Abstract
A complementary DNA library was constructed from the flowers of Chimonanthus praecox, an ornamental perennial shrub blossoming in winter in China. Eight hundred sixty-seven high-quality expressed sequence tag sequences with an average read length of 673.8 bp were acquired. A nonredundant set of 479 unigenes, including 94 contigs and 385 singletons, was identified after the expressed sequence tags were clustered and assembled. BLAST analysis against the nonredundant protein database and nonredundant nucleotide database revealed that 405 unigenes shared significant homology with known genes. The homologous unigenes were categorized according to Gene Ontology hierarchies (biological, cellular, and molecular). By BLAST analysis and Gene Ontology annotation, 95 unigenes involved in stress and defense and 19 unigenes related to floral development were identified based on existing knowledge. Twelve genes, of which 9 were annotated as “cold response,” were examined by real-time RT-PCR to understand the changes in expression patterns under cold stress and to validate the findings. Fourteen genes, including 11 genes related to floral development, were also detected by real-time RT-PCR to validate the expression patterns in the blooming process and in different tissues. This study provides a useful basis for the genomic analysis of C. praecox.
1. Introduction
Chimonanthus praecox (L.) Link, wintersweet, belongs to the Calycanthaceae family. It is a perennial deciduous shrub and blossoms in winter, from late November to March. Its unique flowering time and long blooming period make it one of most popular ornamental plants in China [1]. C. praecox is mainly a garden plant that also provides cut flowers. The flower is strongly fragrant and may be used as a source of essential oil, which has received much attention in New Zealand [2]. C. praecox thrives in cold environments and blooms in low-temperature seasons with little rainfall. The plant is assumed to be rich in genes related to floral development and adversities, especially those responding to environmental stress factors. However, the molecular mechanism that regulates floral development and copes with stresses in C. praecox flowers remains unclear.
Expressed sequence tags (ESTs) have been proven to be an efficient and rapid means to identify novel genes (and proteins) induced by environmental changes or stresses [3–7]. Genes related to flower form, longevity, and scent from roses, Phalaenopsis equestris, and Pandanus fascicularis were identified by ESTs [8–10]. The present study used transcriptomic analysis of C. praecox flowers to identify novel genes induced by environmental changes or related to floral development and ultimately to better understand the physiological and genetic basis of cold acclimation in flowers of woody plants.
2. Materials and Methods
2.1. Complementary DNA (cDNA) Library Construction and Sequencing
The process of C. praecox blossoming includes the following: Stage 1, sprout period; Stage 2, flower-bud period; Stage 3, display-petal period; Stage 4, initiating bloom period; Stage 5, bloom period; Stage 6, wither period [11]. C. praecox flower buds or flowers at the six stages of development (Figure 1) were collected from the nursery at Southwest University, Chongqing, China, for cDNA library construction. The samples were immediately frozen with liquid nitrogen and then refrigerated at −80°C until RNA isolation. Total RNA samples were extracted from these flower buds or flowers using RNA isolation kits (W6771; Watson Biotechnologies Inc.), and RNA quality was detected by BioSpec-mini (Shimadzu). The final RNA sample for the cDNA library construction was bulked by pooling equal amounts of total RNA from each stage.
Figure 1.

Stages of C. praecox blooming. Stage 1, sprout period: bud scales loosen; Stage 2, flower-bud period: flower buds turn green; Stage 3, display-petal period: flower buds enlarge and turn yellow; Stage 4, initiating bloom period: fragrance emerges; Stage 5, bloom period: flowers fully open with strong fragrance; Stage 6, wither period: petals begin withering.
cDNA synthesis from the mixed total RNA and library construction through directional cloning of cDNAs into the λTriplEx2 vector was performed using a SMART cDNA Library Construction Kit (K1051-1) according to the manufacturer's instructions. The size of the insert fragment and the recombinant rate were measured by PCR, as described by Gao and Hu [12], using a random selection of 50 clones. All clones were sequenced from the 5′ end using ABI 3700 (Applied Biosystems).
2.2. Sequence Clustering, Annotation, and Functional Categorization
Poor-quality sequences, sequences with less than 100 bases, and vector sequences were trimmed from the raw sequences using SeqMan II (DNASTAR, Inc., Madison, WI) and manually. The trimmed cDNA sequences were assembled into clusters using the assembly program within SeqMan II set to default parameters. The assembly parameters were set to require a minimum match of 80% over 12 bp to initiate the assembly process. A consensus sequence for unigenes was exported from SeqMan II.
Unigenes (contigs and singletons) were annotated using BLASTX against the NCBI nonredundant protein database with a cut-off E value of the best hit of ≤10−5 [13]. Sequences without a reliable match (>10−5) were subsequently compared with the NCBI nonredundant nucleotide database by performing BLASTN (score >100) for complementary annotation [14]. All well-annotated unigenes were then further classified and mapped to the three Gene Ontology (GO) categories (biological, cellular, and molecular) via AmiGO (http://amigo.geneontology.org/) [15].
2.3. Expression Analysis of Selected Genes Using Quantitative Real-Time RT-PCR
For real-time expression studies, C. praecox seeds were kept under a 16 h light/8 h dark cycle in a growth chamber at 25°C. Small seedlings were subsequently transferred to plastic pots containing a mixture of soilrite and vermiculite (1 : 1). The plantlets were raised to the sixth leaf stage and then subjected to cold treatment (4°C) for 15 min, 1 h, and 6 h. Control plantlets were maintained at 25°C. The tissues were harvested and snap-frozen in liquid nitrogen and kept at −80°C until further use.
The total RNA of flowers at the different developmental stages as well as floral organs dissected from Stage 4 flowers, roots, stems, and leaves were extracted with the above-mentioned method. Equal amounts of DNA-free RNA (5 μg) from different tissues were reverse-transcribed using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa). The primers used for real-time PCR (Table 1) were designed by Primer Premier 5.0 (PREMIER Biosoft, USA). Real-time PCRs were performed in triplicate on 10 μL reactions using 5 μL of SsoFast EvaGreen Supermix (Bio-Rad), 0.5 μL of each primer (final concentration of 500 nM), 3.5 μL of water, and 0.5 μL of cDNA template. Table 1 shows the primer pair-specific temperatures. PCR was carried out using a Bio-Rad CFX96 real-time system machine based on the manufacturer's instructions under the following conditions: denaturation at 95°C for 30 s; 40 cycles of 95°C for 5 s; 59°C for 5 s. Dissociation kinetics was performed using the real-time PCR system at the end of the experiment (60 to 95°C; continuous fluorescence measurement) to check the annealing specificity of the oligonucleotides. A comparative Ct (threshold cycles) method of relative quantification was used to analyze the real-time quantitative RT-PCR data using Bio-Rad CFX Manager Software Version 1.6. Actin and tubulin were used as housekeeping genes for the calculation of relative transcript abundance. The sizes of the amplified products were confirmed via gel electrophoresis. Negative controls with no templates were carried out concurrently.
