Abstract
The polyomaviruses are small DNA viruses that can establish latency in the human host. The name polyomavirus is derived from the Greek roots poly-, which means “many,” and -oma, which means “tumours.” These viruses were originally isolated in mouse (mPyV) and in monkey (SV40). In 1971, the first human polyomaviruses BK and JC were isolated and subsequently demonstrated to be ubiquitous in the human population. To date, at least nine members of the Polyomaviridae family have been identified, some of them playing an etiological role in malignancies in immunosuppressed patients. Here, we describe the biology of human polyomaviruses, their nonmalignant and malignant potentials ability, and their relationship with the host immune response.
1. The Human Polyomaviruses
Polyomavirus is the sole genus of viruses within the Polyomaviridae family. Initially, polyomaviruses were taxonomically classified as a genus of the Papovaviridae family along with papillomaviruses; in 2000, however, the International Committee on Taxonomy of Viruses formally split the two viruses into two new families, Polyomaviridae and Papillomaviridae [1].
The name polyomavirus is derived from the Greek roots poly-, which means “many,” and -oma, which means “tumours.”
The first polyomavirus isolated was the mouse polyomavirus (mPyV) [2] causing the formation of multiple tumour types when inoculated into newborn mice. Subsequently, many polyomaviruses have been found to infect a variety of vertebrate species, including rabbits, rodents, birds, nonhuman primates, and, to date, nine polyomaviruses have been found in humans. The range of host species for each polyomavirus is very narrow, and the productive infection is limited to natural host [3].
Until 2006, BK virus (BKV) and JC virus (JCV), first isolated in 1971 [4, 5], were the only two human polyomaviruses known although some evidence suggested that the simian virus 40 (SV40) could be linked to some human tumours [6].
JCV and BKV usually infect the human population during early childhood, and primary infection is often asymptomatic. These viruses can remain latent in the kidney cells of the host until reactivation which occurs during immunodepression. JCV causes progressive multifocal leukoencephalopathy (PML) in cases of severe immunodeficiency, generally due to HIV infection, whereas BKV causes nephropathy in patients who have undergone to kidney transplants [7, 8]. SV40 does not infect naturally humans, but it was unintentionally introduced in the human population through contaminated polio vaccine in the late 1950s [6].
Lymphotropic polyomavirus (LPV) is another monkey polyomavirus that may also infect humans, since specific antibodies against it are present in the human population, and LPV DNA was detected in blood from immunosuppressed and immunocompetent subjects [9–11].
In 2007, two new human polyomaviruses were independently described: the KI polyomavirus (KIV) at Karolinska Institute and WU polyomavirus (WUV) the at the Washington University [12, 13]. These viruses that are closely related to each other were both identified from nasopharyngeal aspirates from children with respiratory tract infections. So far, no specific diseases have been associated to their infections. In 2008, a fifth polyomavirus, Merkel cell polyomavirus (MCPyV), was isolated from the skin of a patient affected by Merkel Cell carcinoma (MCC) showing its ability to cause most of the Merkel skin cancers [14]. In addition, three other polyomaviruses were isolated from no-tumoral skin, the Human Polyomavirus 6 and 7 (HPyV6, HPyV7), and the Trichodysplasia Spinulosa-associated Virus (TSPyV) [15, 16]. Finally, this year, the last polyomavirus, named Human Polyomavirus 9 (HPyV9), closely related to LPV, was identified from the blood and urine of asymptomatic renal transplant recipients [17] (Table 1).
Table 1.
Human polyomaviruses.
| Nomenclature | Year identified | Prevalence in human population | Disease associations |
|---|---|---|---|
| BKV | 1971 | >90% of adults | Cystitis, polyomavirus-associated nephropathies, ureteral stenosis |
| JCV | 1971 | >70% of adults | Progressive multifocal leukoencephalopathy |
| KIV | 2007 | 55–70% of adults | Not defined |
| WUV | 2007 | 69–80% of adults | Not defined |
| MCPyV | 2008 | 42–70% of adults | Merkel cell carcinoma |
| HPyV6 | 2010 | Not defined | Not defined |
| HPyV7 | 2010 | Not defined | Not defined |
| TSPyV | 2010 | Not defined | Transplant-associated trichodysplasia spinulosum |
| HPyV9 | 2011 | Not defined | Not defined |
BKV: BK virus; JCV: JC virus; KIV: Karolinska Institute polyomavirus; WUV: Washington University polyomavirus; MCPyV: Merkel cell polyomavirus; HPyV6: human polyomavirus 6; HPyV7: human polyomavirus 7; TSPyV: trichodysplasia spinulosum polyomavirus; HPyV9: human polyomavirus 9.
Viruses that belong to the Polyomaviridae family are small and nonenveloped. They have icosahedral capsids, measuring 40.5–44 nm in diameter and circular, double-stranded supercoiled DNA genomes of approximately 5 Kb. The genome can be divided into two regions, one encoding the early proteins (the large and small Tumor Antigens) and another encoding the late, structural proteins Viral Proteins 1–3, which make up the capsid. The late region of BKV, JCV, and SV40 also encodes an agnoprotein, that seems to be implicated in viral maturation and microRNAs, which show a regulatory function in the expression of the Large Tumor Antigen (LTAg) [18, 19]. The early and late regions are separated by a noncoding region, the transcriptional control region (NCCR), which contains the origin of replication and promoters of viral transcription [20] (Figure 1).
Figure 1.
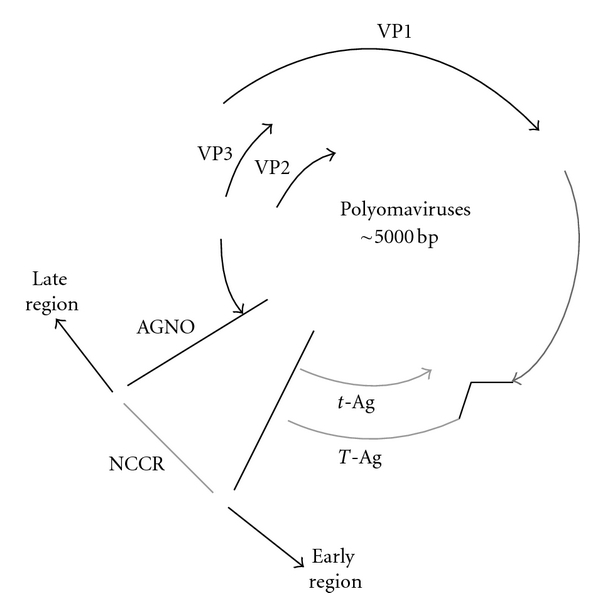
Schematic representation of the polyomaviruses genome organization. The circular double-stranded DNA genome is length about 5kb. The early region encodes the functional proteins LTAg and small T-Antigen, while the late region encodes the structural proteins VP1-3. The genome of JCV, BKV, and SV40 also encodes a small structural protein named Agno. The noncoding control region (NCCR) contains the origin of replication and regulates the replication and transcription of both the early and late genes.
