Abstract
High frequencies of chromosomal anomalies are reported in human and non-human primate in vitro-produced preimplantation embryos. It is unclear why certain embryos develop aneuploidies while others remain euploid. A differential susceptibility to aneuploidy is most likely a consequence of events that occur before oocyte collection. One hypothesis is that the relative transcript levels of cohesins, shugoshins and spindle assembly checkpoint genes are correlated with the occurrence of chromosomal anomalies. Transcript levels of these genes were quantified in individual oocytes that were either mature (group 1, low aneuploidy rate) or immature (group 2, high aneuploidy rate) at retrieval, utilizing TaqMan-based real-time PCR. The transcript level in each oocyte was categorized as absent, below the median or above the median in order to conduct comparisons. Statistically significant differences were observed between group 1 and group 2 for SGOL1 and BUB1. There were more oocytes with SGOL1 expression levels above the median in group 1, while oocytes lacking BUB1 were only observed in group 1. These findings suggest that higher SGOL1 levels in group 1 oocytes could better protect against a premature separation of sister chromatids than in embryos derived from group 2 oocytes. The absence of BUB1 transcripts in group 1 was frequently associated with reduced expression of either mitotic cohesins or shugoshins. We hypothesize that ablation of BUB1 could induce mitotic arrest in oocytes that fail to express a complete complement of cohesins and shugoshins, thereby reducing the number of developing aneuploid preimplantation embryos.
Key words: aneuploidy, chromosomal abnormalities, cohesin, oocytes, spindle assembly checkpoint, shugoshins
Introduction
Numerous chromosomal anomalies occur in human and nonhuman primate in vitro-produced (IVP) preimplantation embryos.1–9 Susceptibility to aneuploidy is likely to occur during oocyte development and maturation prior to oocyte retrieval in human in vitro fertilization (IVF). During this time, oocytes acquire various proteins and transcripts concomitant with follicular growth, enabling them to resume meiosis and support early embryonic development.10 Given that all follicles do not develop synchronously during ovarian hormonal stimulation, it is reasonable to assume that some oocytes and their zygotes may acquire different amounts of transcripts and proteins that, in turn, affect cytogenetic stability.
The fidelity of chromosome segregation is maintained by the coordinated interaction of a multitude of proteins that support chromosome cohesion and a robust spindle assembly checkpoint (SAC). Cohesin multiprotein complexes in humans are composed of four evolutionarily conserved subunits, two structural maintenance of chromosomes proteins (SMC1 and SMC3), a kleisin subunit and a stromal antigen (STAG) subunit.11,12 Unlike SMC3, meiosis- and mitosis-specific variants have been described for kleisin, SMC1 and STAG. Each complex maintains cohesion between replicated chromosomes until one of their subunits is targeted for elimination. The majority of cohesin complexes are removed during prophase in a non-proteolytic manner,13,14 whereas residual cohesins remaining at centromeres are ultimately eliminated during the metaphase-anaphase transition through the separase-mediated proteolytic cleavage of the kleisin subunits REC8 during meiosis or RAD21 during mitosis.15
Premature removal of cohesin complexes surrounding centromeres and other sites during prophase and metaphase is prevented by the presence of Shugoshin-like 1 and Shugoshin-like 2 proteins SGOL1 and SGOL2, respectively.16–18 The SAC, which is primarily composed of the mitotic arrest-deficient (MAD) and budding uninhibited by benzimidazole (BUB) proteins, represents the surveillance mechanism that ensures that proteolytic cleavage of the kleisin subunit is only initiated once all kinetochores are properly attached to spindle microtubules.19,20 This surveillance mechanism regulates the activity of the ubiquitinligase, anaphase-promoting complex (APC) or cyclosome.21 Once all chromosomes are properly attached, cyclin B and securin, the protein inhibiting the enzymatic activity of separase, are targeted for destruction. Separase-mediated cleavage of the kleisin subunit results in the segregation of chromosomes, while the elimination of cyclin B is ultimately responsible for meiotic or mitotic exit.22
The increased frequency of aneuploidy in human conceptuses of older women has been attributed to a decreased expression of SAC genes in oocytes from women of advanced maternal age in comparison to oocytes from younger women.23–26 Because of ethical and practical constraints, however, the oocytes employed in these studies were always discarded, pooled, and only a limited proportion of the genes involved in cytogenetic stability were assessed. As rhesus macaque oocytes and their resulting embryos are as prone to aneuploidy as human oocytes and embryos,2,27 the analysis of transcript levels of a number of genes important for cytogenetic stability in individual rhesus macaque oocytes may provide valuable insights into the etiology of aneuploidy during early development of primates. Moreover, Dupont et al.2 previously reported that rhesus macaque embryos developing from metaphase I (MI) oocytes maturing after retrieval have an increased susceptibility to numerical chromosomal anomalies in comparison to embryos resulting from oocytes that were mature at retrieval. This provides an ideal tool to investigate the gene expression patterns associated with chromosomal anomalies during early development in primate oocytes of differential quality.
Therefore, this study aimed to determine the relative abundance of mRNAs encoding cohesins, shugoshins and components of the SAC (Table 1) in individual rhesus macaque oocytes that were either mature at the time of retrieval or matured within 6 to 8 h after retrieval. Differences in the expression profiles were used to identify gene expression patterns associated with their susceptibility to aneuploidy.
Table 1.
