Abstract
Small RNAs are a commonly used tool for gene silencing and a promising platform for nucleic acid drug development. They are almost exclusively used to silence gene expression post-transcriptionally through degradation of mRNA. Small RNAs, however, can have a broader range of function by binding to Argonaute proteins and associating with complementary RNA targets in the nucleus, including long noncoding RNAs (lncRNAs) and pre-mRNA. Argonaute–RNA complexes can regulate nuclear events like transcription, genome maintenance, and splicing. Thousands of lncRNAs and alternatively spliced pre-mRNA isoforms exist in humans, and these RNAs may serve as natural targets for regulation and therapeutic intervention. This review describes nuclear mechanisms for Argonaute proteins and small RNAs, new pathways for sequence-specific targeting, and the potential for therapeutic development of small RNAs with nuclear targets.
Introduction
Over 40,000 publications related to small interfering RNAs (siRNAs) have been published since the original observation of mammalian RNA interference (RNAi) (Elbashir et al., 2001). Virtually all of these publications, especially those regarding mammalian cells, describe gene silencing through recognition of mRNA in the cytoplasm and post-transcriptional RNAi. However, in nearly every system where RNAi functions, evidence for nuclear RNAi pathways have been uncovered (Malone and Hannon, 2009; MOAZED, 2009; Zhang and Rossi, 2011).
A number of papers have described small RNAs that regulate gene transcription in mammalian cells (Green and Weinberg, 2011). These reports have raised 3 outstanding questions: 1) Do small RNAs naturally regulate processes like transcription and splicing in human cells, 2) what is the mechanism of nuclear RNAi and transcriptional gene silencing in mammals, and 3) can exogenous small RNAs be used to predictably control nuclear events in human cells, potentially leading to novel nucleic acid-based drugs? Answering these questions requires understanding how the RNAi machinery functions in the nucleus. This review discusses common features of characterized nuclear RNAi pathways and focuses on two key components – Argonaute (AGO) proteins and nuclear RNAs.
AGO Proteins: Effectors of Small RNA Activity
The AGO family
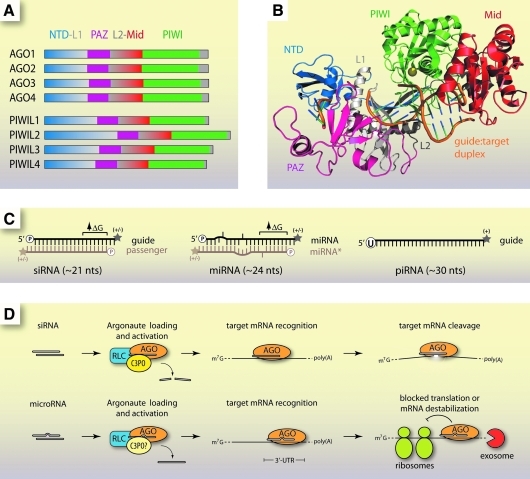
AGO proteins are a highly conserved family found in nearly every life form, from humans to Archaea (Bohmert et al., 1998; Fagard et al., 2000; Song et al., 2004; Tolia and Joshua-Tor, 2007). In humans there are 8 AGO proteins, 4 from the AGO clade (AGO1-4) and 4 from the PIWI clade (PIWIL1-4) (Fig. 1A) (Carmell et al., 2002; Sasaki et al., 2003). Known AGO proteins range from 1 in the fission yeast Schizosaccharomyces pombe to 27 in the worm Caenorhabditis elegans (Tolia and Joshua-Tor, 2007).
FIG. 1.
The structural and functional features of canonical RNA interference (RNAi) in animals. (A) The human Argonaute (AGO) family consists of 8 proteins containing conserved PAZ, Mid, and PIWI domains essential for small RNA binding and function in RNAi pathways. (B) Crystal structure of an Argonaute protein from Thermus thermophilus in complex with a DNA–RNA duplex (Wang et al., 2008). Protein domains are color-coded and labeled. (C) The structural features of common small RNAs in animals cells. A 5′ phosphate is indicated by an encircled “P,” 2′-O-methylation by a star (“+” or “−” indicates presence or absence), and ↑ΔG indicates higher free energy at that end of the duplex. (D) Canonical RNAi pathways in the cytoplasm. Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are loaded into Argonaute and activated by accessory factors like the RISC-loading complex and C3PO in animal cells. The activated complexes then recognize and bind complementary RNAs. Perfect target complementarity (siRNA) can result in cleavage and degradation, whereas imperfect complementarity (miRNA) results in translational inhibition or mRNA destabilization.
AGO protein structure
AGO proteins contain the characteristic PAZ (Piwi-AGO-Zwille) and PIWI domains. The overall structural fold of AGO proteins from Archaea (crystal structures of full-length mammalian AGOs have not been reported) is that of a crescent shape formed by the N-terminal, PAZ, and Mid domains (Song et al., 2004). The PAZ domain recognizes and binds the 3′ end of small RNAs (Yan et al., 2003; Lingel et al., 2004; Ma et al., 2004). The PIWI domain is an RNase H-like fold that harbors “slicer” activity for cleavage of target RNA substrates (Song et al., 2004; Wang et al., 2008). However, not all AGO proteins are cleavage competent. For instance, in humans, slicer activity has only been demonstrated for AGO2 even though AGO3 also possesses the characteristic aspartate-aspartate-histidine catalytic triad (Meister et al., 2004; Hock and Meister, 2008).