The primary infection of polyomaviruses is usually asymptomatic, probably occurring via respiratory and/or feco-oral transmission and establishing a latent phase of infection in the host. JCV and BKV persist in the kidney, while lymphoid tissue and the central nervous system (CNS) [21–23] have been indicated as possible sites of latency. HPyV6, HPyV7, TSPyV probably persist in the skin and in the lymphocytes, but little is known about the cell tropism of these most recent discovered viruses [24].
Under normal conditions polyomaviruses probably develop mechanisms to evade immune recognition and enable their latency into their human hosts, but these mechanisms are unknown. Very recently, Bauman and colleagues demonstrated that a viral miRNA identical in sequence between JCV and BKV is involved in immune regulation, resulting in the escape of the infected cells from the natural killer cells-mediated killing. In particular, it has been showed that the miRNA of polyomaviruses targets and downregulates the stress-induced ligand ULBP3, which is normally recognized by the killer receptor NKG2D, expressed by NK cells and other immune cells [25]. Consequently, the reduction of ULBP3 results in decreased killing of the infected cells [26].
Thus, reactivation of the infection is closely linked to the impairment of the immunological state of the host. There are two possible outcomes to infection of a cell by polyomaviruses. A productive infection occurs when polyomaviruses infect permissive cells: in this case, the virus entry into target cells is characterized by nuclear DNA replication and assembly of the viral capsid followed by the cell lysis. On the contrary, nonproductive infection is established when the virus infects nonpermissive cells blocking the viral DNA replication and triggering cell transformation (oncogenesis) [27].
2. Seroprevalence Polyomaviruses in the Human Population
Polyomaviruses are widely distributed in the worldwide population, and their prevalence has been thoroughly investigated. First, it should be emphasized the high degree of identity between the genome and aminoacidic sequences of the human polyomaviruses VP1. For instance, the closely related viruses BKV and JCV share 78.2% identity of the VP1 amino acid sequences and even greater identity (81.3%) is shared between BKV and SV40. KIV and WUV show 66% of sequence homology and are phylogenetically compared with LPV. They are more distant to JCV and BKV presenting only 28% of amino acid identity [28–30]. Consequently, results of seroepidemiological studies to determine the prevalence of antibodies against specific human polyomaviruses could be highly heterogeneous considering their possibility to cross-react with all polyomaviruses species. In addition, the methods used to measure the specific seroreactivity to human polyomaviruses are different, including virus particles antigen assay, enzyme, linked immunosorbent assay (ELISA), hemoagglutination inhibition assay, and immune electron microscopy [31].
In general, serological population surveys for the detection of antibodies indicates that seroconversion to human polyomaviruses takes place early in life, 5 to 7 years for BKV, with conversion to JCV occurring later. Little is known about the occurrence of the exposure for the other newly discovered human polyomaviruses [30, 32–35]. Kean and colleagues determined the seroprevalence for LPV (14%), MCPyV isolate 350 (23–34%), KIV (56%), and WUV (54%) in a group of 721 young children, confirming that also, for these viruses, the seroconversion may occur during the childhood [9]. Serological evidence of exposure to MCPyV in childhood has been also reported by Chen et al. who observed a seroprevalence of 35% in children 4 to 13 years of age and by Tolstov et al. who described a seroprevalence of 34% in subjects under the age of 21 [36, 37]. The seroprevalence of TSPyV was estimated as 5% among children aged 1–4 years, rising to 48% at 6–10 years [38].
The reported adult levels of seroprevalence vary between 40 and 95% for BKV, between 32 and 95% for JCV, between 54% and 91% for both KIV and WUV, and between 25 and 88% for MCPyV [9, 35, 39–43]. LPV and LPV-related virus were reported to be present in the human population at lower level, about 15–20% of adults [9]. Only few seroprevalence data are reported for SV40 testing around 2–10% in the different geographic areas [44, 45].
3. Tumor Association of Human Polyomaviruses
Research on the role of polyomaviruses in development of neoplasms began in 1953 when Gross isolated a virus, from a mice lymphoma, that induced an identical tumour when infected into newborn mice [46]. The virus, identified as murine polyomavirus (mPyV), became the archetypal member of Polyomaviridae family [2].
The ability of the polyomavirus SV40 to transform different cell types from many species and to cause tumours in animals model was described in 1960 [47]. To date, the transforming properties of SV40 have been well characterized and they have been used as models to study the potential oncogenicity of all other human polyomaviruses.
Polyomaviruses have been found associated with specific tumor types such as brain and bone tumors, mesotheliomas, and lymphomas and with kidney diseases. Specifically, they can reproduce different types of brain tumours (e.g., medulloblastomas, neuroblastomas, astrocytomas, and neuroectodermal primitive tumours) when intracerebrally inoculated in rodents and monkeys [48, 49].
It has been demonstrated by several in vitro studies that the main factor implicated in cell transformation and tumour development is the early protein LTAg. LTAg is a multifunctional protein, fundamental for the viral life cycle of polyomaviruses, because it regulates the viral genome replication and gene expression [50]. The early regulatory protein LTAg is functionally divided in several domains, defined, from the N-terminal to the C-terminal, as follows: the DNaJ domain, linking to the cellular factor HSc70; the LXCXE motif, that specifically binds and inactivates the Rb family members; the NLS domain, that is necessary for the nuclear localization of the protein; the Helicase domain; finally, the p53 binding domain [27, 51–53] (Figure 2). All these domains cooperate in binding to and inactivating cellular proteins that prevent transition into S phase. Consequently, the virus, that needs the S phase in order to replicate, inhibits itself replication.
Figure 2.
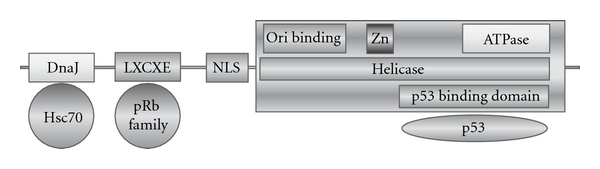
Polyomaviruses LT Ag structure. The functional domains are defined as follows: DnaJ domain that binds cellular Hsc70, LXCXE motif that binds the proteins belonging to the pRb family, NLS domain, that is the nuclear organization signal, Helicase domain, and p53 binding domain, that binds the p53 cellular suppressor protein.