Selected proteins associated with chromosome segregation
| Cohesin Complex Subunits | |
| RAD21 | Cohesin complex subunit protein that causes dissociation of the cohesin complex after its cleavage by separase at mitotic anaphase onset. |
| REC8 | Meiosis-specific paralogue of RAD21. |
| STAG1, STAG2 | Cohesin complex subunit protein that causes dissociation of the cohesin complex after its phosphorylation by aurora B and Plk1 kinases during the prophase pathway. |
| STAG3 | Protein that replaces mitotic STAG1 and STAG2 during meiosis. |
| SMC3 | Cohesin complex subunit protein during mitosis and meiosis. |
| SMC1A | Cohesin complex subunit protein during mitosis. |
| SMC1B | Meiosis specific paralogue of SMC1A. |
| Spindle Assembly Checkpoint proteins | |
| MAD2L1 | Protein that inhibits APCcdc20 when not all kinetochores are attached to microtubules. |
| BUB1B | Protein that inhibits APCcdc20 when not all kinetochores are attached to microtubules. |
| BUB1 | Protein that helps to recruit shugoshins to kinetochores. |
| BUB3 | Protein that facilitates binding of BUB1 to kinetochores. |
| Shugoshins | |
| SGOL1 and SGOL2 | Proteins that protects centromeric cohesion from phosphorylation during the prophase pathway and from separase during meiosis I. |
Results
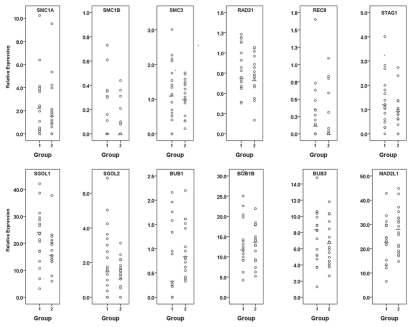
Three rhesus macaque females provided a cohort of oocytes, including oocytes that were mature at retrieval (extruded polar body present, group 1) and others that matured shortly thereafter (6–8 h post-retrieva, group 2). In total, 36 individual oocytes were analyzed for cohesins, shugoshins and SAC gene expression profiles. Normalized expression levels for each individual oocyte from groups 1 and 2 are presented in Figure 1 and Table 2. Spearman rank-order correlation coefficient analysis established significant (α = 0.05) relationships between transcript levels of group 1 and group 2 oocytes (Table 2). A significant influence of female on transcript levels was observed. However, because data were non-normally distributed and included a large number of zero values, expression profiles were subsequently categorized as absent, below the median or above the median. χ2 analysis was then performed to compare obtained frequencies. Analysis of additional transcripts regulating mitochondrial function, including FIS1, OPA1 and TFAM, demonstrated no overall significant transcript level differences between the two groups (Sup. Fig. 1).
Figure 1.
Scatterplots of normalized gene expression levels for each transcript in individual oocytes mature at retrieval (group 1) or maturing post-retrieval (group 2). Bars represent the median level.
Table 2.
Significant Correlation Coefficients between transcript expression levels in group 1 and group 2 oocytes
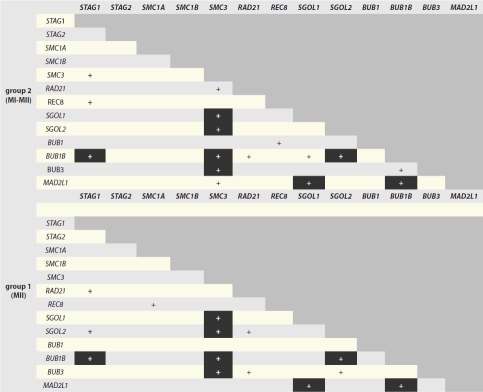 |
Highlighted correlations are conserved between groups.
Cohesin transcripts.
Meiosis-specific cohesins SMC1B and REC8 and the mitotic/meiotic cohesin STAG2 were sporadically observed in group 1 and group 2 oocytes (Fig. 1). All oocytes from both groups expressed RAD21, whereas four oocytes from group 1 and five oocytes from group 2 failed to express at least one of the mitotic cohesins (SMC1A, SMC3 or STAG1), although no statistically significant differences for the incidence of transcript expression were observed between the two groups (Fig. 2). Conserved correlations were observed between STAG1 and BUB1B expression between both groups, while SMC3 correlated with shugoshin (SGOL1, SGOL2) and BUB1B and BUB3 expression in both groups (Table 2).
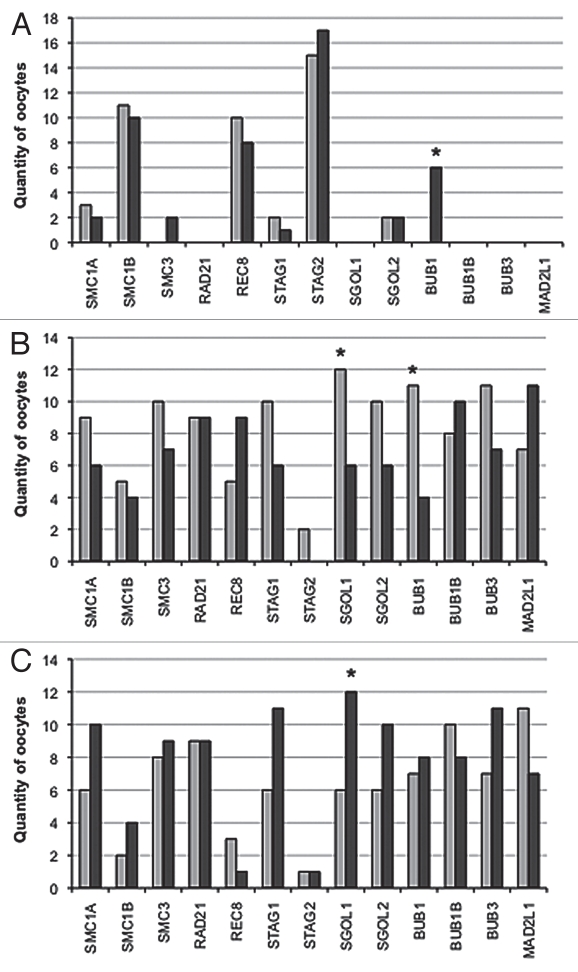
Figure 2.