Table 1.
Forward (F) and reverse (R) primers used in real-time PCR.
| Gene code | Annotation (sequence homology) | Primer sequence (5′→3′) | Tm (°C) a | Cycles | Product length |
|---|---|---|---|---|---|
| Cp88 | Membrane channel protein | F: ATGTGGACTTTTTGCTGGAGTTTTT | 59 | 40 | 90 |
| R: GCAATACAAGCATTTACCCAGTCCT | |||||
| Cp173 | Low temperature and salt responsive protein | F: CTGACGCTCTTTGGATGGCTACC | 59 | 40 | 172 |
| R: ACGAATCAACGGTCACAAACACG | |||||
| Cp215 | Putative abscisic acid-induced protein | F: TGCTCTTTACTTGCTGGCTCGTCT | 59 | 40 | 193 |
| R: TCTCGCCCATTCTCTTCTATGTATTTC | |||||
| Cp274 | Glutathione S-transferase | F: ATGTGCTAAATCTCATTCCAGTCTTCC | 59 | 40 | 85 |
| R: GCCGTGATATCCTGCGAAACTAG | |||||
| Cp359 | 1,4-Alpha-glucan-maltohydrolase | F: TATTATCTGGCAAGGTTCCACTCGTT | 59 | 40 | 86 |
| R: TGTGTTGTAGAACCCAGCAGTCAGC | |||||
| Cp364 | Cold acclimation Protein WCOR413-like protein beta form | F: TCCAATAAGCCGAATCAAAATACAGT | 59 | 40 | 84 |
| R: GTCTGTCTTCATCGCCAAATACTCC | |||||
| Cp375 | 3-Oxoacyl-[acyl-carrier-protein] synthase | F: GTAACTGCGCCGAAACGAGAAAAAG | 59 | 40 | 78 |
| R: CCCAAAGACAGAAACCAGCCCC | |||||
| Cp440 | Putative multiple stress-responsive zinc-finger protein | F: GTTTCCCGATTTTGTGTGGCG | 59 | 40 | 183 |
| R: AGTTGTTGATGCAGAGAATTGGTCCT | |||||
| Cp465 | Catalase | F: CATCGTTCGCTTCTCTACCGTTATT | 59 | 40 | 118 |
| R: TGTTTCCGACCAGATCAAAGTTACC | |||||
| Cp197 | AGL9.2 | F: TGAGAATAAGATAAATCGGCAGGTGAC | 59 | 40 | 128 |
| R: TTTTCCTCTGCTGGAGAAGACAACG | |||||
| Cp203 | LLP-B3 protein | F: GCGATGTGAAACTTGTCAGTAGCCC | 59 | 40 | 171 |
| R: AGTTTGGCACAATCAGGACGAGGTT | |||||
| Cp268 | Caffeoyl-CoA O-methyl-transferase-like protein | F: AACACAAATGGAGAAGAGAAAACCC | 59 | 40 | 193 |
| R: CATCGGCAGAGGTCGTCATAAGA | |||||
| Cp297 | SRG1-like protein | F: TAAGAACACCGTCCCATCTCGCTAC | 59 | 40 | 143 |
| R: GAGTGCAGTCTCTCCAATTCATCAG | |||||
| Cp328 | STYLOSA protein | F: GGGGAACCAACAAGAACAACAGAT | 59 | 40 | 161 |
| R: GCCCAGTGGCACCATTTAGAAGAT | |||||
| Cp330 | RAC-like G-protein Rac1 | F: ACCGCTTCTACCGCCTTTTCTCC | 59 | 40 | 166 |
| R: AGGTCTTTCCAACAGCACCATCTCC | |||||
| Cp360 | Peroxisomal fatty acid betao-xidation multifunctional protein AIM1 | F: TATGCTGGTGTTTTGAAAGAGGCG | 59 | 40 | 193 |
| R: CTATGCCAGACCCCATCAAACCTC | |||||
| Cp383 | Secondary cell wall-related glycosyltransferase family 47 | F: CAGGCGACACCCCCTCCTCTAAC | 59 | 40 | 150 |
| R: TCAAGGCATCAGAGTTACGCACAAAG | |||||
| Cp423 | Putative polyketide synthase | F: GAGTTCAGACACTGTATGCCCTTGG | 59 | 40 | 165 |
| R: TTGCTTGTAAAATTCGTCCTTCGTT | |||||
| Cp436 | Allergen-like protein BRSn20 | F: CTCTTATCGCTCTCTGCTTCCTTCC | 59 | 40 | 176 |
| R: GTTGCTGTTAACGTCTCTGCATTCC | |||||
| Cp458 | MADS-box protein 9 | F: TGGTTGAGTATGATGTAGAGAAGCGAC | 59 | 40 | 98 |
| R: ACCCATCATCTGAAGGTGTGCTATT | |||||
| Cp82 | Dormancy related protein | F: TGGAATAGAAAGAGCAAATACAGCG | 59 | 40 | 172 |
| R: TACAAAAGGCTCAATGGCGTCC | |||||
| Cp24 | No hits | F: GCAGTTTACATACTACGGGAGAGGCT | 59 | 40 | 106 |
| R: CGGCTTACGGAATCGTCATCAC | |||||
| Cp64 | Putative protein | F: CCAATCACTCTCCCTGAGGATGTAT | 59 | 40 | 159 |
| R: TTCACCGACTCCTTGTTCTTTTAGC | |||||
| Internal control | Actin | F: GTTATGGTTGGGATGGGACAGAAAG | 59 | 40 | 199 |
| R: GGGCTTCAGTAAGGAAACAGGA | |||||
| Internal control | Tublin | F: TAGTGACAAGACAGTAGGTGGAGGT | 59 | 40 | 139 |
| R: GTAGGTTCCAGTCCTCACTTCATC |
aAnnealing temperature.
3. Results
3.1. General Characteristics of the cDNA Library and the Ests
A cDNA library constructed from C. praecox flowers at different stages of development was used as a source of ESTs. The titers of the primary library and amplified library were 1.4×106 and 1.0×1010 pfu/mL, respectively, with a recombinant rate of 96% for the original library. The sizes of the inserts ranged from 0.5 to 2.5 kb, and the average insert size was estimated to be 1.1 kb by PCR amplification of inserts from 50 randomly selected clones. These results indicate that our cDNA library was qualified.
In total, 896 random cDNA clones were successfully sequenced to generate ESTs. Trimming of the short sequences (<100 bp), vector sequences, and poor-quality sequences resulted in 867 high-quality ESTs, constituting a total of 584,201 bases in the C. praecox sequence. The average read length of these ESTs was 673.8 bp. All 867 sequences were deposited in GenBank under accession numbers DW222667 to DW223533. The clustering of ESTs generated 94 contigs (containing 2 or more ESTs) and 385 singletons (containing only 1 EST), yielding 479 unigenes. The redundancy of the library was calculated as 44.8% [(1 − Number of Unigenes/Number of ESTs) × 100%]. Figure 2 shows the distribution of ESTs in unigenes after clustering. Forty-two contigs had more than 2 ESTs, with the largest one containing 77 ESTs.