Transgenic mice with the polyomaviruses LTAg gene under the control of the early viral promoter have developed brain tumours, such as neuroblastomas and medulloblastomas [54–56]. The transforming activity of LTAg protein is due to the fact that it can bind and inactivate cellular tumour suppressor proteins, such as p53 and pRb [57, 58]. Besides these mechanisms of tumourigenicity, which have been analyzed in depth, it has also been shown that LTAg is able to deregulate the cell cycle also by interacting with beta-catenin, a component of the Wnt signal pathway, and enhancing the expression of c-myc [59]. LTAg can bind IRS-1, which is translocated in the cell nucleus, where it cooperates in the process of malignant transformation [60]. Finally, LTAg can bind HSc70, which affects the cell cycle.
Polyomaviruses LTAg is also able to promote mutagenic events and chromosomal instability in B lymphocytes and morphological changes, such as multinuclei, aneuplodia, and polyplodia in transgenic mice [61, 62].
LTAg DNA belonging to JCV, BKV, and SV40 has been found in different types of human tumours, such as brain tumours (medulloblastomas, astrocytomas, ependymomas, oligodendrogliomas, and glioblastomas), bone tumours, colorectal carcinoma, urinary tract tumours, and lymphomas. Moreover, the LTAg, expressed protein has also been identified and localized in almost all the types of above-mentioned tumours [63–67], showing that the virus is able to express the oncogenic protein in the host and to cooperate in cell transformation.
The transforming properties of the very recently discovered KIV and WUV have been tested only by a few groups, but the results are preliminary. On the contrary, MCPyV, since its discovery, has been seriously thought as a candidate etiological agent in the development of Merkel Cell Carcinoma (see below) (Table 2).
Table 2.
Polyomaviruses-associated tumors.
| Virus | Cancer | Viral product |
|---|---|---|
| JCV | brain tumors | DNA, RNA, protein |
| Lymphoma (Hodgkin disease) | DNA, protein | |
| Leukemias | DNA | |
| Colorectal carcinoma | DNA, protein | |
| Gastric cancer | DNA | |
| Lung cancer | DNA, RNA, protein | |
| Esophageal carcinomas | DNA, protein | |
| Prostate cancer | DNA, protein | |
| Tongue carcinoma | DNA, protein | |
|
| ||
| BKV | Brain tumors | DNA, RNA, protein |
| Bone tumors | DNA, RNA | |
| Insulinomas | DNA | |
| Kaposi's sarcoma | DNA | |
| Urinary tract tumors/bladder tumors | DNA, RNA, protein | |
| Adrenal adenoma | DNA | |
| Genital tumors | DNA | |
| Renal carcinoma | DNA | |
| Prostate cancer | DNA, protein | |
|
| ||
| SV40 | Brain tumors | DNA, RNA, protein |
| Mesotheliomas | DNA, RNA, protein | |
| Bone tumors | DNA | |
| Lymphomas | DNA | |
| Leukemias | DNA | |
| Urotheliomas | DNA | |
| Breast cancer | DNA, protein | |
|
| ||
| MCPyV | Merkel cell carcinoma | DNA, protein |
|
| ||
| KIPyV | Not done | — |
|
| ||
| WUPyV | Not done | — |
4. Immunity to Polyomaviruses-Induced Tumours: The Polyomavirus Mouse Model
The role of the immune surveillance in polyomaviruses reactivation is essential, since the virus-induced pathology mainly occurs in immunocompromised hosts. However, except for molecular-epidemiological studies, there is little information about the role of innate immunity related to polyomaviruses, and how the innate immune system handles the virus, whereas more information could be obtained regarding the adaptive immunity.
In addition, because polyomaviruses have a narrow host range that restricts productive infection to their natural hosts, the only tractable system for studying polyomaviruses pathogenesis and immunity is the mPyV infection model. Since its discovery and given the ease of genome manipulation and expansion in tissue cultures, the mPyV has been used as infection model for the human polyomaviruses. When inoculated into immunocompromised adult mice or newborn mice of different inbred strains, polyomavirus induces multiple tumours that may arise from more than a dozen different cell types [68, 69].
The main study on the role of the innate immune response to the polyomaviruses infection in the contest of susceptibility to virus-induced tumours has been published several years ago, by Velupillai and colleagues [70, 71]. They studied two different inbred strains of mice, infected with mPyV. The first strain, known as BR mice, was resistant to the tumour, because of an antitumor immune response mediated by CD8+ T cells specific for a peptide derived from the viral middle T-antigen. The effective production of IL-12 and of IFN-gamma allowed an effective cell-mediated immunity. The second strain, indeed, called PE mice, was highly susceptible to the tumour and transmitted its susceptibility as a dominant trait in crosses with BR mice. This susceptibility was shown to be due to the absence of a type 1 cytokine response, leading to a failure to sustain virus-specific cytotoxic T-lymphocytes. PE mice, in fact, secreted IL-10, with no detectable production of IFN-gamma, and, only after administration of IFN-gamma and recombinant IL-12 to newborn mice, cytotoxic activity was restored and a reduction in tumours frequency was achieved [72].
In addition, Lowe and colleagues demonstrated the importance of the innate immunity in increasing the immune response against SV40 LTAg, using mice model, affected with SV40 induced-pulmonary metastasis. They reported upregulation of expression of inflammatory cytokines, such as TNFalpha, IL4, IL2, and RANTES, after tumorigenic growth and observed that it was correlated with beneficial reactions. Probably, these inflammatory mediators induced NK cells activation that led to the destruction of malignant cells [73].
The role of NK cells and γδT cells in killing tumor cells was also investigated by Mishra in colleagues in 2010. They observed that mice lacking αβT cells are protected from the formation of tumors induced by the mPyV if they have γδT cells and NK cells. In addition, mice lacking both cells develop the tumors earlier than mice that have only NK cells. Additional experiments showed that NK cells and γδT cells mount antitumor but not antiviral responses, since they do not have any effects on the amount of persisting virus [74].
Many studies have indicated that multiple components of the immune system contribute to limiting mPyV replication, including early induction of antibodies, recruitment of CD4+ and CD8+ T cells, and generation of humoral immunity.
Firstly, it has been observed that congenitally thymus-deprived nude mice have increased susceptibility to virus oncogenesis, demonstrating that T cells prevent the tumours induction. Then, the results were enriched by the observation that knockout mice with complete lack of major histocompatibility complex (MHC) class I molecules were mPyV tumour-susceptible. Consequently, MHC-class I-restricted CD8+ T cells are thought to be the primary effectors-cell population, during polyomaviruses infection [75, 76].