Incidence of relative gene expression levels in individual rhesus macaque oocytes mature at retrieval (group 1; dark gray bars) or maturing post-retrieval (group 2; light gray bars). (A) Quantity of oocytes lacking each specific transcript; (B) Quantity of oocytes with specific transcript expression levels below the median; (C) Quantity of oocytes with specific transcript expression levels above the median.*represents a statistically significant difference between group 1 and group 2 (α = 0.05).
Shugoshins transcripts.
More oocytes expressed high SGOL1 levels in group 1 than group 2 (χ2 = 4.0, df = 1.0, p ≤ 0.05; see Fig. 2C). In both groups, SGOL2 expression was significantly correlated with BUB1B and SMC3, and SGOL1 levels were correlated with MAD2L1 and SMC3 expression (Table 2).
Spindle assembly checkpoint transcripts.
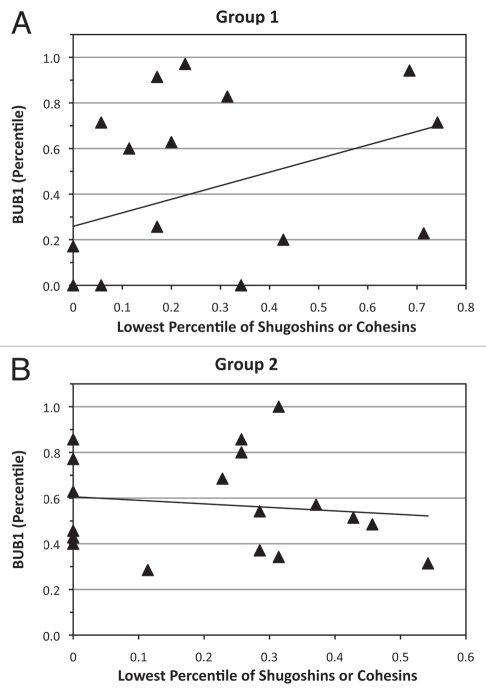
All oocytes expressed BUB1B, BUB3 and MAD2L1 transcripts. Except for BUB1, all SAC genes showed significant correlations with some cohesin and shugoshin genes (Table 2). BUB1B expression was significantly correlated with STAG1, SMC3, SGOL2 and MAD2L1 in both groups. Six oocytes in group 1 failed to express BUB1 in comparison to none in group 2 (p ≤ 0.05; see Fig. 2A). In addition, there were significantly more oocytes with BUB1 expression below the median in group 2 than group 1 (χ2 = 9.3, df = 2.0, p ≤ 0.05; Fig. 2b). Spearman correlation coefficient analysis between BUB1 expression and the lowest value of either cohesin or shugoshin expression showed a significant correlation in group 1 oocytes (Fig. 3A); (rho = 0.60, p = 0.0009). No such association was observed in group 2 oocytes (Fig. 3B).
Figure 3.
Correlation curves for BUB1 compared with minimal expression of shugoshins or mitotic cohesins (STAG1, SMC1A, SMC3, RAD21, SGOL1 and SGOL2). (A) significant positive correlation (rho = 0.60, p < 0.01) within group 1 oocytes. (B) Absence of a significant correlation (rho = -0.20, p = 0.43) in group 2 oocytes.
Discussion
Susceptibility to aneuploidy is likely to occur during oocyte development and maturation, prior to oocyte retrieval. It is reasonable to assume that some oocytes acquire different transcript and protein levels that, in turn, affect their cytogenetic stability. The fidelity of chromosome segregation is maintained by the coordinated interaction of a number of proteins that support chromosome cohesion and a robust spindle assembly checkpoint. Previous reports support the hypothesis that faulty SAC and cohesin activites predispose oocytes to premature chromosome separation and aneuploidy,28,29 while conflicting studies exist for mouse oocytes.30–35 However, any discrepancy may reflect the reduced incidence of chromosomal instability in the mouse36 or the use of discard oocytes collected from infertile human IVF patients. The present study assessed the expression levels of cohesins, shugoshins and SAC genes in individual rhesus macaque oocytes displaying differential maturational status upon retrieval after controlled ovarian hormonal stimulation. This study capitalized on the model in which rhesus macaque embryos derived from oocytes that extruded their polar body after retrieval (group 2 in the present study) produce a higher frequency of chromosomal anomalies in comparison to embryos generated from oocytes that were mature (MII) at retrieval (group 1 in the present study).2
RAD21, BUB1B, BUB3 and MAD2L1 were expressed in all oocytes analyzed. Categorization of the expression profiles as absent or below or above the median demonstrated that only BUB1 and SGOL1 transcripts were different between the two groups, where increased expression of the SGOL1 transcript was observed more frequently in oocytes from group 1 than those in group 2. In addition, the incidence of oocytes lacking BUB1 was significantly more frequent in group 1. Functional studies have shown that loss of cohesins may result in a premature separation of sister chromatids, cell cycle delay, cell cycle arrest and apoptosis.37–40 In the present study, two females produced oocytes in both groups that failed to express at least one of the mitotic cohesins (SMC1A, SMC3 or STAG1). Oocytes lacking mitotic cohesin transcripts were observed in four instances in group 1 and in five occasions in group 2. Most of the affected oocytes were contributed by one female, which suggests that, at the oocyte transcript level, rhesus macaque females respond differently to controlled ovarian stimulation. It is common in human IVF to pool all retrieved oocytes together, regardless of maturational status, to optimize embryo numbers. However, maturational status at retrieval appears to be a strong indicator of individual stimulation response, and therefore susceptibility to aneuploidy.