Figure 2.
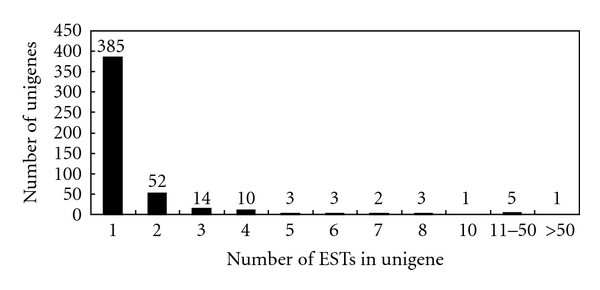
Distribution of C. praecox ESTs among unigenes.
3.2. Functional Annotation and Classification of C. praecox Unigenes
The 479 unigenes were compared with the nonredundant protein and nucleotide sequences database in NCBI using BLAST. Four hundred five unique sequences, corresponding to 84.6% of all the unigenes, shared significant homology with sequences in the public databases. Of these, 266 were similar to genes of known functions, whereas 139 were similar to putative uncharacterized proteins (Table 2). The remaining 74 unigenes (15.4%) had only very weak or no matches and were considered as novel genes in C. praecox flowers.
Table 2.
Overview of C. praecox flowers ESTs.
| Items | Number |
|---|---|
| Total sequenced cDNA | 896 |
| Total number of ESTs analyzed | 867 |
| Total reading valid length (bp) | 584,201 |
| Average EST length (bp) | 673.8 |
| Unigenes | 479 |
| Contigs | 94 |
| Singletons | 385 |
| Redundancy | 44.8 |
Table 3 summarizes the highly expressed genes that contained more than 5 ESTs in one contig. The first and third most abundant ESTs were homologous to lipid transfer protein (LTP); the second most abundant ESTs encoded protein related to the adenine nucleotide translocator and then seed-specific protein, mannose-specific lectin, and LEA III protein (Table 3). These ESTs possibly corresponded to the most abundantly expressed genes in C. praecox flowers. Two hundred eighty-one ESTs were found to have more than five copies (Table 3), 385 had single copies of ESTs, and the remaining ESTs contained two to four copies in C. praecox flowers.
Table 3.
The 15 most abundant ESTs in the C. praecox flowers cDNA library.
| Gene code | Length(bp) | Putative function | E-value | Number of ESTs |
|---|---|---|---|---|
| Cp1 | 1319 | lipid transfer protein | 1.00E − 34 | 77 |
| Cp2 | 2124 | adenine nucleotide translocator | 9.00E − 80 | 47 |
| Cp4 | 1024 | lipid transfer protein | 2.00E − 35 | 37 |
| Cp6 | 1144 | hypothetical protein | 1.00E − 34 | 19 |
| Cp7 | 710 | seed specific protein Bn15D18B | 4.00E − 19 | 18 |
| Cp20 | 896 | mannose specific lectin | 5.00E − 28 | 17 |
| Cp16 | 593 | putative LEA III protein isoform 2 | 6.00E − 10 | 10 |
| Cp10 | 900 | glutathione S-transferase GST 22 | 4.00E − 80 | 8 |
| Cp11 | 928 | palmitoyl-acyl carrier protein thioesterase | 7.00E − 22 | 8 |
| Cp14 | 1531 | hypothetical protein | 2.00E − 79 | 8 |
| Cp17 | 717 | stearoyl acyl carrier protein desaturase | 2.00E − 31 | 7 |
| Cp26 | 1074 | aquaporin | 8.00E − 97 | 7 |
| Cp23 | 696 | S28 ribosomal protein | 4.00E − 20 | 6 |
| Cp24 | 574 | No hits | NULL | 6 |
| Cp27 | 652 | proline-rich protein | 1.00E − 30 | 6 |
|
| ||||
| Total | 14882 | 15 | 281 | |
The database-matched 405 unigenes were found to have BLAST hits with 99 organisms, among which the highest number was from Arabidopsis thaliana (32.6%; 132 unigenes), followed by Oryza sativa (24.0%; 97 unigenes) and Nicotiana tabacum (4.2%; 17 unigenes) (Table 4). The remaining 67 unigenes (16.5%) had BLAST hits with a single organism only. The extensive distribution of the matched organisms may be attributed to the fact that the genome of C. praecox significantly differed from the genomes of model plants, as well as the fact that the relative plants' genomes have not yet been widely studied.
Table 4.
Statistics of organism origin of matched-function homologs.
| Organism name | No. of unigenes | Percentage (%)a |
|---|---|---|
| Arabidopsis thaliana | 132 | 32.6% |
| Oryza sativa (japonica cultivar-group) | 97 | 24.0% |
| Nicotiana tabacum | 17 | 4.2% |
| Glycine max | 8 | 2.0% |
| Lycopersicon esculentum | 7 | 1.7% |
| Gossypium hirsutum | 6 | 1.5% |
| Cucumis sativus | 5 | 1.2% |
| Capsicum annuum | 5 | 1.2% |
| Pisum sativum | 4 | 1.0% |
| Solanum tuberosum | 4 | 1.0% |
| Petunia x hybrida | 4 | 1.0% |
| Nicotiana attenuata | 3 | 0.7% |
| Helianthus annuus | 3 | 0.7% |
| Beta vulgaris | 3 | 0.7% |
| Calycanthus floridus var. glaucus | 3 | 0.7% |
| Ipomoea batatas | 3 | 0.7% |
| Hyacinthus orientalis | 3 | 0.7% |
| Persea americana | 3 | 0.7% |
| Ricinus communis | 3 | 0.7% |
| Lily mottle virus | 3 | 0.7% |
| Medicago sativa | 2 | 0.5% |
| Zea mays | 2 | 0.5% |
| Cicer arietinum | 2 | 0.5% |
| Triticum aestivum | 2 | 0.5% |
| Hevea brasiliensis | 2 | 0.5% |
| Asparagus officinalis | 2 | 0.5% |
| Panax ginseng | 2 | 0.5% |
| Malus x domestica | 2 | 0.5% |
| Elaeis oleifera | 2 | 0.5% |
| Pinus taeda | 2 | 0.5% |
| Spinacia oleracea | 2 | 0.5% |
| The other 67 organisms | 1 | 16.5% |
aProportion from unigenes with BLAST hits.
The initial annotations were further simplified into plant-specific annotations (plant GO slim; http://amigo.geneontology.org/) to obtain additional insights into the putative functions of unigenes. Of the 479 C. praecox unigenes, 364 were assigned GO terms in any category (biological, cellular, and molecular). Figures 3, 4, and 5 classify the unigenes according to terms in the biological process ontology, molecular function ontology, and cellular component ontology, respectively.
Figure 3.