Since polyomaviruses infection is persistent in the host, the immune system has to face with repetitive antigen encounter. In this situation of chronic antigen exposure, host mechanisms may come into play to intentionally downregulate CTL activity to prevent widespread destruction of antigen-bearing cells. Thus, peripheral tolerance mechanisms are engaged to deregulate antigen-specific CD8+ T cell responses. Many models of tolerance come from pathogenic persistent viruses in humans, including HCV, EBV, HIV, and HBV [77–80]. In these cases, CD8+ T cells gradually lack functional competence and weaken their cytotoxicity. However, the premature termination of antiviral CD8+ T cell function for oncogenic viruses can cause serious consequences. In transgenic mice, in fact, T-cell response is compromised by either central or peripheral tolerance [81, 82].
The role of CD4+ T lymphocytes in immunity to virus-induced tumours has been less studied, but it is thought to provide help for MHC-class I-restricted CD8+ cytotoxic (antitumour) T lymphocytes. Kennedy et al. evaluated the role of T-cell subsets in tumour immunity induced by recombinant SV40 T-Ag within an experimental murine pulmonary metastasis model. By depleting mice of either CD4+ or CD8+ T cells, indications were found that CD4+ T cells but not CD8+ T cells were critical in the production of antibodies to LTAg and in tumour immunity. Then, it was shown that IgG1 was the dominating IgG subclass indicating that Th2 type T-helper cells were involved [83]. CD4+ T cells are also required for recruitment of naive antiviral CD8+ T cells during persistent mPyV infection [84]. Two dominant mPyV-specific CD4+ T cells populations are present, one directed toward an epitope derived from the LTAg and the other from the VP1, that differed quantitatively and qualitatively during the course of infection. They are both stably maintained during persistent infection, but the VP1-specific CD4+ T-cell response is higher than that against LTAg. In addition, mPyV-specific CD4+ T cells, although not essential for expansion of CD8+ T cells during acute infection, are indispensable for de novo priming of antiviral naive CD8+ T cells in persistently infected hosts [85].
The potential cytotoxic activity of polyomavirus-specific CD4+ T cells has been studied in BKV-seropositive subjects, who also showed BKV-specific CD4+ and CD8+ cells. After expansion in culture, in fact, the majority of the BKV-specific CD4+ T cells was shown to express CD40, to secrete both IFN-gamma and TNF-alpha, to contain both granzyme A and granzyme B, and to degranulate/mobilize CD107 in response to antigen-specific stimulation [86].
Moreover, tumour induction was found to be age dependent and this age-related resistance to tumour induction was immunologically mediated as animal models that have been immunocompromised by X-irradiation become susceptible to tumour induction.
5. Potential Clinical Application of Immunotherapy
One of the strategies that has been used for adoptive immunotherapy of cancer identifies, isolates, and expands in vitro CD8+ T-cells responsive to human tumours antigens. However, few immunotherapeutic approaches that target the recruitment of tumor reactive CD8+ T cells have been effective against solid tumors. Surely, tumors of the CNS provide a unique challenge to immunotherapy due to more stringent regulation of lymphocyte circulation and the potential for the negative side effects induced by T-cell effector functions. In 2008, a mouse model of autochthonous brain cancer to assess adoptive immunotherapeutic approaches of polyomavirus-induced disease was examined. This model consisted of SV40 LTAg transgenic mice SV11 which develop spontaneous choroid plexus tumors due to expression of full-length LTAg and lack the endogenous CD8+ T cells that in nontransgenic mice respond to the three different LTAg epitopes, I, II/III, and IV. In the model, donor T cells against the immunodominant epitope IV are subject to the potential effects of peripheral tolerance and the immunosuppressive tumor environment following adoptive transfer into SV11 mice with both minimal disease and advanced stage tumors. The performed experiments demonstrated that LTAg-specific CD8+ T cells from immune donors accumulated at the tumor site and are associated with reduced tumor burden and extension of the lifespan. In addition, it has been shown that IFN-gamma component donor cells play a major role in the immune-mediated control of established autochthonous tumors in CNS [87, 88].
Many other studies are needed to confirm the feasibility of immunotherapy approaches to polyomavirus-induced tumors.
6. A Special Focus on the Merkel Cell Polyomavirus
Feng and colleagues have published the identification of Merkel Cell Polyomavirus (MCPyV) DNA in the in Merkel Cell Carcinoma samples (MCC) in 2008. The innovative method used consists in generating cDNAs library from target cell transcripts, in pyrosequencing the DNA and finally in screening all the sequencing-data in order to identify non human transcripts. The detected non human sequence was a fusion transcript between an unknown virus LTAg and a human receptor tyrosine phosphatase. Further investigation by the same group led to the sequence of the complete genome of this previously unknown polyomavirus, the MCPyV [14].
MCC is a neuroectodermal tumour arising from mechanoreceptor Merkel Cells that are present in the skin of the limbs and face and around hair follicles [89]. It is a rare and aggressive skin cancer, unusual before the age of 50, with an incidence rate of 0.44 cases per 100,000 subjects in the USA; however, its incidence is dramatically increasing, tripling from 1986 to 2001. The risk factors for the development of MCC comprise excessive UV light exposure, age >50 years old and immunosuppressive state of the host [89–92].
The paper by Feng and colleagues reported the detection of sequences in 8 of 10 (80%) MCC tumours, but only in 9 of 84 (10,7%) control tissues, including skin tissue. In addition, the ability of the MCPyV to integrate into the human genome was described: in six of eight positive tissues, the MCPyV genome was integrated within the host genome in a clonally pattern; metastatic cells also carried the same integration pattern. On the contrary, genome was present in the episomal form in all the other nontumour positive tissues. All the above-mentioned virus characteristics led the scientific community to state that MCPyV may be a candidate causative agent in the etiopathogenesis of MCC.
Moreover, it was observed in a subsequent study that the LTAg amplified from 9 out of 9 MCC tissues had a mutation, resulting in a truncated protein, that lacked the helicase domain. On the contrary, the wild-type form of LTAg was detected in non-MCC tissues [14, 93–96].
Since the initial report, many other studies, have been published around the world, confirming the presence of this polyomavirus in MCC. In particular, they refer that the range of virus recovering from MCC fresh, frozen, formalin-fixed, or paraffin-embedded tumour samples varies between 24% and 100%, even if the data cannot be compared because of the different methodologies employed [97].
