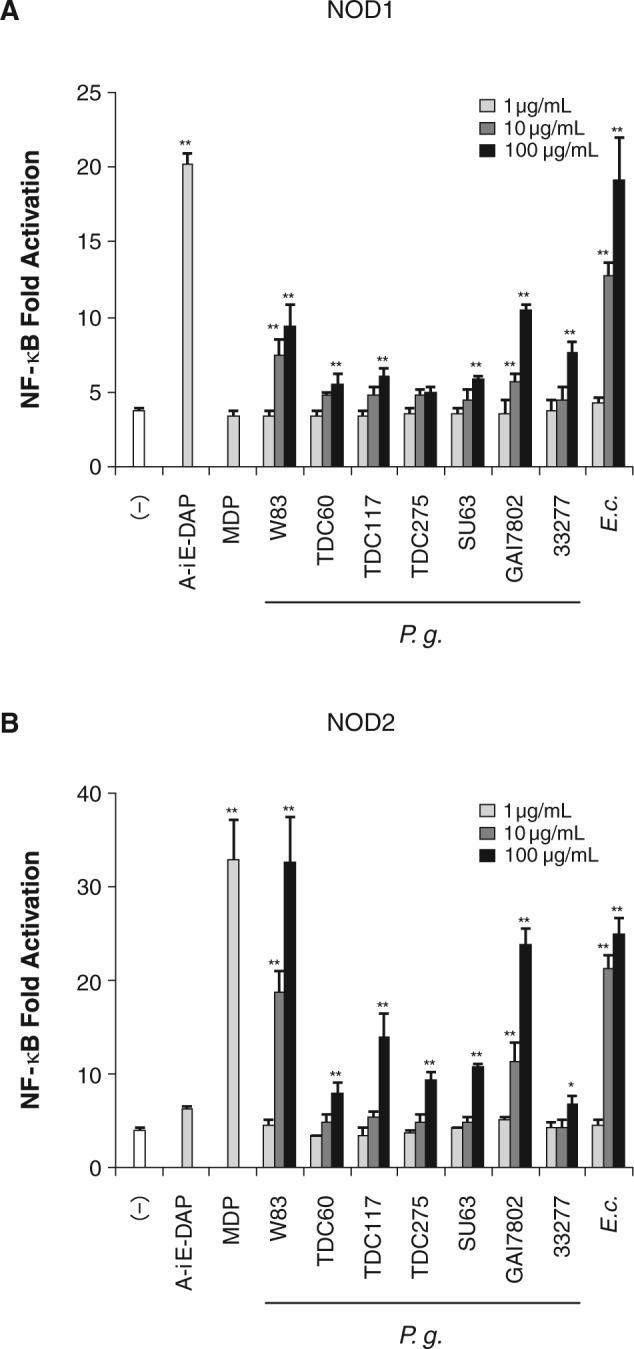
Activations of NOD1 and NOD2 by strains of P. gingivalis. HEK293T cells were transfected for 24 hrs with pUNO/hNod1 (A) or pUNO/hNod2 plasmid (B) together with NF-κB-firefly luciferase and renilla luciferase reporter plasmid for 24 hrs, and the cells were stimulated with heat-killed P. gingivalis strains: W83, TDC60, TDC117, TDC275, SU63, GAI7802, and ATCC 33277. Luciferase activities were measured 12 hrs after stimulation and expressed as means ± SD of triplicate experiments. The results are representative of 3 different experiments. *P < 0.05, **P < 0.01 vs. (-).
To further address the role of NODs in induction of the inflammatory cytokine IL-8 in periodontal cells, we stimulated HSC-2 cells with sPGN. Before the stimulation, HSC-2 cells were pre-treated with IFN-γ for 3 days. IFN-γ treatment significantly increased the response to A-iE-DAP (Fig. 4A). The sPGNs from E. coli, A. actinomycetemcomitans, and F. nucleatum stimulated HSC-2 cells to induce IL-8 more strongly than did those from P. gingivalis and A. viridans (Fig. 4B).