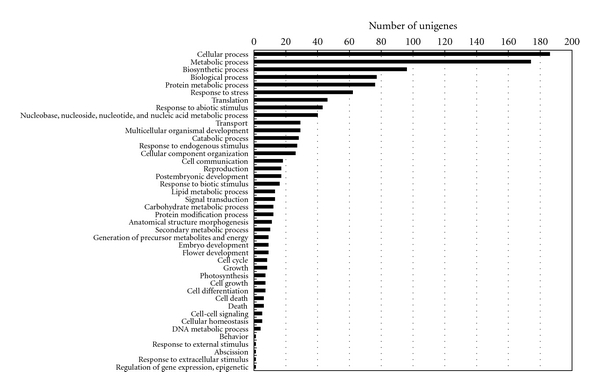
GO classification of the ESTs based on their biological functions in the C. praecox flower cDNA library.
Figure 4.
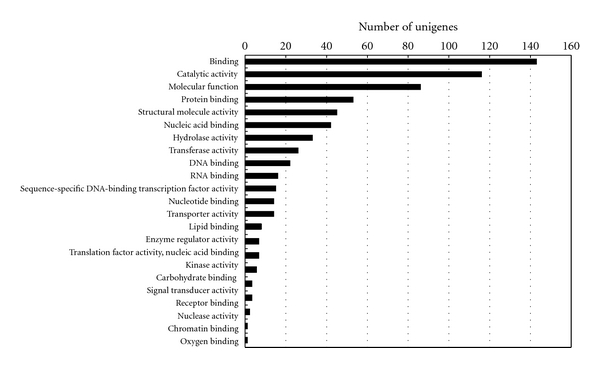
GO classification of the ESTs based on their molecular functions in the C. praecox flower cDNA library.
Figure 5.
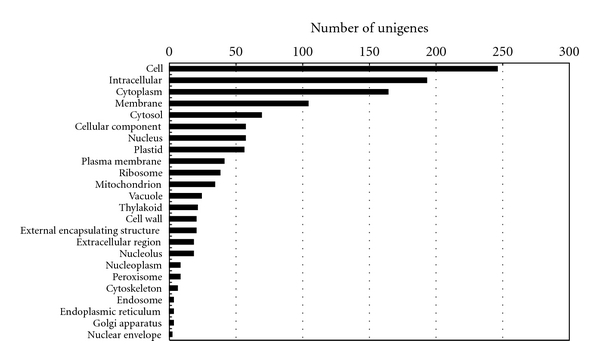
GO classification of the ESTs based on their cellular components in the C. praecox flower cDNA library.
3.3. Sequences Related to Stress and Defense
Among the ESTs that matched genes with known or putative functions, approximately 95 unigenes (291 ESTs) involved in stress and defense accounted for 19.8% (95/479) of all unigenes and 33.6% (291/867) of all ESTs. Table 5 shows the nonredundant ESTs that share similarities with genes related to defense and stress response according to GO classifications and previously published data. As expected, the ESTs involved in stress and defense were highly abundant in the library because the C. praecox thrives and blossoms in winter, thereby confirming previous reports of related transcripts with higher levels of defense in developing flowers [16–20].
Table 5.
Sequences related to stress and defense.
| Gene code | Putative function | E-value | Matches no. | Number of ESTs |
|---|---|---|---|---|
| Cp465 | catalase | 1.00E − 148 | BAC79443.1 | 1 |
| Cp346 | transporter | 1.00E − 138 | AAP13421.1 | 2 |
| Cp382 | peroxidase | 1.00E − 125 | AAK52084.1 | 1 |
| Cp374 | GMPase | 1.00E − 122 | AAT58365.1 | 1 |
| Cp333 | putative aldolase | 1.00E − 122 | AAM64281.1 | 1 |
| Cp397 | chitinase | 1.00E − 105 | AAF04454.1 | 1 |
| Cp452 | cysteine synthase | 3.00E − 99 | BAA05965.1 | 1 |
| Cp314 | tryptophan synthase beta chain | 2.00E − 94 | AAN15574.1 | 1 |
| Cp357 | Stromal cell-derived factor 2-like protein | 2.00E − 93 | XP_481233.1 | 1 |
| Cp191 | glutathione S-transferase GST 23 | 5.00E − 92 | AAG34813.1 | 2 |
| Cp25 | dehydroascorbate reductase | 2.00E − 89 | AAL71857.1 | 3 |
| Cp438 | ras-related protein RAB8-3 | 9.00E − 89 | BAB84324.1 | 1 |
| Cp459 | GTP-binding protein | 1.00E − 86 | AAN31076.1 | 1 |
| Cp340 | putative 3-Hydroxyisobutyryl-coenzyme A hydrolase | 9.00E − 84 | AAN41356.1 | 1 |
| Cp379 | cell-autonomous heat shock cognate protein 70 | 1.00E − 82 | gAAN86276.1 | 1 |
| Cp39 | auxin-binding protein | 3.00E − 80 | AAB51240.1 | 4 |
| Cp10 | glutathione S-transferase GST 22 | 4.00E − 80 | AAG34812.1 | 8 |
| Cp304 | putative 2-Nitropropane dioxygenase | 2.00E − 79 | AAL34288.1 | 3 |
| Cp21 | glutathione S-transferase GST 22 | 2.00E − 78 | AAG34812.1 | 3 |
| Cp311 | GTP-binding protein | 6.00E − 73 | AAM12880.1 | 1 |
| Cp353 | patatin-like protein | 1.00E − 73 | CAB16788.1 | 2 |
| Cp274 | glutathione S-transferase | 1.00E − 72 | AAF61392.1 | 1 |
| Cp359 | 1,4-Alpha-glucan-maltohydrolase | 1.00E − 71 | CAH60892.1 | 1 |
| Cp59 | BTF3b-like transcription factor | 2.00E − 68 | AAT67244.1 | 3 |
| Cp76 | NDKB_FLABI Nucleoside diphosphate kinase B | 2.00E − 63 | P47920 | 1 |
| Cp166 | putative tropinone reductase | 6.00E − 59 | AAM62552.1 | 1 |
| Cp313 | putative glutathione S-transferase T3 | 5.00E − 56 | AAG16758.1 | 1 |
| Cp156 | protolysis and peptidolysis | 6.00E − 56 | CAA50022.1 | 1 |
| Cp120 | putative pyruvate dehydrogenase E1 alpha subunit | 8.00E − 56 | XP_467697.1 | 1 |
| Cp80 | sadtomato protein | 7.00E − 55 | AAS77347.1 | 1 |
| Cp440 | putative multiple stress-responsive zinc-finger protein | 6.00E − 54 | BAD35553.1 | 1 |
| Cp175 | epoxide hydrolase | 6.00E − 50 | AAB02006.1 | 1 |
| Cp364 | cold acclimation protein WCOR413-like protein beta form | 4.00E − 49 | AAG13394.1 | 1 |
| Cp471 | pathogenesis-related protein 4B | 1.00E − 48 | eCAA41438.1 | 1 |
| Cp145 | Thioredoxin | 4.00E − 44 | CAA77847.1 | 1 |
| Cp406 | auxin-responsive family protein | 1.00E − 43 | NP_565113.1 | 1 |
| Cp38 | basic PR-1 protein precursor | 3.00E − 43 | AAU20808.1 | 4 |
| Cp215 | putative abscisic acid-induced protein | 4.00E − 41 | BAD37454.1 | 1 |
| Cp66 | GLRX_RICCO Glutaredoxin | 5.00E − 40 | sp|P55143| | 2 |
| Cp235 | histone H2A | 5.00E − 38 | CAA07234.1 | 1 |
| Cp111 | topoisomerase 6 subunit B | 5.00E − 37 | CAC24690.1 | 1 |
| Cp232 | cysteine proteinase inhibitor-like protein | 2.00E − 36 | BAB03156.1 | 2 |
| Cp220 | glyoxalase I family protein | 2.00E − 35 | NP_973579.1 | 2 |
| Cp214 | acyl-CoA-binding protein | 3.00E − 35 | CAA70200.1 | 1 |
| Cp27 | proline-rich protein | 1.00E − 30 | AAF78903.1 | 6 |