The Mid domain specifically binds to the 5′ phosphate of the loaded siRNA guide strand (Parker et al., 2005; Ma et al., 2005). A recent crystal structure of the human AGO2 MID domain revealed a similar fold to that of archaeal and bacterial homologues, as well as the structural basis for the 5' nucleotide A/U preference observed for eukaryotic microRNAs (miRNAs) (Frank et al., 2010). The siRNA strand stretches along a positively charged groove formed by the N-terminal, PAZ, and Mid domains, allowing base-pairing to a complementary RNA and access of the PIWI domain for cleavage (Fig. 1B) (Wang et al., 2008).
Classes of small RNA partners for AGO
Small RNAs that bind AGO proteins fall into 3 main classes: siRNAs, miRNAs, and PIWI-interacting RNAs (piRNAs). They function in different RNAi pathways and exhibit distinct physical properties (Fig. 1C). All three have been reported to function at both transcriptional and post-transcriptional levels (BARTEL, 2009; MOAZED, 2009; Siomi et al., 2011).
Small interfering RNAs are ∼21 nucleotides (nts) long and are composed of perfectly complementary strands with 2-nt overhangs at the 3′ end and a terminal 5′ phosphate (Elbashir et al., 2001). At the 3′ termini they may have a 2' hydroxyl, as in humans, or a 2′-O-methyl group, as in fruit flies (Yu et al., 2005; Horwich et al., 2007). The strand that directs cleavage is called the guide, while the strand that is released during AGO loading is called the passenger.
MicroRNAs are abundant, small RNAs derived from longer primary transcripts (pri-miRNAs) which fold into hairpins. Pri-miRNA hairpins are processed into shorter precursors (pre-miRNAs) by the RNase III endonuclease Drosha before undergoing further cleavage by Dicer, another RNase III enzyme, and final loading into AGO proteins (BARTEL, 2004). miRNAs share some traits with siRNAs. They possess 3′ dinucleotide overhangs (Lee et al., 2003). The 5′ end is phosphorylated and the 3′ end can be a 2′ hydroxyl or, especially in plants, a 2′-O-methyl (Denli et al., 2004; Gregory et al., 2004; Yu et al., 2005). During loading, the strand that is removed is called miRNA* instead of the passenger (BARTEL, 2009). miRNAs are not perfectly complementary strands but instead contain bulges and mismatches (BARTEL, 2009). miRNAs can act to block translation or cause instability of targeted mRNAs (Saxena et al., 2003; Guo et al., 2010).
PIWI-interacting RNAs are distinct from siRNAs or miRNAs. They do not require Dicer for biogenesis, are 2′-O-methylated at the 3′ termini, and often include uracil at the 5′ end (Aravin et al., 2006; Gunawardane et al., 2007; Ghildiyal and Zamore, 2009). It is not known exactly how piRNAs are produced or whether they exert their function through cleavage of their target RNAs, which are typically transposon-containing transcripts (Tolia and Joshua-Tor, 2007; Malone and Hannon, 2009).
AGO function
AGO proteins are essential for RNAi in the cytoplasm (Fig. 1D) (Liu et al., 2004). When a guide RNA is perfectly complementary to its RNA target, a slicer-competent AGO, such as AGO2 in humans, will catalyze RNA cleavage (Zamore et al., 2000; Liu et al., 2004; Meister et al., 2004). If imperfect complementarity is encountered, cleavage typically does not occur, because the mismatched bases disrupt catalytic interactions at AGO's active site (Wang et al., 2008; BARTEL, 2009).
While purified or recombinant human AGO2–siRNA complexes are sufficient to carry out target RNA cleavage (Rand et al., 2004), other proteins facilitate function inside the cell. Loading of AGO with siRNA requires the RISC-loading complex, composed of Dicer and TRBP (R2D2 in Drosophila), and activation requires the C3PO complex for efficient passenger strand removal (MacRae et al., 2008; Liu et al., 2009). In humans C3PO may be the sole loader and activator of AGO2 (Ye et al., 2011).
Accessory proteins can also help localize RNA–AGO complexes. When mRNAs are bound by miRNPs they are transported to cytoplasmic p-bodies where mRNAs may be degraded or isolated to suppress translation (Eulalio et al., 2007; Kulkarni et al., 2010). This localization is mediated by proteins like GW182, which bind AGO through GW-repeat motifs known as AGO hook domains (Till et al., 2007; Takimoto et al., 2009). Other proteins have been copurified with AGO and identified by mass spectrometry, but clear roles in RNAi have not yet been demonstrated (Hock et al., 2007; Landthaler et al., 2008).