The prevalence of MCPyV in normal tissues or tumours other than MCC are more contrasting. The virus genome has been detected in skin samples with variably frequency ranging from 10% to 30%, whereas other studies reported prevalence rates of McCoy DNA up to 96%, in cutaneous swabs [98]. The virus was also searched in samples from neuroendocrine tumours from a variety of anatomic sites, colorectal cancer, prostate cancer, hematopoietic neoplasm, central nervous system tumours, mesothelioma, and many others, but none of the studies showed an association between virus and disease. In addition, the reported numbers of virus genome copies per MCC tumour cell are much higher (range 0.8–173/cell) [95, 99] than the highest value obtained for the other MCPyV genome-positive samples (0.01–0.0001 genome copies/cell). Finally, the antibody titres against MCPyV are much higher in MCC patients compared to healthy controls [100, 101]
The proposed mechanism by which MCPyV induces MCC in virus-positive tumours is based on the activity of the truncated LTAg: it retains the region with oncogenic potential such as the DnaJ domain and the retinoblastoma binding pocket but lacks the domain supporting the viral DNA replication. This is important because the loss of replication activity upon integration in MCC demonstrates that is not simply a “passenger virus” that happens to grow well in MCC cells [93, 96, 102]. Moreover, the immunoprecipitation assays have shown that truncated LTAg is able to bind to Rb protein, a critical step into carcinogenesis event. The production of LTAg is necessary for the proliferation of MCPyV-positive MCC, since it has been demonstrated that the silencing of the virus gene by short-hairpin RNA causes growth arrest and/or cell death of the specific cell lines. Thus, it has been proposed that MCPyV plays an important role in the MCC pathogenesis by means of viral integration into the host and specific mutations in the LTAg region that retains the Rb-binding capacity, eliminating the possibility of viral replication.
7. Immunobiology of MCPyV-Positive MCC
As for the other human polyomaviruses, a strong link between MCPyV-positive MCC and the immune system has been suggested. Patients with immune system impairment, such as HIV-related immunodeficiency, drugs-associated immunosuppression, or forms of chronic leukaemia, are at up to 50-fold increased risk of developing MCC [89, 90]. Additionally, it has been shown that MCC tumours regress following improvement in immune function.
As already reported above, the high and wide seroprevalence of MCPyV suggests that the exposure to the virus is a common event in the human population and that the capsid virus proteins are well recognized by the host immune system. However, MCPyV-positive MCC patients show a higher titer of antibodies against VP1, compared to the healthy population or to MCPyV-negative MCC patients, probably due to the exposure to an higher MCPyV viral load. In addition, the presence of a strong humoral immune response to MCPyV has been associated with a better prognosis of the tumours, perhaps showing that host immune system plays a critical role also in the remission of the infection [103].
Interestingly, Paulson and colleagues collected sera from 205 MCC cases and from 530 healthy controls and screened them for T-Ag reactivity, demonstrating that IgG antibody reactivity to the MCPyV LTAg and st-Ag oncoproteins was significatively associated with MCC. In addition, they observed rapid changes in LTAg titre corresponding to the different disease burden: titres were highest when patients were sickest, suggesting that antibodies to LTAg are not protective against disease progression. The authors hypothesized that the rapid turnover of IgG antibodies recognizing LTAg in MCC patient may be due to the instability within the B-cell population, caused by ineffective priming within the tumour microenvironment [104]. In contrast to capsid proteins that are highly visible to the humoral immune system, T-antigen oncoproteins are not present in viral particles, are only expressed after viral entry into host cells, are located in the nucleus [105], and are thus less likely to trigger an antibody response except in the setting of dying or diseased tissue (such as a tumour that persistently expresses the LTAg).
Very recently, it was observed that MCPyV-specific T-helper cells secrete the IL13, IL10 cytokines, and IFN-gamma, that have a strong tumour-suppressing and antiviral functions [106].
The specific immune response mediated by T cells has also been documented in MCPyV-positive MCC patients. A study published recently has shown that intratumoral infiltration of CD8+ lymphocytes is a factor of improved prognosis of the disease. The same study showed also an overexpression of genes encoding cytotoxic granules such as granzymes, chemokine CCL19, and many lymphocyte-activation molecules in patients with improved survival [107].
8. Conclusions
The recent identification of many components of the Polyomaviridae family that are able to infect humans represents a strong incentive for the scientific community to improve and increase the research on the oncogenic potentialities of these viruses. The ubiquity and persistent nature of polyomaviruses make them very challenging in order to define the mechanisms of their pathogenicity. In particular, since the association between the viral aetiology of the tumours and the state of the host immune system is well established, it is urgent to focus future studies on the nature of the relationship between the host immune system and polyomaviruses infection.
Acknowledgments
This work was supported by NIMH Grant no. MH072528 awarded to P. Ferrante and Italian Ministry grant RC 11/11 awarded to M. Comar. The authors wish to thank Dr. Lucia Signorini, University of Milano, Italy, for her precious technical support and all the personnel of the Laboratory of Translational Research, University of Milano, Italy.
References
- 1. http://ICTVonline.org.