| Cp20 | lectin | 5.00E − 28 | AAD45250.1 | 17 |
| Cp134 | metallothionein-like protein | 6.00E−27 | CAB52585.1 | 1 |
| Cp127 | Gip1-like protein | 2.00E − 24 | CAD10106.1 | 1 |
| Cp296 | bZIP transcription factor | 2.00E − 24 | AAN61914.1 | 2 |
| Cp375 | 3-Oxoacyl-[acyl-carrier-protein] synthase | 2.00E − 23 | AAC78479.1 | 1 |
| Cp104 | metallothionein-like protein type 2 | 4.00E − 18 | CAB77242.1 | 2 |
| Cp372 | allyl alcohol dehydrogenase | 5.00E − 17 | BAA89423.1 | 1 |
| Cp36 | metallothionein-like protein type 2 | 1.00E − 13 | AAV97748.1 | 5 |
| Cp173 | low temperature and salt responsive protein | 1.00E − 12 | BAC23051.1 | 1 |
| Cp472 | cystatin | 2.00E − 07 | AAO18638.1 | 1 |
| Cp88 | membrane channel protein | 3.00E − 06 | CAB83138.1 | 2 |
| Also related to development | ||||
| Cp391 | actin | 1.00E − 166 | AAN40685.1 | 1 |
| Cp402 | xyloglucan endotransglycosylase precursor | 1.00E − 110 | AAC09388.1 | 1 |
| Cp26 | aquaporin | 8.00E − 97 | CAE53881.1 | 7 |
| Cp381 | 4-Coumarate-CoA ligase-like protein | 2.00E − 89 | AAP03021.1 | 1 |
| Cp461 | glyceraldehyde-3-phosphate dehydrogenase | 1.00E − 86 | CAC88118.1 | 1 |
| Cp291 | 3-Ketoacyl-CoA thiolase | 6.00E − 84 | CAA47926.1 | 1 |
| Cp378 | probable ubiquitin-conjugating enzyme E2 | 2.00E − 83 | AAC32141.1 | 1 |
| Cp400 | auxin response factor 4 | 5.00E − 81 | BAD19064.1 | 1 |
| Cp288 | expansin | 4.00E − 80 | AAO15998.1 | 2 |
| Cp226 | calmodulin | 3.00E − 79 | AAB68399.1 | 4 |
| Cp323 | putative calmodulin | 3.00E − 73 | CAC84563.1 | 2 |
| Cp298 | putative adrenal gland protein AD-004 | 2.00E − 65 | XP_477670.1 | 1 |
| Cp326 | UDP-glucose:salicylic acid glucosyltransferase | 2.00E − 65 | AAF61647.1 | 1 |
| Cp58 | actin-depolymerizing factor 1 | 1.00E − 63 | AAK72617.1 | 2 |
| Cp167 | ABC transporter | 4.00E − 60 | AAP80385.1 | 1 |
| Cp286 | putative CCR4-associated factor | 4.00E − 60 | AAN13153.1 | 1 |
| Cp368 | MYB8 protein | 1.00E − 60 | CAD87008.1 | 1 |
| Cp324 | actin depolymerizing factor | 2.00E − 59 | AAD23407.1 | 1 |
| Cp450 | TAF9 | 1.00E − 58 | AAR28026.1 | 1 |
| Cp405 | myb family transcription factor-like | 2.00E − 52 | BAD29385.1 | 1 |
| Cp77 | putative actin-depolymerizing factor 1 | 4.00E − 48 | XP_475079.1 | 1 |
| Cp121 | TAF10 | 3.00E − 41 | AAR28030.1 | 1 |
| Cp347 | putative transcription factor | 2.00E − 38 | XP_479103.11 | 2 |
| Cp4 | lipid-transfer protein | 2.00E − 35 | AAS13435.1 | 37 |
| Cp1 | lipid transfer protein | 1.00E − 34 | CAA63340.1 | 77 |
| Cp275 | lipid transfer protein precursor | 4.00E − 30 | AAL27855.1 | 2 |
| Cp9 | lipid transfer protein 1 precursor | 3.00E − 25 | AAT45202.1 | 1 |
| Cp216 | lipid-transfer protein | 3.00E − 24 | NP_915960.1 | 4 |
| Cp90 | zinc finger homeobox family protein | 3.00E − 22 | NP_565088.1 | 1 |
| Cp41 | lipid transfer protein | 3.00E − 18 | BAC77694.1 | 1 |
| Cp13 | lipid transfer protein isoform 4 | 1.00E − 16 | AAO33394.1 | 1 |
| Cp338 | polyprotein | 2.00E − 16 | CAD92110.1 | 1 |
| Cp82 | dormancy related protein, putative | 6.00E − 15 | AAG51119.1 | 1 |
| Cp8 | lipid-transfer protein | 7.00E − 14 | AAS13435.1 | 1 |
| Cp141 | lipid-transfer protein | 4.00E − 13 | AAS13435.1 | 1 |
| Cp211 | neutral/alkaline invertase 1 | 2.00E − 12 | AAV28809.1 | 1 |
| Cp451 | putative MYB family transcription factor | 8.00E − 11 | AAD23043.1 | 1 |
| Cp34 | Lea5 protein | 3.00E − 10 | CAA86851.1 | 4 |
| Cp16 | putative LEA III protein isoform 2 | 6.00E − 10 | CAC39110.1 | 10 |
Twelve unigenes (22 ESTs) related to cold stress tolerance were found in the library; these were classified as “response to cold” according to GO terms [e.g., β-amylase, acyl-CoA-binding protein, 3-hydroxyisobutyryl-coenzyme A hydrolase, catalase, low-temperature and salt-responsive protein, glutathione S-transferase (GST), membrane channel protein, and abscisic-acid-induced protein] and “cold acclimation” (e.g., fatty acid biosynthesis 1 and WCOR413-like protein). In this class, the most abundant sequences encode GST. Five GSTs encoded by 16 ESTs were identified in the library. GST genes exhibited a diverse range of responses to jasmonates, salicylic acid, ethylene, as well as oxidative stress in Arabidopsis [21], and were induced by heavy metals and hypoxic stress in rice roots [22]. In the current study, however, not enough cold-related unigenes were obtained, as expected. Some unknown functional genes related to cold stress tolerance likely exist in this library.