Figure 4.
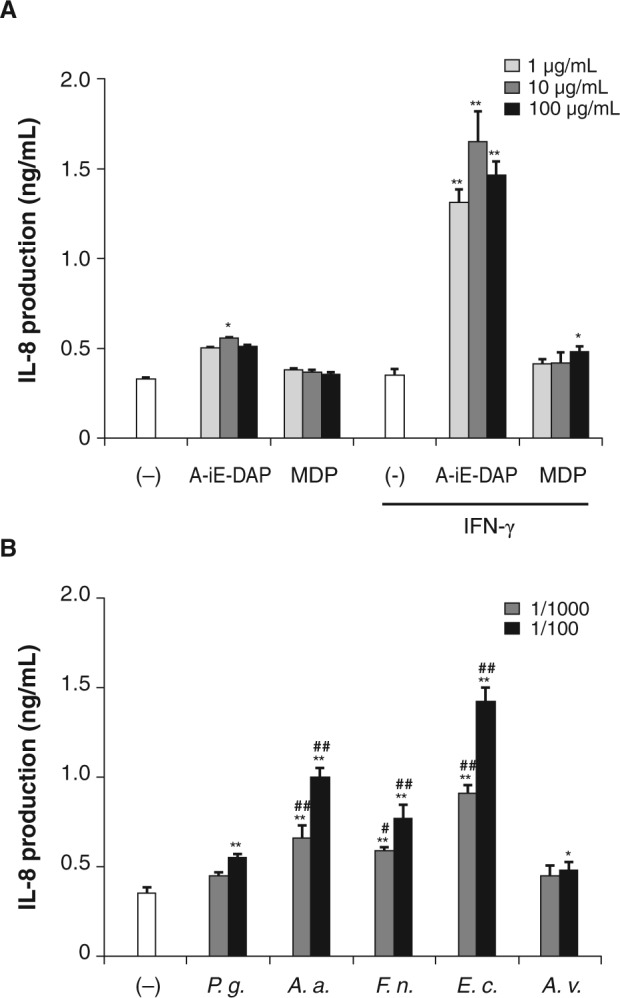
Production of IL-8 in oral epithelial cells. (A) HSC-2 cells were stimulated with A-iE-DAP or MDP in a serum-starved condition after treatment with or without IFN-γ. At 2 hrs after stimulation, the cell culture medium was supplemented with 10% FBS by the addition of FBS and further incubated for 22 hrs. The levels of IL-8 in culture medium were measured. (B) IFN−γ-primed HSC-2 cells were stimulated with sPGNs in a serum-starved condition. At 2 hrs after stimulation, the cell culture medium was supplemented with 10% FBS by the addition of FBS and further incubated for 22 hrs. The levels of IL-8 in culture medium were measured. Data are expressed as means ± SD of triplicate experiments. The results are representative of 3 different experiments. P.g., P. gingivalis ATCC 33277; A.a., A. actinomycetemcomitans; F.n., F. nucleatum; E.c., E. coli; A.v., A. viridans. *P < 0.05, **P < 0.01 vs. (-); #P < 0.05, ##P < 0.01 vs. P.g.