The failure to express shugoshins, like cohesin deficiencies, may result in premature sister chromatid separation. Shugoshins protect cohesin multiprotein complexes from non-proteolytic removal during the prophase pathway and from proteolytic cleavage during the metaphase-anaphase transition of the first meiotic division.41,42 All rhesus macaque oocytes in this study expressed SGOL1, while two oocytes in both groups failed to express SGOL2. Literature suggests that SGOL2 in mammals, in contrast to SGOL1, is required for proper chromosome segregation during meiosis I, while it is dispensable during mitosis.42,43 Although these reports would suggest that failure to express SGOL2 in rhesus macaque MII oocytes should not alter chromosome dynamics during subsequent development, this hypothesis remains to be confirmed. Significantly, there were more oocytes expressing SGOL1 above the median in group 1 than in group 2. Consequently, embryos generated from MII oocytes at the time of retrieval (group1) may have a lower susceptibility for premature chromosome segregation during prophase as a result of the maintenance of SGOL1 levels. Analysis of additional components regulating localization of these proteins and chromosome segregation, including PP2A,44 the anaphase-promoting complex and cdc2045 as well as protein levels for the various components investigated, would further elucidate the susceptibility of late-maturing oocytes to aneuploidy.
A fine balance in the expression of SAC components appears to provide multiple levels of control to circumvent aneuploidy.32–35 With the exception of BUB1, transcriptional profiles of the SAC genes analyzed in this study did not significantly differ between the two groups of oocytes. Several studies have demonstrated multiple facets of BUB1's involvement in spindle checkpoint regulation. While the small kinase regulates the activity of the APC, it also recruits other SAC proteins and shugoshins to kinetochores.46–48 Many studies concur that ablation of BUB1 using RNA interference (RNAi) results in mitotic arrest after a premature separation of the sister chromatids.48,49 The absence of BUB1 in group 1 oocytes in the present study was strongly correlated with the absence or low expression of one of the mitotic cohesins STAG1, SMC1A and SMC3 or shugoshins SGOL1 and SGOL2, which supports the hypothesis that BUB1 may monitor cohesion between chromosomes prior to the metaphase-anaphase transition.
Shugoshin-depletednHeLA cells likewise undergo mitotic arrest, consistent with this hypothesis.48,50,51 In the present study, oocytes with cohesin or shugoshin insufficiencies in group 1, in contrast to oocytes in group 2, may be better equipped to undergo mitotic arrest, thereby preventing embryonic development that would result in an aneuploidy. Therefore, transcription of BUB1 may be regulated by the integrity of either cohesins or shugoshins in the metaphase II spindle. As the metaphase II spindle assembles in oocytes that extrude a polar body after retrieval (group 2), the proper assembly of cohesins and shugoshins may not be appropriately monitored at the time of RNA processing. Consequently, downregulation of BUB1 could be reduced in these oocytes. Indeed, there were no oocytes lacking BUB1 transcripts in group 2 oocytes, supporting this hypothesis.
The underlying cause predisposing some rhesus macaque and human embryos to become aneuploid while others remain euploid is unknown. Oocytes from women of advanced maternal age display more meiotic errors, are hypothesized to contain fewer SAC proteins and have been shown to display downregulated proapoptotic TAp73,52 suggesting aged eggs are able to avoid checkpoint control and continue development. Moreover, in vitro maturation of immature oocytes has been associated with an increased incidence of aneuploidy53 irrespective of maturation time prior to IVF,54 spindle abnormalities55 and alterations in gene expression of a number of functional processes, including the cell cycle.56 A limited number of genes associated with cytogenetic stability have been assessed in human oocytes.23–26 These studies are complicated by the fact that the oocytes collected are of poor quality (discarded) and retrieved from infertile patients undergoing IVF treatments. The present study assessed the transcriptional profile of a comprehensive array of cohesins, shugoshins and genes involved in the SAC in oocytes retrieved from young fertile monkeys after controlled ovarian hormonal stimulation. We report that oocytes with differential aneuploidy rates display different gene expression patterns that may be associated with their susceptibility to chromosomal errors. Likewise, the absence or presence of various transcripts that regulate cytogenetic stability could indicate varying susceptibility and likely reflects the heterogeneous nature of the oocytes retrieved, as some oocytes may be aspirated from good quality, as well as more atretic, follicles.57 The differential expression of SGOL1 and BUB1 highlighted in this study provides a potential mechanism as to why embryos developing from MII oocytes with delayed polar body extrusion are at a higher risk to become chromosomally abnormal in comparison to embryos developing from MII oocytes mature at retrieval. However, since it was beyond the scope of this study to assess protein levels or post-translational modifications, these conclusions need to be confirmed. Nevertheless, this study highlights that human oocytes that fail to complete maturation prior to retrieval should be considered for use in human IVF with caution, as gene expression perturbations could increase the susceptibility to aneuploidy and impair early embryonic development.