- 2.Stewart SE, Eddy BE. Neoplasms in mice inoculated with a tumor agent carried in tissue culture. Journal of the National Cancer Institute. 1958;20(6):1223–1243. doi: 10.1093/jnci/20.6.1223. [DOI] [PubMed] [Google Scholar]
- 3.Major EO. Human polyomavirus. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 4th edition. 2001. pp. 2175–2196. [Google Scholar]
- 4.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. The Lancet. 1971;1(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 5.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. The Lancet. 1971;1(7712):1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 6.Shah K, Nathanson N. Human exposure to SV40: review and comment. American Journal of Epidemiology. 1976;103(1):1–12. doi: 10.1093/oxfordjournals.aje.a112197. [DOI] [PubMed] [Google Scholar]
- 7.Brooks BR, Walker DL. Progressive multifocal leukoencephalopathy. Neurologic Clinics. 1984;2(2):299–313. [PubMed] [Google Scholar]
- 8.Berger JR, Major EO. Progressive multifocal leukoencephalopathy. Seminars in Neurology. 1999;19(2):193–200. doi: 10.1055/s-2008-1040837. [DOI] [PubMed] [Google Scholar]
- 9.Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathogens. 2009;5(3) doi: 10.1371/journal.ppat.1000363. Article ID e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.zur Hausen H, Gissmann L. Lymphotropic papovaviruses isolated from African green monkey and human cells. Medical Microbiology and Immunology. 1979;167(3):137–153. doi: 10.1007/BF02121180. [DOI] [PubMed] [Google Scholar]
- 11.Delbue S, Tremolada S, Branchetti E, et al. First identification and molecular characterization of lymphotropic polyomavirus in peripheral blood from patients with leukoencephalopathies. Journal of Clinical Microbiology. 2008;46(7):2461–2462. doi: 10.1128/JCM.00381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allander T, Andreasson K, Gupta S, et al. Identification of a third human polyomavirus. Journal of Virology. 2007;81(8):4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaynor AM, Nissen MD, Whiley DM, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathogens. 2007;3(5, article no. e64):0595–0604. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host and Microbe. 2010;7(6):509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathogens. 2010;6(7) doi: 10.1371/journal.ppat.1001024. Article ID e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scuda N, Hofmann J, Calvignac-Spencer S, et al. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. Journal of Virology. 2011;85(9):4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan CS, Sung CK, Pack CD, et al. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387(1):157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. Journal of Virology. 1984;51(2):458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura T, Kunitake T, Guo J, Tominaga T, Kawabe K, Yogo Y. Transmission of the human polyomavirus JC virus occurs both within the family and outside the family. Journal of Clinical Microbiology. 1994;32(10):2359–2363. doi: 10.1128/jcm.32.10.2359-2363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorries K, Vogel E, Gunther S, Czub S. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology. 1994;198(2):59–70. doi: 10.1006/viro.1994.1008. [DOI] [PubMed] [Google Scholar]
- 23.Dorries K. Human Polyomavirus: Molecular and Clinical Prespectives. 2001. Latent and persistent polyomavirus infection; pp. 197–235. [Google Scholar]
- 24.Moens U, Ludvigsen M, Van Ghelue M. Human polyomaviruses in skin diseases. Pathology Research International. 2011;2011:12 pages. doi: 10.4061/2011/123491. Article ID 123491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nature Reviews Immunology. 2003;3(10):781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 26.Bauman Y, Nachmani D, Vitenshtein A, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host and Microbe. 2011;9(2):93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 27.White MK, Khalili K. Polyomaviruses and human cancer: molecular mechanisms underlying patterns of tumorigenesis. Virology. 2004;324(1):1–16. doi: 10.1016/j.virol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Johnson EM. Structural evaluation of new human polyomaviruses provides clues to pathobiology. Trends in Microbiology. 2010;18(5):215–223. doi: 10.1016/j.tim.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles WA. Human Polyomaviruses. Molecular and Clinical Perspectives. 2001. The epidemiology of BK virus and the occurence of antigenic and genomic subtypes; pp. 527–559. [Google Scholar]
- 30.Knowles WA, Pipkin P, Andrews N, et al. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. Journal of Medical Virology. 2003;71(1):115–123. doi: 10.1002/jmv.10450. [DOI] [PubMed] [Google Scholar]
- 31.Rekvig OP, Bendiksen S, Moens U. Immunity and autoimmunity induced by polyomaviruses: clinical, experimental and theoretical aspects. Advances in Experimental Medicine and Biology. 2006;577:117–147. doi: 10.1007/0-387-32957-9_9. [DOI] [PubMed] [Google Scholar]
- 32.Dei R, Marmo F, Corte D. Age-related changes in the prevalence of precipitating antibodies to BK virus in infants and children. Journal of Medical Microbiology. 1982;15(3):285–291. doi: 10.1099/00222615-15-3-285. [DOI] [PubMed] [Google Scholar]
- 33.Flaegstad T, Traavik T, Kristiansen BE. Age-dependent prevalence of BK virus IgG and IgM antibodies measured by enzyme-linked immunosorbent assays (ELISA) Journal of Hygiene. 1986;96(3):523–528. doi: 10.1017/s0022172400066328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolt A, Sasnauskas K, Koskela P, Lehtinen M, Dillner J. Seroepidemiology of the human polyomaviruses. Journal of General Virology. 2003;84(6):1499–1504. doi: 10.1099/vir.0.18842-0. [DOI] [PubMed] [Google Scholar]
- 35.Viscidi RP, Rollison DE, Sondak VK, et al. Age-specific seroprevalence of merkel cell polyomavirus, BK virus, and JC virus. Clinical and Vaccine Immunology. 2011;18(10):1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Hedman L, Mattila PS, et al. Serological evidence of Merkel cell polyomavirus primary infections in childhood. Journal of Clinical Virology. 2011;50(2):125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Tolstov YL, Knauer A, Chen JG, et al. Asymptomatic primary merkel cell polyomavirus infection among adults. Emerging Infectious Diseases. 2011;17(8):1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen T, Mattila PS, Jartti T, Ruuskanen O, Söderlund-Venermo M, Hedman K. Seroepidemiology of the newly found trichodysplasia spinulosa-associated polyomavirus. Journal of Infectious Diseases. 2011;204(10):1523–1526. doi: 10.1093/infdis/jir614. [DOI] [PubMed] [Google Scholar]
- 39.Eash S, Manley K, Gasparovic M, Querbes W, Atwood WJ. The human polyomaviruses. Cellular and Molecular Life Sciences. 2006;63(7-8):865–876. doi: 10.1007/s00018-005-5454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalianis T, Ramqvist T, Andreasson K, Kean JM, Garcea RL. KI, WU and Merkel cell polyomaviruses: a new era for human polyomavirus research. Seminars in Cancer Biology. 2009;19(4):270–275. doi: 10.1016/j.semcancer.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathogens. 2009;5(9) doi: 10.1371/journal.ppat.1000578. Article ID e1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viscidi RP, Clayman B. Serological cross reactivity between polyomavirus capsids. Advances in Experimental Medicine and Biology. 2006;577:73–84. doi: 10.1007/0-387-32957-9_5. [DOI] [PubMed] [Google Scholar]