Discussion
PGN of A. actinomycetemcomitans has been reported to contain meso-DAP, which is typically observed in most Gram-negative bacteria such as E. coli (Schleifer and Kandler, 1972; Barnard and Holt, 1985). PGNs of P. gingivalis and F. nucleatum have been reported to contain L,L-DAP and meso-Lanthionine, respectively, instead of meso-DAP (Vasstrand et al., 1979; Barnard and Holt, 1985). A. viridans has L-Lys-type PGN, which is typical for Gram-positive bacteria (Schleifer and Kandler, 1972). In the present study, there were differences in NOD1-stimulatory activities among the species of bacteria. A. actinomycetemcomitans showed the strongest activity, the same as that of E. coli, followed by F. nucleatum, and P. gingivalis showed the weakest activity among these Gram-negative bacteria. A. viridans did not activate NOD1. These results are consistent with the results of a previous study that used synthetic muramyl tripeptides (MTP) for stimulation of HEK cells expressing NOD1 (Girardin et al., 2003c). In that study, it was shown that NOD1-stimulatory activity of L.L-DAP containing MTP (MTPL,L-DAP) was significantly weaker than that of MTPmeso-DAP or MTPmeso-Lanthionine. Thus, it is likely that the NOD1-stimulatory activities of periodontal pathogens are greatly influenced by the nature of the third amino acid of the stem peptide in their PGN.
All PGNs contain an MDP structure in the muropeptide; however, the NOD2-stimulatory activities differed among bacterial strains. A. actinomycetemcomitans showed the strongest activity, similar to that of A. viridians, followed by E. coli and F. nucleatum, and P. gingivalis showed the weakest activity. It has been reported that MTPL-Lys could activate HEK cells expressing NOD2 as strongly as could MDP, but MTPmeso-DAP, MTPmeso-Lanthionine, and MTPL,L-DAP could not activate these cells (Girardin et al., 2003c). These results suggested that MDP, but not MTP, is critical for NOD2-stimulatory activities in PGNs of Gram-negative bacteria. A study on stem peptides in PGN of E. coli demonstrated that most of the stem peptides were tripeptides or tetrapeptides, and that NOD2-stimulatory dipeptides accounted for only 2.1% of all stem peptides (Glauner et al., 1988). The percentage of dipeptides in muropeptides was reported to differ depending on the species or strains of bacteria (Antignac et al., 2003). E. coli and A. actinomycetemcomitans might contain large amounts of dipeptides compared with P. gingivalis. The strong NOD2-stimulatory activities of P. gingivalis strains W83 and GAI7802 might be explained by this possibility. P. gingivalis has been classified into virulent and avirulent strains based on its ability to form necrotic abscesses in an animal model (Grenier and Mayrand, 1987). Inoculation of strain W83 in mice induced abscesses, secondary lesions, sepsis, and death of animals with higher frequency than did strain ATCC 33277 (Neiders et al., 1989). In the present study, strain W83 exhibited stronger NOD1- and NOD2-stimulatory activities than did strain ATCC 33277. Further experiments may be necessary to demonstrate the relationship between NODs-stimulatory activities and the heterogeneity of virulence among P. gingivalis strains.
HSC-2 cells responded well to A-iE-DAP to produce IL-8 after IFN-γ treatment. The sPGNs from A. actinomycetemcomitans and E. coli induced IL-8 production in HSC-2 cells. P. gingivalis consistently induced a weaker response, but it should be noted that the differences between P. gingivalis and the other bacteria in IL-8 expression were not as great as the differences in NF-κB activation.
We carried out muramidase treatment to obtain sPGNs. Both NOD1- and NOD2-stimulatory activities of insoluble PGNs were increased after treatment with muramidase (data not shown). Lysozyme confers muramidase activity in saliva and plays a role in bacterial killing by destroying the bacterial cell wall. Therefore, lysozyme might facilitate recognition of bacteria by NOD1 and NOD2 in addition to bactericidal functions.
P. gingivalis, as well as A. actinomycetemcomitans and F. nucleatum, has been reported to invade host epithelial cells (Meyer et al., 1991; Lamont et al., 1995; Han et al., 2000; Jandik et al., 2008). A previous study revealed that intracellular recognition by NOD1 and NOD2 is critical for clearance of the intracellular bacterium Listeria monocytogenes (Kobayashi et al., 2005). In the present study, we demonstrated that NOD1- and NOD2-stimulatory activities of P. gingivalis were weaker than those of other periodontal pathogens. In addition, P. gingivalis LPS has a unique structure of lipid A, different from that of enterobacterial LPS (Ogawa, 1993), and acts as an antagonist for human lipid A receptor TLR4 (Yoshimura et al., 2002). These weak immunogenic properties of P. gingivalis might be one of the strategies of this bacterium for escape from innate immunity and survival in the periodontal pocket (Darveau et al., 1998).