Materials and Methods
Unless otherwise stated, chemicals and reagents were purchased from Sigma-Aldrich.
Controlled hormonal ovarian stimulation.
All procedures were performed according to the institutional animal care and animal use committee (IACUC) protocols approved at the Oregon National Research Primate Center. The ovarian stimulation protocol has been described previously and was initiated on one of the first three days of menstruation.58 Briefly, adult, female rhesus monkeys previously exhibiting normal menstrual cycles received recombinant human follicle-stimulating hormone (rhFSH, Organon; 30 IU per injection, twice daily, i.m.) for 7 consecutive days, followed by 2 d of rhFSH and recombinant human luteinizing hormone (rhLH, EMD Serono; 30 IU rhFSH and 30 IU rhLH per injection, twice daily, i.m.). On the eighth day of the rhFSH treatment, animals also received the gonadotropin-releasing hormone (GnRH) antagonist, acyline (NICHD; 0.075 mg/kg animal body weight), to prevent a spontaneous LH surge. On the final day of rhFSH treatment, 32–33 h before follicular aspiration, oocyte maturation was induced with a single injection of recombinant human chorionic gonadotropin (rhCG, EMD Serono; 1,000 IU, i.m.).
Oocyte collection, RnA extraction and cDnA synthesis.
The procedures for oocyte recovery have been described previously in references 59–61. Briefly, fluid from ovarian follicles was collected in TALP-Hepes containing 0.3% bovine serum albumin (BSA). Oocytes were separated from the aspirated follicular fluid using a cell strainer (Becton-Dickinson; Falcon, 70 µm pore size, #364195). Following denudation from their surrounding granulosa cells using 0.03% hyaluronidase, the oocytes were rinsed in TALP-Hepes containing 0.3% BSA and categorized according to their meiotic status (MII, MI and GV). After classification, all oocytes were placed in TALP-medium culture drops supplemented with BSA62 and incubated in 5% CO2 in air at 37°C until the time they would normally be inseminated, 6–8 h post-retrieval, at which point the maturity of the oocytes was reevaluated. Equivalent numbers of metaphase II (MII) oocytes mature at retrieval (group 1) and MI at retrieval (group 2) per female were randomly selected for the current study. To guarantee the removal of all of the granulosa cells, oocytes were repeatedly aspirated with plastic stripper tips (Mid Atlantic Diagnostics, #MXL3-125) and rinsed in calcium/magnesium-free phosphate buffered saline drops (PBS, Invitrogen, #14190-144) supplemented with 1 mg/ml polyvinyl alcohol (PVA).
RnA isolation and reverse transcription.
Individual oocytes collected at 6–8 h post-retrieval were transferred to 0.2 ml PCR tubes and stored in lysis buffer (Stratagene) at -80°C until further processed. Following thawing on ice and according to the manufacturer's instructions, RNA from the rhesus macaque oocytes was extracted using the Absolutely RNA Nanoprep Kit (Stratagene, #400753). Recovered RNA was reverse transcribed into cDNA using random hexamers and Superscript III reverse transcriptase (Invitrogen, #18080-051) following the manufacturers protocol.
Primer design for Taqman gene expression assays.
Sequencing of PCR products was undertaken in rhesus macaque testes and colon cDNA prior to Taqman primer design. The targets of interest included SMC1A, SMC1B, SMC3, STAG1, STAG2, STAG3, RAD21, REC8, SGOL1, SGOL2, MAD2L1, BUB1, BUB1B and BUB3 (Table 1). Each PCR product was common between various transcript variants and highly conserved between humans and rhesus macaques, according to the National Center for Biotechnology (NCBI) GenBank database. Following sequencing, a consensus sequence was created within which repetitive sequences and single nucleotide polymorphisms (SNPs) were identified using Repeat Masker (Institute for Systems Biology) and Blast SNP (NCBI) prior to being uploaded using the File Builder Software (Applied Biosystems) to facilitate the generation of custom TaqMan gene expression assays. The product size of the amplicons and sequences of the designed primers and probes are specified in Table 3.
Table 3.