Another class of genes involved in “response to absence of light,” “low light intensity,” and “response to red or blue light” according to GO terms was also represented in the library. Eight unigenes (10 ESTs) were identified, including acyl-CoA-binding protein, sadtomato protein, and catalase, among others.
Of the stress- and defense-related unigenes, 39 were possibly related to development. Nine types of LTPs or LTP precursors were encoded by the highest number of ESTs in this study (Table 5). Research has suggested different functions for LTPs in the physiology of plants, such as cutin synthesis, β-oxidation, somatic embryogenesis, pollen development, allergenics, plant signaling, and plant defense [23–25], but the true physiological role of LTPs in C. praecox flowers has yet to be determined. Fourteen ESTs encoding two kinds of LEA proteins, groups III and V, were identified. The presence of LEA proteins correlates well with freezing, water deficit, and salt stress [26–28], probably through the prevention of enzyme aggregation [29], and likely plays a similar role in C. praecox. Some other development-related unigenes were also found, such as MYB, calmodulin, actin, CCR4-associated factor, ubiquitin-conjugating enzyme E2, and ABC transporter, which are involved in transcription factor activity, signal transduction, cell structure, nucleotide metabolism, protein metabolic process, and transporter activity, respectively.
3.4. Sequences Related to Floral Development
Table 6 shows the 19 unigenes related to floral development. Five of these were homologous to MADS box transcription factor genes. Plant life critically depends on the function of MADS box genes encoding MADS domain transcription factors, which are present to a limited extent in nearly all major eukaryotic groups but constitute a large gene family in land plants [30].
Table 6.
Sequences related to floral development.
| Gene code | Putative function | E-value | Matches no. | Number of ESTs |
|---|---|---|---|---|
| Cp360 | Peroxisomal fatty acid beta-oxidation multifunctional protein AIM1 | 1.00E − 108 | XP_464920.1 | 1 |
| Cp268 | caffeoyl-CoA O-methyl-transferase-like protein | 1.00E − 107 | CAB80122.1 | 2 |
| Cp237 | MADS box protein | 1.00E − 106 | AAQ83835.1 | 2 |
| Cp116 | MADS box transcription factor AP3-1 | 2.00E − 95 | AAF73928.1 | 1 |
| Cp330 | RAC-like G-protein Rac1 | 1.00E − 93 | AAD47828.2 | 1 |
| Cp217 | beta-D-glucosidase | 2.00E − 92 | AAQ17461.1 | 1 |
| Cp413 | leucine-rich repeat transmembrane protein kinase | 6.00E − 86 | NP_192248.2 | 1 |
| Cp408 | isoflavone reductase related protein | 3.00E − 85 | AAC24001.1 | 1 |
| Cp423 | putative polyketide synthase | 8.00E − 76 | NP_919053.1 | 1 |
| Cp335 | AGL 6 | 3.00E − 73 | AAY25580.1 | 2 |
| Cp395 | isopentenyl pyrophosphate:dimethyllallyl pyrophosphate isomerase | 2.00E − 70 | BAB09611.1 | 1 |
| Cp297 | SRG1-like protein | 1.00E − 61 | CAB81342.1 | 3 |
| Cp383 | secondary cell wall-related glycosyltransferase family 47 | 3.00E − 49 | AAX33321.1 | 1 |
| Cp197 | AGL9.2 | 4.00E − 42 | AAX15924.1 | 1 |
| Cp436 | allergen-like protein BRSn20 | 1.00E − 37 | AAM62935.1 | 1 |
| Cp407 | caffeate O-methyltransferase | 8.00E − 23 | AAV36331.1 | 1 |
| Cp203 | LLP-B3 protein | 1.00E − 22 | AAN76546.1 | 1 |
| Cp328 | STYLOSA protein | 1.00E − 15 | CAF18245.1 | 1 |
| Cp458 | MADS-box protein 9 | 9.00E − 15 | AAQ72497.1 | 1 |
MADS box genes control diverse developmental processes in flowering plants—ranging from root to flower and fruit development—and they especially control the processes of the transition from vegetative to reproductive development and establishment of floral organ identity [31]. The present study has also identified six unigenes related to secondary metabolism that are probably involved in floral color and fragrance.
The role of chilling temperature in dormancy in vegetative buds and induction of flowering has been investigated in many temperate-region species, particularly in A. thaliana and Populus spp. [32, 33]. The physiological processes of dormancy release and induction of flowering competence rely on longer-term chilling temperature and a period of vernalization, respectively [34, 35]. The current study has identified dormancy and vernalization-related genes; however, only one unigene (Cp82; E-value = 6.00E −15) was annotated as a dormancy-related protein in the library The possible reason was only a small-scale sequencing in our study or the processes of dormancy release and induction of flowering competence in C. praecox only last a short period.
3.5. Expression Analysis of Cold-Responsive and Floral Development-Related Genes
Real-time RT-PCR was performed for 12 unigenes, including 9 selected from the GO Slim annotation belonging to “response to cold” (Cp88, Cp173, Cp215, Cp274, Cp359, Cp364, Cp375, Cp440, and Cp465), 1 annotated as a dormancy-related protein (Cp82), and 2 without functional annotation (Cp24 and Cp64), to analyze the changes in their expression due to cold stress (Figure 6). The data revealed that Cp82 responded to cold stress immediately after treatment and reached peak expression levels as early as 15 min. Cp24 (no annotation), Cp88 (membrane channel protein), Cp274 (GST), Cp364 (cold acclimation protein WCOR413-like protein), and Cp440 (multiple stress-responsive zinc-finger protein) were upregulated after 15 min of treatment and reached their expression peaks at 1 h. Cp173 (low-temperature and salt-responsive protein), Cp215 (abscisic acid-induced protein), Cp359 (1,4-alpha-glucan-maltohydrolase), Cp375 (3-oxoacyl-[acyl-carrier-protein] synthase), and Cp465 (catalase) displayed later responses and reached their peak expression levels at 6 h. Cp64 (no annotation) exhibited minor changes in its transcript level.
Figure 6.