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by grants from the Takeda Science Foundation and the Sumitomo Foundation.
References
- Antignac A, Rousselle JC, Namane A, Labigne A, Taha MK, Boneca IG. (2003). Detailed structural analysis of the peptidoglycan of the human pathogen Neisseria meningitidis. J Biol Chem 278:31521-31528. [DOI] [PubMed] [Google Scholar]
- Barnard MR, Holt SC. (1985). Isolation and characterization of the peptidoglycans from selected Gram-positive and Gram-negative periodontal pathogens. Can J Microbiol 31:154-160. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. (2003). An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 4:702-707. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. (1998). Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun 66:1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge BL, Chang YS, Gage D, Tomasz A. (1992). Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A J Biol Chem 267:11248-11254. [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, et al. (2003a). Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. (2003b). Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278:8869-8872. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Hervé M, Blanot D, Boneca IG, Philpott DJ, et al. (2003c). Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem 278:41702-41708. [DOI] [PubMed] [Google Scholar]
- Glauner B, Höltje JV, Schwarz U. (1988). The composition of the murein of Escherichia coli. J Biol Chem 263:10088-10095. [PubMed] [Google Scholar]
- Grenier D, Mayrand D. (1987). Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, et al. (2000). Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandik KA, Belanger M, Low SL, Dorn BR, Yang MC, Progulske-Fox A. (2008). Invasive differences among Porphyromonas gingivalis strains from healthy and diseased periodontal sites. J Periodontal Res 43:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. (2004). Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637-649. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Golenbock D, Silverman N. (2005). Peptidoglycan recognition by the Drosophila Imd pathway. J Endotoxin Res 11:383-389. [DOI] [PubMed] [Google Scholar]
- Kim YG, Park JH, Shaw MH, Franchi L, Inohara N, Núñez G. (2008). The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28:246-257. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Núñez G, et al. (2005). Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307:731-734. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. (1995). Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães JG, Philpott DJ, Nahori MA, Jehanno M, Fritz J, Le Bourhis L, et al. (2005). Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep 6:1201-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DH, Sreenivasan PK, Fives-Taylor PM. (1991). Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect Immun 59:2719-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, et al. (1989). Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res 24:192-198. [DOI] [PubMed] [Google Scholar]
- Nishihara T, Koseki T. (2004). Microbial etiology of periodontitis. Periodontol 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- Ogawa T. (1993). Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett 332:197-201. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J, Reynolds HS, Genco RJ. (1980). Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25:134-144. [DOI] [PubMed] [Google Scholar]
- Stewart-Tull DE. (1980). The immunological activities of bacterial peptidoglycans. Annu Rev Microbiol 34:311-340. [DOI] [PubMed] [Google Scholar]
- Sugawara Y, Uehara A, Fujimoto Y, Kusumoto S, Fukase K, Shibata K, et al. (2006). Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res 85:524-529. [DOI] [PubMed] [Google Scholar]
- Vasstrand EN, Hofstad T, Endresen C, Jensen HB. (1979). Demonstration of lanthionine as a natural constituent of the peptidoglycan of Fusobacterium nucleatum. Infect Immun 25:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. (2004). Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166-1174. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. (1999). Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol 163:1-5. [PubMed] [Google Scholar]
- Yoshimura A, Kaneko T, Kato Y, Golenbock DT, Hara Y. (2002). Lipopolysaccharides from periodontopathic bacteria Porphyromonas gingivalis and Capnocytophaga ochracea are antagonists for human toll-like receptor 4. Infect Immun 70:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]