Sequences of real-time PCR primers, TaqMan probe and product size
| Gene | Forward Primer | Reverse Primer | Probe | Product size |
| SGOL1 | GCA AA C GCA GGT CTT TTA TAG CT | GTT GTC TTG GTA ATT TTT CAG CAG TGT | AA G TGT TGG TGA TTA TTT G | 80 |
| SGOL2 | TGT CAG CTA GCA AGA AGA AA C GAA T | ATT GTT GTC ACA CTG CAT TTT CTC T | ATG GAA CCC TTG CAA ATG | 128 |
| BUB1 | GCA AGT GCA TGA CTG TGA AA T CA | GCC AA G CCA GCA GAT AAA TCA TC | TTG GAA ACG GAT TTT TG | 111 |
| BUB1B | AAA GAC AGC AA G AGC TCC AA T CAT | GCA TCT GTT GAG GAA ATG GAT TTT GG | CAA GGC TCC AA G CCA G | 95 |
| BUB3 | CCA GTC AA T GCC ATT TCT TTT CAC A | ACC GAT GGA ATT GGC ACA GT | TTG CCA CAG GTG GTT CT | 119 |
| RAD21 | TGA AGA ATT TCA TGA CTT TGA TCA GCC A | CTG CTG GGC CAC ATC GA | CTG CCT GAC TTA GAT GAC A | 64 |
| REC8 | CCA GCT GCA GAT CCG AA T AGA TAT | GCC AGG TGG TTA GGA AGC A | CTG GGT AGC TCA GTC TCC | 66 |
| SMC1α | GCC GCA TTG CCT TTG GA | CAC CAG AGA TCA CTC CTG ACT TC | CAG CGC CAC AA G ACA GT | 87 |
| SMC1β | ACA GAT GAA GAA AGA CTG GCA TTT GA | ACT TCT CTA TTC GTT TTT TAT GAT CTT CTA TTT GT | TTA GAT TTC CCT GAA CTT CTC | 109 |
| SMC3 | AGC CCT GGA ATA TAC CAT TTA CAA TCA G | GCA TCC TGC TGA GCA TCT CTT AA | CAG AAA GCT CAT CAA GTT T | 126 |
| STAG1 | AGA GTT GCT ATT AGA AGA ACC TGT TCA | CTT GAC GAA TTG TAC AAA CCA TTA GCT | ACG ATC AGA CAT TGC TTC C | 98 |
| STAG2 | ATC AGT TGA AAA CAA GAG AA G CCA TTG | GCG GAT TAG GCT CTT TAA AA G CAA A | CCA TGC TAC ACA AA G ATG | 78 |
| STAG3 | TCT TTC CTC ACC GAC AGC TAT TTA AAA | GCC CTT TCA GGG CCT TCA | ATA AGC ACC GAG AA G TC | 97 |
| MAD2L1 | TCT CAG AAA GCT ATC CAG GAT GAA AT | TCT TCC CAT TTT TCA GGT ACA ACC A | ATG AA C AGG AAA CTT CC | 149 |
Multiplex target specific preamplification of cDNA.
A multiplex PCR amplification of the 14 cDNA targets was performed in a final reaction volume of 25 µl using PreAmplification Mix (Applied Biosystems, #4364130) according to manufacturer's instructions. Due to its high abundance, the endogenous control 18S rRNA was not preamplified. Reaction mixtures were initially held at 95°C during 10 min before 14 preamplification cycles of 15 sec at 95°C and 4 min at 60°C were executed. The linearity of preamplification was tested following quantitative real-time PCR of unamplified and preamplified 0.1 ng/µl rhesus macaque testes cDNA. Resulting CT values were used to calculate ΔΔCT values, indicative of the linearity of the preamplification. ΔΔCT values approximating zero were considered linear. All transcripts were successfully linearly amplified, with the exception of STAG3, which was excluded from further analysis.
quantitative real-time PCR.
All preamplified samples were analyzed in triplicate in a 10 µl final reaction volume using the 7900HT Fast Real-Time PCR System. The reaction mixture contained 1xTaqMan Universal Master Mix (Applied Biosystems, #4304437), 1xTaqMan Gene Expression Assay (Applied Biosystems) and 2.5 µl of cDNA. After an initial hold at 95°C for 10 min, reaction mixtures underwent 40 cycles of 15 sec at 95°C and 1 min at 60°C. Standard curves were generated using preamplificed testis cDNA. 18S rRNA Gene Expression Assay primers were used for normalization (Applied Biosystems, #Hs03928985_g1).
Statistical analysis.
Gene expression profiles per group were expressed as the mean ± the standard deviation (SD). As the data were not normally distributed, all gene expression data were transformed by categorization as absent, below median or above median expression, then analyzed using a Pearson χ2. Spearman rank-order correlation coefficients and p-values were calculated between genes for each maturational group. Spearman correlation coefficient analysis was repeated for BUB1 and the lowest value for either cohesins or shugoshins per oocyte (STAG1, SMC1A, SMC3, RAD21, SGOL1 and SGOL2). All differences with p-values ≤ 0.05 were considered to be statistically significant.
Acknowledgements
We would like to acknowledge Dr. Susan Land for her valuable advice. Additionally, we would like to thank Organon, The Netherlands for their generous support by providing human recombinant FSH hormones.
Disclosure of Potential Conflicts of Interest
The authors report no conflict of interest.
Financial Support
Supported by National Institutes of Health grants HD045966, RR015395 and HD046553; by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, DHHS; and by the Oregon National Primate Research Center, RR00163.
Author Contributions
C.D., conception and project design, data acquisition, analysis and interpretation, drafted manuscript; A.H., project design, data interpretation, drafted manuscript; D.R.A., funding, data interpretation, reviewing of manuscript, final manuscript approval; M.B.Z., funding, provided samples for analysis; C.A.B., funding, project design, reviewing of manuscript, final manuscript approval.