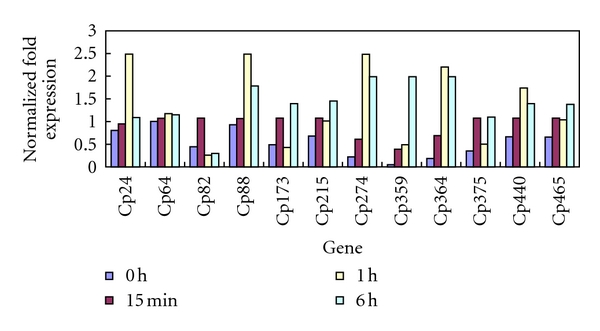
Expression analysis of 12 genes under cold stress (4°C).
Real-time RT-PCR was also applied to validate the expression patterns of 11 genes related to floral development (Cp197, Cp203, Cp268, Cp297, Cp328, Cp330, Cp360, Cp383, Cp423, Cp436, and Cp458), Cp24, Cp64, and Cp82 (Figure 7). The expression patterns of these 14 genes were analyzed in roots, stems, leaves, outer tepals, middle tepals, inner tepals, stamina, and pistils. The results showed clear differences in their expression. The genes related to floral development, except for Cp203 (LLP-B3 protein) and Cp297 (SRG1-like protein), increased by more than twofold in middle tepals. Cp24 and Cp203 presented an active expression in stamina but showed a very slight accumulation in other tissues. Cp268 (caffeoyl-CoA O-methyl-transferase-like protein) and Cp458 (MADS box protein 9) presented a peak in middle tepals but were only slightly or not expressed at all in roots, stems, leaves, and pistils. Cp197 (AGL9.2) was higher in all reproductive organs and presented its peak in middle tepals but was not detected in roots, stems, and leaves. The expression of Cp328 (STYLOSA protein) and that of Cp383 (secondary cell wall-related glycosyltransferase family 47) were not detected in roots; moreover, Cp328 increased by more than fourfold in middle tepals, whereas Cp383 did in pistils and stems. The accumulation of Cp82 (dormancy-related protein) transcripts was higher in stems, leaves, stamina, and pistils. The other genes expressed in all the detected tissues, but Cp64 and Cp297 (SRG1-like protein) presented lower levels of expression. Cp360 (peroxisomal fatty acid β-oxidation multifunctional protein AIM1) increased by more than twofold in roots, middle tepals, and pistils.
Figure 7.
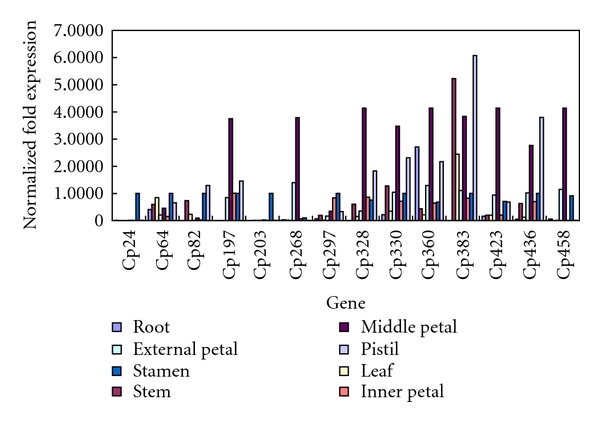
Expression analysis of 14 genes in different tissues of C. praecox.
Quantitative real-time PCR methods were used to validate the transcript levels of the 14 genes further during the blooming process in C. praecox (Figure 8). The results showed that all these genes were not detected or very slightly expressed in Stage 1 and that Cp64, Cp82, Cp268, Cp297, Cp330, Cp360, and Cp383 had almost no accumulation in Stage 2. The transcript accumulations of Cp203 and Cp458 were sharply elevated to the highest level in Stage 2 but dramatically decreased at the subsequent stages of floral development. The expressions of Cp24, Cp82, and Cp436 presented a peak in Stage 4. The transcript accumulations of the other 9 genes increased during the six developmental stages and reached their peak in Stage 6. The expression of Cp297(SRG1-like protein) was significantly high in Stage 6 but very low in the other stages, and it was associated with flower senescence. The SRG1 gene is reportedly expressed in senescing organs of Arabidopsis plants [36].
Figure 8.
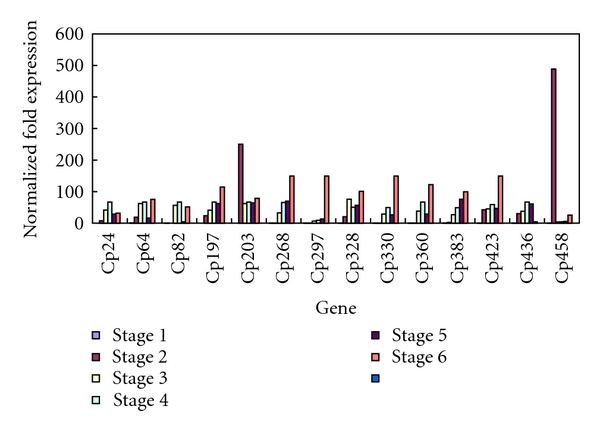
Expression analysis of 14 genes during C. praecox flowering.
The current study found few references about bud dormancy in C. praecox. The expression pattern of Cp82 was very attractive. However, the results indicate that Cp82 is not certainly related to flower-bud dormancy. The dormancy-related protein CAA93825.1, which matched Cp82 (E-value = 1e−15), has been reported to play a role in dormancy breaking and in the germination of Trollius ledebourii seeds [37]. These data warrant further research into the relativity of Cp82 to dormancy breaking and germination in C. praecox seeds.
4. Conclusions
A cDNA library was constructed to generate an EST collection from C. praecox, thereby providing a preliminary view into the genomic properties of this species. This collection of high-quality ESTs represents the first EST data set for C. praecox. Eight hundred sixty-seven valid EST sequences were generated, and 479 unigenes were assembled, among which 266 unigenes (55.53%) were identified according to their significant similarities with proteins of known functions. The EST sequences have been deposited in GenBank under accession numbers DW222667 to DW223533. Stress response genes and floral development-related genes were also identified. This study evaluated the expression patterns of 23 genes, including 2 novel ones, using real-time RT-PCR. Further investigations in this direction would help in the discovery of promising candidates with a key role in the development of stress tolerance for woody plants to bloom.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant nos. 31070622 and 30872063) and the Fundamental Research Funds for the Central Universities (Grant nos. XDJK2009A004 and XDJK2010C071).