- 43.Moens U, Van Ghelue M, Johannessen M. New Research on Oncogenic Viruses. 2007. Human polyomaviruses: molecular mechanisms for transformation and their association with cancer; pp. 1–63. [Google Scholar]
- 44.Martini F, Corallini A, Balatti V, Sabbioni S, Pancaldi C, Tognon M. Simian virus 40 in humans. Infectious Agents and Cancer. 2007;2(1, article 13) doi: 10.1186/1750-9378-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens U, Johannessen M, Barcena-Panero A, Gerits G, van Ghelue M. Emerging polyomaviruses in the human population. Reviews in Infection. 2010;1:59–93. [Google Scholar]
- 46.Gross L. A filterable agent recovered from AK leukemic extracts, causing salivary gland carcinomas in C3H mice. Proceedings of the Society for Experimental Biology and Medicine. 1953;83:414–431. doi: 10.3181/00379727-83-20376. [DOI] [PubMed] [Google Scholar]
- 47.Sweet BH, Hilleman MR. The vacuolating virus, S.V. 40. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- 48.Zu Rhein GM. Studies of JC virus-induced nervous system tumors in the Syrian hamster: a review. Progress in Clinical and Biological Research. 1983;105:205–221. [PubMed] [Google Scholar]
- 49.Walker DL, Padgett BL, ZuRhein GM. Human papovavirus (JC): induction of brain tumors in hamsters. Science. 1973;181(4100):674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- 50.Imperiale MJ, Major EO. Fields Virology. 2007. Polyomaviridae; pp. 2263–2298. [Google Scholar]
- 51.Moens U, Van Ghelue M, Johannessen M. Oncogenic potentials of the human polyomavirus regulatory proteins. Cellular and Molecular Life Sciences. 2007;64(13):1656–1678. doi: 10.1007/s00018-007-7020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moens U, van Ghelue M, Johannessen M. Human polyomaviruses: molecular mechanisms for transformation and their association with cancers. In: Tunely EI, editor. New Research on Oncogenic Viruses. New York, NY, USA: Nova Science Publishers; 2007. pp. 2–63. [Google Scholar]
- 53.Gjoerup O, Chang Y. Update on human polyomaviruses and cancer. Advances in Cancer Research. 2010;106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- 54.Small JA, Khoury G, Jay G. Early regions of JC virus and BK virus induce distinct and tissue-specific tumors in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(21):8288–8292. doi: 10.1073/pnas.83.21.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franks RR, Rencic A, Gordon J, et al. Formation of undifferentiated mesenteric tumors in transgenic mice expressing human neurotropic polyomavirus early protein. Oncogene. 1996;12(12):2573–2578. [PubMed] [Google Scholar]
- 56.Gordon J, Del Valle L, Otte J, Khalili K. Pituitary neoplasia induced by expression of human neurotropic polyomavirus, JCV, early genome in transgenic mice. Oncogene. 2000;19(42):4840–4846. doi: 10.1038/sj.onc.1203849. [DOI] [PubMed] [Google Scholar]
- 57.Khalili K, Del Valle L, Otte J, Weaver M, Gordon J. Human neurotropic polyomavirus, JCV, and its role in carcinogenesis. Oncogene. 2003;22(33):5181–5191. doi: 10.1038/sj.onc.1206559. [DOI] [PubMed] [Google Scholar]
- 58.Poulin DL, Kung AL, DeCaprio JA. p53 targets simian virus 40 large T antigen for acetylation by CBP. Journal of Virology. 2004;78(15):8245–8253. doi: 10.1128/JVI.78.15.8245-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gan DD, Reiss K, Carrill T, et al. Involvement of Wnt signaling pathway in murine medulloblastoma induced by human neurotropic JC virus. Oncogene. 2001;20(35):4864–4870. doi: 10.1038/sj.onc.1204670. [DOI] [PubMed] [Google Scholar]
- 60.Del Valle L, Wang JY, Lassak A, et al. Insulin-like growth factor I receptor signaling system in JC virus T antigen-induced primitive neuroectodermal tumors—medulloblastomas. Journal of NeuroVirology. 2002;8(2):138–147. doi: 10.1080/13550280290101111. [DOI] [PubMed] [Google Scholar]
- 61.Lazutka JR, Neel JV, Major EO, et al. High titers of antibodies to two human polyomaviruses, JCV and BKV, correlate with increased frequency of chromosomal damage in human lymphocytes. Cancer Letters. 1996;109(1-2):177–183. doi: 10.1016/s0304-3835(96)04448-5. [DOI] [PubMed] [Google Scholar]
- 62.Neel JV, Major EO, Awa AA, et al. Hypothesis: "Rogue cell"-type chromosomal damage in lymphocytes is associated with infection with the JC human polyoma virus and has implications for oncopenesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(7):2690–2695. doi: 10.1073/pnas.93.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arrington AS, Lednicky JA, Butel JS. Molecular characterization of SV40 DNA in multiple samples from a human mesothelioma. Anticancer Research. 2000;20(2 A):879–884. [PubMed] [Google Scholar]
- 64.Caldarelli-Stefano R, Vago L, Omodeo-Zorini E, et al. Detection and typing of JC virus in autopsy brains and extraneural organs of AIDS patients and non-immunocompromised individuals. Journal of NeuroVirology. 1999;5(2):125–133. doi: 10.3109/13550289909021994. [DOI] [PubMed] [Google Scholar]
- 65.Caldarelli-Stefano R, Boldorini R, Monga G, Meraviglia E, Zorini EO, Ferrante P. JC virus in human glial-derived tumors. Human Pathology. 2000;31(3):394–395. doi: 10.1016/s0046-8177(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 66.Krynska B, Del Valle L, Croul S, et al. Detection of human neurotropic JC virus DNA sequence and expression of the viral oncogenic protein in pediatric medulloblastomas. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):11519–11524. doi: 10.1073/pnas.96.20.11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Mattei M, Martini F, Corallini A, et al. High incidence of BK virus large-T-antigen-coding sequences in normal human tissues and tumors of different histotypes. International Journal of Cancer. 1995;61(6):756–760. doi: 10.1002/ijc.2910610603. [DOI] [PubMed] [Google Scholar]
- 68.Dawe CJ, Freund R, Mandel G. Variations in polyoma virus genotype in relation to tumor induction in mice. Characterization of wild type strains with widely differing tumor profiles. American Journal of Pathology. 1987;127(2):243–261. [PMC free article] [PubMed] [Google Scholar]
- 69.Lukacher AE. Pathogen-host standoff: immunity to polyomavirus infection and neoplasia. Immunologic Research. 2004;29(1–3):139–150. doi: 10.1385/IR:29:1-3:139. [DOI] [PubMed] [Google Scholar]
- 70.Velupillai P, Yoshizawa I, Dey DC, et al. Wild-derived inbred mice have a novel basis of susceptibility to polyomavirus-induced tumors. Journal of Virology. 1999;73(12):10079–10085. doi: 10.1128/jvi.73.12.10079-10085.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velupillai P, Carroll JP, Benjamin TL. Susceptibility to polyomavirus-induced tumors in inbred mice: role of innate immune responses. Journal of Virology. 2002;76(19):9657–9663. doi: 10.1128/JVI.76.19.9657-9663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drake DR, III, Knoepp L, Actor JK, Lukacher AE. Patterns of expression of viral and cytokine gene transcripts during mouse polyoma virus infection. Combinatorial Chemistry and High Throughput Screening. 2000;3(4):329–341. doi: 10.2174/1386207003331562. [DOI] [PubMed] [Google Scholar]