Supplementary Material
References
- 1.Benadiva CA, Kligman I, Munné S. Aneuploidy 16 in human embryos increases significantly with maternal age. Fertil Steril. 1996;66:248–255. [PubMed] [Google Scholar]
- 2.Dupont C, Bavister BD, Armant DR, Brenner CA. Rhesus macaque embryos derived from MI oocytes maturing after retrieval display high rates of chromosomal anomalies. Hum Reprod. 2009;24:929–935. doi: 10.1093/humrep/den429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupont C, Froenicke L, Lyons LA, Bavister BD, Brenner CA. Chromosomal instability in rhesus macaque preimplantation embryos. Fertil Steril. 2009;91:1230–1237. doi: 10.1016/j.fertnstert.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magli MC, Gianaroli L, Munné S, Ferraretti AP. Incidence of chromosomal abnormalities from a morphologically normal cohort of embryos in poor-prognosis patients. J Assist Reprod Genet. 1998;15:297–301. doi: 10.1023/A:1022596528036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Márquez C, Sandalinas M, Bahçe M, Alikani M, Munné S. Chromosome abnormalities in 1,255 cleavage-stage human embryos. Reprod Biomed Online. 2000;1:17–26. doi: 10.1016/S1472-6483(10)61988-8. [DOI] [PubMed] [Google Scholar]
- 6.Munné S, Alikani M, Tomkin G, Grifo J, Cohen J. Embryo morphology, developmental rates and maternal age are correlated with chromosome abnormalities. Fertil Steril. 1995;64:382–391. [PubMed] [Google Scholar]
- 7.Pellicer A, Rubio C, Vidal F, Mínguez Y, Giménez C, Egozcue J, et al. In vitro fertilization plus preimplantation genetic diagnosis in patients with recurrent miscarriage: an analysis of chromosome abnormalities in human preimplantation embryos. Fertil Steril. 1999;71:1033–1039. doi: 10.1016/S0015-0282(99)00143-0. [DOI] [PubMed] [Google Scholar]
- 8.Rubio C, Simón C, Vidal F, Rodrigo L, Pehlivan T, Remohí J, et al. Chromosomal abnormalities and embryo development in recurrent miscarriage couples. Hum Reprod. 2003;18:182–188. doi: 10.1093/humrep/deg015. [DOI] [PubMed] [Google Scholar]
- 9.Voullaire L, Collins V, Callaghan T, McBain J, Williamson R, Wilton L. High incidence of complex chromosome abnormality in cleavage embryos from patients with repeated implantation failure. Fertil Steril. 2007;87:1053–1058. doi: 10.1016/j.fertnstert.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 10.Watson AJ. Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J Anim Sci. 2007;85:1–3. doi: 10.2527/jas.2006-432. [DOI] [PubMed] [Google Scholar]
- 11.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 12.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 13.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe Y. Modifying sister chromatid cohesion for meiosis. J Cell Sci. 2004;117:4017–4023. doi: 10.1242/jcs.01352. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y. Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol. 2005;17:590–595. doi: 10.1016/j.ceb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Hoyt MA, Totis L, Roberts BTS. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 20.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 21.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 22.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/S1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 23.Steuerwald N, Cohen J, Herrera RJ, Sandalinas M, Brenner CA. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol Hum Reprod. 2001;7:49–55. doi: 10.1093/molehr/7.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Steuerwald NM, Bermúdez MG, Wells D, Munné S, Cohen J. Maternal age-related differential global expression profiles observed in human oocytes. Reprod Biomed Online. 2007;14:700–708. doi: 10.1016/S1472-6483(10)60671-2. [DOI] [PubMed] [Google Scholar]
- 25.Wells D, Bermúdez MG, Steuerwald N, Malter HE, Thornhill AR, Cohen J. Association of abnormal morphology and altered gene expression in human preimplantation embryos. Fertil Steril. 2005;84:34355. doi: 10.1016/j.fertnstert.2005.01.143. [DOI] [PubMed] [Google Scholar]
- 26.Wells D, Bermudez MG, Steuerwald N, Thornhill AR, Walker DL, Malter H, et al. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum Reprod. 2005;20:1339–1348. doi: 10.1093/humrep/deh778. [DOI] [PubMed] [Google Scholar]
- 27.Nichols SM, Gierbolini L, Gonzalez-Martínez JA, Bavister BD. Effects of in vitro maturation and age on oocyte quality in the rhesus macaque Macaca mulatta. Fertil Steril. 2010;93:1591–1600. doi: 10.1016/j.fertnstert.2008.12.141. [DOI] [PubMed] [Google Scholar]
- 28.Mailhes JB. Faulty spindle checkpoint and cohesion protein activities predispose oocytes to premature chromosome separation and aneuploidy. Environ Mol Mutagen. 2008;49:642–658. doi: 10.1002/em.20412. [DOI] [PubMed] [Google Scholar]
- 29.McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabé AM, Helmhart W, Kudo NR, et al. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol. 2009;19:369–380. doi: 10.1016/j.cub.2009.01.064. [DOI] [PubMed] [Google Scholar]
- 30.Duncan FE, Chiang T, Schultz RM, Lampson MA. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod. 2009;81:768–776. doi: 10.1095/biolreprod.109.077909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol. 2008;316:397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun SC, Lee SE, Xu YN, Kim NH. Perturbation of Spc25 expression affects meiotic spindle organization, chromosome alignment and spindle assembly checkpoint in mouse oocytes. Cell Cycle. 2010;9:4552–4559. doi: 10.4161/cc.9.22.13815. [DOI] [PubMed] [Google Scholar]