References
- 1.Zhao KG, Zhou MQ, Chen LQ, Zhang D, Robert GW. Genetic diversity and discrimination of Chimonanthus praecox (L.) link germplasm using ISSR and RAPD markers. HortScience. 2007;42(5):1144–1148. [Google Scholar]
- 2.Feng JQ. New Zealand flower industry—with special reference to wintersweet introduction and commercialization. Journal of Beijing Forestry University. 2007;29(supplement 1):4–8. [Google Scholar]
- 3.Wei H, Dhanaraj AL, Rowland LJ, Fu Y, Krebs SL, Arora R. Comparative analysis of expressed sequence tags from cold-acclimated and non-acclimated leaves of Rhododendron catawbiense Michx. Planta. 2005;221(3):406–416. doi: 10.1007/s00425-004-1440-1. [DOI] [PubMed] [Google Scholar]
- 4.Mahalingam R, Gomez-Buitrago A, Eckardt N, et al. Characterizing the stress/defense transcriptome of Arabidopsis . Genome Biology. 2003;4(3, article R20) doi: 10.1186/gb-2003-4-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iturriaga G, Cushman MAF, Cushman JC. An EST catalogue from the resurrection plant Selaginella lepidophylla reveals abiotic stress-adaptive genes. Plant Science. 2006;170(6):1173–1184. [Google Scholar]
- 6.Zhang J, John UP, Wang Y, et al. Targeted mining of drought stress-responsive genes from EST resources in Cleistogenes songorica. Journal of Plant Physiology. 2011;168(15):1844–1851. doi: 10.1016/j.jplph.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Ozgenturk NO, Oru F, Sezerman U, et al. Generation and analysis of expressed sequence tags from Olea europaea L. Comparative and Functional Genomics. 2010;2010:9 pages. doi: 10.1155/2010/757512. Article ID 757512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Channelière S, Rivière S, Scalliet G, et al. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Letters. 2002;515(1–3):35–38. doi: 10.1016/s0014-5793(02)02413-4. [DOI] [PubMed] [Google Scholar]
- 9.Vinod MS, Sankararamasubramanian HM, Priyanka R, Ganesan G, Parida A. Gene expression analysis of volatile-rich male flowers of dioecious Pandanus fascicularis using expressed sequence tags. Journal of Plant Physiology. 2010;167(11):914–919. doi: 10.1016/j.jplph.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Tsai WC, Hsiao YY, Lee SH, et al. Expression analysis of the ESTs derived from the flower buds of Phalaenopsis equestris . Plant Science. 2006;170(3):426–432. [Google Scholar]
- 11.Wu CL, Hu NZ. Studies on the flower form and blooming characteristics of the wintersweet. Acta Horticulturae Sinica. 1995;22(3):277–282. [Google Scholar]
- 12.Gao QK, Hu C. Construction of a cDNA library of host recognition kairomone for telenomus theophilae. Insect Science. 2002;9(1):35–39. [Google Scholar]
- 13.Ogihara Y, Mochida K, Nemoto Y, et al. Correlated clustering and virtual display of gene expression patterns in the wheat life cycle by large-scale statistical analyses of expressed sequence tags. Plant Journal. 2003;33(6):1001–1011. doi: 10.1046/j.1365-313x.2003.01687.x. [DOI] [PubMed] [Google Scholar]
- 14.Sterky F, Regan S, Karlsson J, et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbon S, Ireland A, Mungall CJ, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández P, Paniego N, Lew S, Hopp HE, Heinz RA. Differential representation of sunflower ESTs in enriched organ-specific cDNA libraries in a small scale sequencing project. BMC Genomics. 2003;4, article no. 40 doi: 10.1186/1471-2164-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lotan T, Ori N, Fluhr R. Pathogenesis-related proteins are developmentally regulated in tobacco flowers. The Plant cell. 1989;1(9):881–887. doi: 10.1105/tpc.1.9.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neale AD, Wahleithner JA, Lund M, et al. Chitinase, β-1,3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell. 1990;2(7):673–684. doi: 10.1105/tpc.2.7.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Q, Kawata EE, Morse MJ, Wu HM, Cheung AY. A flower-specific cDNA encoding a novel thionin in tobacco. Molecular and General Genetics. 1992;234(1):89–96. doi: 10.1007/BF00272349. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA. Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell. 1993;5(2):203–213. doi: 10.1105/tpc.5.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Molecular Biology. 2002;49(5):515–532. doi: 10.1023/a:1015557300450. [DOI] [PubMed] [Google Scholar]
- 22.Moons A. Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Letters. 2003;553(3):427–432. doi: 10.1016/s0014-5793(03)01077-9. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho ADO, Gomes VM. Role of plant lipid transfer proteins in plant cell physiology—a concise review. Peptides. 2007;28(5):1144–1153. doi: 10.1016/j.peptides.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Chen G, Hao X, et al. CaMF2, an anther-specific lipid transfer protein (LTP) gene, affects pollen development in Capsicum annuum L. Plant Science. 2011;181(4):439–448. doi: 10.1016/j.plantsci.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Kiełbowicz-Matuk A, Rey P, Rorat T. The organ-dependent abundance of a Solanum lipid transfer protein is up-regulated upon osmotic constraints and associated with cold acclimation ability. Journal of Experimental Botany. 2008;59(8):2191–2203. doi: 10.1093/jxb/ern088. [DOI] [PubMed] [Google Scholar]
- 26.Ndong C, Danyluk J, Wilson KE, Pocock T, Huner NPA, Sarhan F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiology. 2002;129(3):1368–1381. doi: 10.1104/pp.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dramé KN, Clavel D, Repellin A, Passaquet C, Zuily-Fodil Y. Water deficit induces variation in expression of stress-responsive genes in two peanut (Arachis hypogaea L.) cultivars with different tolerance to drought. Plant Physiology and Biochemistry. 2007;45(3-4):236–243. doi: 10.1016/j.plaphy.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Park SC, Kim YH, Jeong JC, et al. Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta. 2011;233(3):621–634. doi: 10.1007/s00425-010-1326-3. [DOI] [PubMed] [Google Scholar]
- 29.Grelet J, Benamar A, Teyssier E, Avelange-Macherel MH, Grunwald D, Macherel D. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiology. 2005;137(1):157–167. doi: 10.1104/pp.104.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gramzow L, Theissen G. A hitchhiker’s guide to the MADS world of plants. Genome Biology. 2010;11(6, article no. 214) doi: 10.1186/gb-2010-11-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker A, Theißen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution. 2003;29(3):464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 32.Chao WS, Foley ME, Horvath et al. DP. Signals regulating dormancy in vegetative buds. International Journal of Plant Developmental Biology. 2007;1(1):49–56. [Google Scholar]
- 33.Dennis ES, Peacock WJ. Epigenetic regulation of flowering. Current Opinion in Plant Biology. 2007;10(5):520–527. doi: 10.1016/j.pbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Chouard P. Vernalization and its relations to dormancy. Annual Review of Plant Physiology. 1960;11:191–238. [Google Scholar]
- 35.Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends in Plant Science. 2007;12(5):217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Callard D, Axelos M, Mazzolini L. Novel molecular markers for late phases of the growth cycle of arabidopsis thaliana cell-suspension cultures are expressed during organ senescence. Plant Physiology. 1996;112(2):705–715. doi: 10.1104/pp.112.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey PC, Lycett GW, Roberts JA. A molecular study of dormancy breaking and germination in seeds of Trollius ledebouri. Plant Molecular Biology. 1996;32(3):559–564. doi: 10.1007/BF00019110. [DOI] [PubMed] [Google Scholar]