- 73.Lowe DB, Shearer MH, Aldrich JF, Winn RE, Jumper CA, Kennedy RC. Role of the innate immune response and tumor immunity associated with simian virus 40 large tumor antigen. Journal of Virology. 2010;84(19):10121–10130. doi: 10.1128/JVI.01095-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mishra R, Chen AT, Welsh RM, Szomolanyi-Tsuda E. NK cells and gammadelta T cells mediate resistance to polyomavirus-induced tumors. PLoS Pathogens. 2010;6(5):p. e1000924. doi: 10.1371/journal.ppat.1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swanson PA, Pack CD, Hadley A, et al. An MHC class Ib-restricted CD8 T cell response confers antiviral immunity. Journal of Experimental Medicine. 2008;205(7):1647–1657. doi: 10.1084/jem.20080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swanson PA, Lukacher AE, Szomolanyi-Tsuda E. Immunity to polyomavirus infection: the polyomavirus-mouse model. Seminars in Cancer Biology. 2009;19(4):244–251. doi: 10.1016/j.semcancer.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. Journal of Experimental Medicine. 2001;194(10):1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Baarle D, Hovenkamp E, Callan MFC, et al. Dysfunctional Epstein-Barr virus (EBV)-specific CD8+ T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2001;98(1):146–155. doi: 10.1182/blood.v98.1.146. [DOI] [PubMed] [Google Scholar]
- 79.Kostense S, Ogg GS, Manting EH, et al. High viral burden in the presence of major HIV-specific CD8+ T cell expansions: evidence for impaired CTL effector function. European Journal of Immunology. 2001;31(3):677–686. doi: 10.1002/1521-4141(200103)31:3<677::aid-immu677>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 80.Appay V, Nixon DF, Donahoe SM, et al. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. Journal of Experimental Medicine. 2000;192(1):63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spencer JV, Braciale TJ. Incomplete CD8+ T lymphocyte differentiation as a mechanism for subdominant cytotoxic T lymphocyte responses to a viral antigen. Journal of Experimental Medicine. 2000;191(10):1687–1698. doi: 10.1084/jem.191.10.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reignat S, Webster GJM, Brown D, et al. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. Journal of Experimental Medicine. 2002;195(9):1089–1101. doi: 10.1084/jem.20011723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kennedy RC, Shearer MH, Watts AM, Bright RK. CD4+ T lymphocytes play a critical role in antibody production and tumor immunity against simian virus 40 large tumor antigen. Cancer Research. 2003;63(5):1040–1045. [PubMed] [Google Scholar]
- 84.Kemball CC, Pack CD, Guay HM, et al. The antiviral CD8+ T cell response is differentially dependent on CD4+ T cell help over the course of persistent infection. Journal of Immunology. 2007;179(2):1113–1121. doi: 10.4049/jimmunol.179.2.1113. [DOI] [PubMed] [Google Scholar]
- 85.Lin E, Kemball CC, Hadley A, et al. Heterogeneity among viral antigen-specific CD4+ T cells and their de novo recruitment during persistent polyomavirus infection. Journal of Immunology. 2010;185(3):1692–1700. doi: 10.4049/jimmunol.0904210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou W, Sharma M, Martinez J, et al. Functional characterization of BK virus-specific CD4+ T cells with cytotoxic potential in seropositive adults. Viral Immunology. 2007;20(3):379–388. doi: 10.1089/vim.2007.0030. [DOI] [PubMed] [Google Scholar]
- 87.Yorty JL, Tevethia SS, Schell TD. Rapid accumulation of adoptively transferred CD8+ T cells at the tumor site is associated with long-term control of SV40 T antigen-induced tumors. Cancer Immunology, Immunotherapy. 2008;57(6):883–895. doi: 10.1007/s00262-007-0424-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tatum AM, Mylin LM, Bender SJ, et al. CD8+ T cells targeting a single immunodominant epitope are sufficient for elimination of established SV40 T antigen-induced brain tumors. Journal of Immunology. 2008;181(6):4406–4417. doi: 10.4049/jimmunol.181.6.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. Journal of Investigative Dermatology. 2007;127(9):2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- 90.Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: a population based study. Journal of Cutaneous Pathology. 2010;37(1):20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 91.Hodgson NC. Merkel cell carcinoma: changing incidence trends. Journal of Surgical Oncology. 2005;89(1):1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 92.Heath M, Jaimes N, Lemos B, et al. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. Journal of the American Academy of Dermatology. 2008;58(3):375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Houben R, Shuda M, Weinkam R, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. Journal of Virology. 2010;84(14):7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shuda M, Feng H, Hyun JK, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shuda M, Arora R, Hyun JK, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. International Journal of Cancer. 2009;125(6):1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. Journal of Clinical Investigation. 2011;121(9):3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Current Oncology Reports. 2011;13(6):488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dworkin AM, Tseng SY, Allain DC, Iwenofu OH, Peters SB, Toland AE. Merkel cell polyomavirus in cutaneous squamous cell carcinoma of immunocompetent individuals. The Journal of Investigative Dermatology. 2009;129(12):2868–2874. doi: 10.1038/jid.2009.183. [DOI] [PubMed] [Google Scholar]
- 99.Loyo M, Guerrero-Preston R, Brait M, et al. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. International Journal of Cancer. 2010;126(12):2991–2996. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Foulongne V, Kluger N, Dereure O, et al. Merkel cell polyomavirus in cutaneous swabs. Emerging Infectious Diseases. 2010;16(4):685–687. doi: 10.3201/eid1604.091278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Faust H, Pastrana DV, Buck CB, Dillner J, Ekström J. Antibodies to Merkel cell polyomavirus correlate to presence of viral DNA in the skin. Journal of Infectious Diseases. 2011;203(8):1096–1100. doi: 10.1093/infdis/jiq173. [DOI] [PubMed] [Google Scholar]
- 102.Houben R, Adam C, Baeurle A, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. International Journal of Cancer. 2012;130(4):847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 103.Touzé A, Le Bidre E, Laude H, et al. High levels of antibodies against merkel cell polyomavirus identify a subset of patients with merkel cell carcinoma with better clinical outcome. Journal of Clinical Oncology. 2011;29(12):1612–1619. doi: 10.1200/JCO.2010.31.1704. [DOI] [PubMed] [Google Scholar]
- 104.Paulson KG, Carter JJ, Johnson LG, et al. Antibodies to merkel cell polyomavirus T antigen oncoproteins reflect tumor burden in merkel cell carcinoma patients. Cancer Research. 2010;70(21):8388–8397. doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakamura T, Sato Y, Watanabe D, et al. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology. 2010;398(2):273–279. doi: 10.1016/j.virol.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 106.Kumar A, Chen T, Pakkanen S, et al. T-helper cell-mediated proliferation and cytokine responses against recombinant merkel cell polyomavirus-like particles. PLoS ONE. 2011;6(10):7 pages. doi: 10.1371/journal.pone.0025751. Article ID e25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paulson KG, Iyer JG, Tegeder AR, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. Journal of Clinical Oncology. 2011;29(12):1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]