- 33.Wei L, Liang XW, Zhang QH, Li M, Yuan J, Li S, et al. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle. 2010;9:1112–1121. doi: 10.4161/cc.9.6.10957. [DOI] [PubMed] [Google Scholar]
- 34.Zhang QH, Wei L, Tong JS, Qi ST, Li S, Ou XH, et al. Localization and function of mSpindly during mouse oocyte meiotic maturation. Cell Cycle. 2010;9:2230–2236. doi: 10.4161/cc.9.11.11895. [DOI] [PubMed] [Google Scholar]
- 35.Zhu JL, Lin SL, Li M, Ouyang YC, Hou Y, Schatten H, et al. Septin2 is modified by SUMOylation and required for chromosome congression in mouse oocytes. Cell Cycle. 2010;9:1607–1616. doi: 10.4161/cc.9.8.11463. [DOI] [PubMed] [Google Scholar]
- 36.Bean CJ, Hassold TJ, Judis L, Hunt PA. Fertilization in vitro increases non-disjunction during early cleavage divisions in a mouse model system. Hum Reprod. 2002;17:2362–2367. doi: 10.1093/humrep/17.9.2362. [DOI] [PubMed] [Google Scholar]
- 37.Ghiselli G. SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol Cancer. 2006;5:52. doi: 10.1186/1476-4598-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/S0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 39.Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell. 2001;1:759–770. doi: 10.1016/S1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 40.Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, et al. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–218. doi: 10.1016/S0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 41.Gómez R, Valdeolmillos A, Parra MT, Viera A, Carreiro C, Roncal F, et al. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007;8:173–180. doi: 10.1038/sj.embor.7400877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, et al. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 43.Llano E, Gomóz R, Gutiérrez-Caballero C, Herrán Y, Sánchez-Martín M, Vázquez-Quiñones L, et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008;22:2400–2413. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suja JA. Fighting of Casein kinase 1 and PP2A/Shugoshin for cohesins during meiosis I. Cell Cycle. 2010;9:2929–2930. doi: 10.4161/cc.9.15.12696. [DOI] [PubMed] [Google Scholar]
- 45.Vogt E, Kirsch-Volders M, Parry J, Eichenlaub-Ritter U. Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat Res. 2008;651:14–29. doi: 10.1016/j.mrgentox.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 46.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 47.Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3 and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gjoerup OV, Wu J, Chandler-Militello D, Williams GL, Zhao J, Schaffhausen B, et al. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci USA. 2007;104:8334–8339. doi: 10.1073/pnas.0703164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Guglielmino MR, Santonocito M, Vento M, Ragusa M, Barbagallo D, Borzi P, et al. TAp73 is downregulated in oocytes from women of advanced reproductive age. Cell Cycle. 2011;10:3253–3256. doi: 10.4161/cc.10.19.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magli MC, Ferraretti AP, Crippa A, Lappi M, Feliciani E, Gianaroli L. First meiosis errors in immature oocytes generated by stimulated cycles. Fertil Steril. 2006;86:629–635. doi: 10.1016/j.fertnstert.2006.02.083. [DOI] [PubMed] [Google Scholar]
- 54.Strassburger D, Goldstein A, Friedler S, Raziel A, Kasterstein E, Mashevich M, et al. The cytogenetic constitution of embryos derived from immature (metaphase I) oocytes obtained after ovarian hyperstimulation. Fertil Steril. 2010;94:971–978. doi: 10.1016/j.fertnstert.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 55.Cekleniak NA, Combelles CM, Ganz DA, Fung J, Albertini DF, Racowsky C. A novel system for in vitro maturation of human oocytes. Fertil Steril. 2001;75:1185–1193. doi: 10.1016/S0015-0282(01)01789-7. [DOI] [PubMed] [Google Scholar]
- 56.Jones GM, Cram DS, Song B, Magli MC, Gianaroli L, Lacham-Kaplan O, et al. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Hum Reprod. 2008;23:1138–1144. doi: 10.1093/humrep/den085. [DOI] [PubMed] [Google Scholar]
- 57.Chaffin CL, Stouffer RL. Role of gonadotrophins and progesterone in the regulation of morphological remodelling and atresia in the monkey peri-ovulatory follicle. Hum Reprod. 2000;15:2489–2495. doi: 10.1093/humrep/15.12.2489. [DOI] [PubMed] [Google Scholar]
- 58.Zelinski-Wooten MB, Hutchison JS, Hess DL, Wolf DP, Stouffer RL. Follicle stimulating hormone alone supports follicle growth and oocyte development in gonadotrophin-releasing hormone antagonist-treated monkeys. Hum Reprod. 1995;10:1658–1666. doi: 10.1093/oxfordjournals.humrep.a136151. [DOI] [PubMed] [Google Scholar]
- 59.Bavister BD, Boatman DE, Collins K, Dierschke DJ, Eisele SG. Birth of rhesus monkey infant after in vitro fertilization and nonsurgical embryo transfer. Proc Natl Acad Sci USA. 1984;81:2218–2222. doi: 10.1073/pnas.81.7.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bavister BD, Boatman DE, Leibfried L, Loose M, Vernon MW. Fertilization and cleavage of rhesus monkey oocytes in vitro. Biol Reprod. 1983;28:983–999. doi: 10.1095/biolreprod28.4.983. [DOI] [PubMed] [Google Scholar]
- 61.Wolf DP, Vandevoort CA, Meyer-Haas GR, Zelinski-Wooten MB, Hess DL, Baughman WL, et al. In vitro fertilization and embryo transfer in the rhesus monkey. Biol Reprod. 1989;41:335–346. doi: 10.1095/biolreprod41.2.335. [DOI] [PubMed] [Google Scholar]
- 62.Bavister BD, Yanagimachi The effects of sperm extracts and energy sources on the motility and acrosome reaction of hamster spermatozoa in vitro. Biol Reprod. 1977;16:228–237. doi: 10.1095/biolreprod16.2.228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.