Natural Roles in the Nucleus for AGO and the Small RNAs
Regulation of DNA methylation in plants
Plants use siRNAs to guide de novo methyltransferases to homologous DNA and establish sequence-specific DNA methylation (Wassenegger et al., 1994; Matzke et al., 2009). This pathway is called RNA-directed DNA methylation (RdDM). To initiate silencing, AGO4 is loaded with siRNA and the ribonucleoprotein (RNP) complex is directed to chromatin by complementarity to nascent noncoding RNA (ncRNA) synthesized by Pol V and sometimes Pol II (Zilberman et al., 2003; Wierzbicki et al., 2008, 2009; He et al., 2009a) (Fig. 2A). AGO4 in turn interacts with Pol V or Pol II through polymerase subunits that contain AGO hook domains (He et al., 2009b).
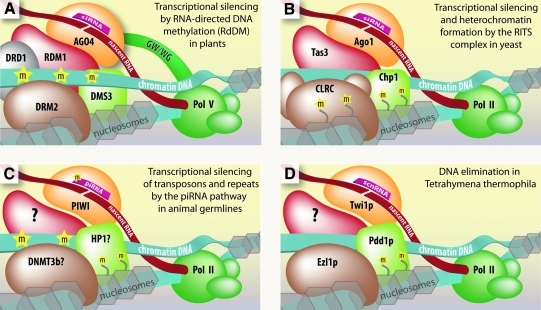
FIG. 2.
Current models for the molecular mechanisms of transcriptional regulation and genome maintenance by endogenous small RNAs in the nucleus. (A) RNA-directed DNA methylation (RdDM) in plants. AGO4 binds siRNAs and is recruited to the chromatin by complementarity to nascent RNA produced by AGO4-associated Pol V or other polymerases. The DNA methyltransferase DRM2 and a chromatin interacting and remodeling complex termed DDR are also recruited. (B) Heterochromatin formation and silencing by the RNA-induced transcriptional silencing (RITS) complex in yeast. RITS is comprised of Ago1, Tas3, and Chp1, which binds H3K9me nucleosomes. This complex is targeted primarily to centromeric repeats by Ago1-bound siRNA, which interacts with nascent RNA. The CLRC complex is also recruited to deposit histone H3–lysine 9 methyl (H3K9me) marks. (C) Transposon gene silencing in animal germlines. piRNAs target a silencing complex to transposable and repeat sequences to block their expression. PIWI-bound piRNAs have been proposed to guide PIWI and silencing factors to genomic targets by interaction with complementary chromatin-associated or nascent RNA. Factors implicated include heterochromatin protein 1 (HP1) and de novo DNA methyltransferase (DNMT3b), as well as deposition of DNA and nucleosome methylation marks. (D) DNA elimination in Tetrahymena uses scan RNAs (scnRNAs) to guide Twi1p to nascent RNA, leading to the elimination of the associated DNA at that site. This process involves H3K9 and H3K27 methylation and recruitment of chromatin modification and remodeling proteins.
In addition to AGO4 and Pol V, the de novo DNA methyltransferase DRM2 and the DDR complex are also recruited (Law et al., 2010). The DDR complex is composed of DDR1, DMS3, and RDM1 proteins. DDR1 and DMS3 are both chromatin remodelers (Kanno et al., 2004, 2008). RDM1 has single-stranded methyl DNA binding properties and associates with AGO4 and DRM2, leading to the suggestion that it bridges AGO4-chromatin-DRM2 interactions (Gao et al., 2010).
Silencing of centromeric repeats in yeast
In the yeast S. pombe the RNA-induced transcriptional silencing (RITS) complex guides transcriptional gene silencing (Volpe et al., 2002; Verdel et al., 2004). Pericentromeric DNA repeats are the main target of RITS and are silenced by the establishment and maintenance of heterochromatin through histone H3–lysine 9 methylation (H3K9me) (Nakayama et al., 2001; Volpe et al., 2002).
Centromeric repeats require ncRNA transcription for silencing to occur (Reinhart and Bartel, 2002). Silencing is triggered by siRNAs generated from nascent ncRNAs, which are subsequently loaded into AGO1 (Verdel et al., 2004). Nascent ncRNAs in turn serve as platforms for binding and recruitment of RITS by base-pairing interactions with the complementary AGO1-bound siRNAs (Motamedi et al., 2004; Buhler et al., 2006; Iida et al., 2008) (Fig. 2B).
Tas3 and Chp1 are also recruited to the chromatin (Verdel et al., 2004). Tas3 has an Ago-hook domain that interacts with AGO1 directly, while Chp1 is a chromodomain protein that binds H3K9me marks (Sadaie et al., 2004; Till et al., 2007). Thus, Tas3, and Chp1 bridge interactions between AGO–siRNA complexes and chromatin and contribute to specificity by recognizing H3K9me. Chromatin and RNA-associated RITS also recruits the CLRC complex. CLRC contains an H3K9 methyltransferase enzyme called Clr4, among other subunits, to help establish and maintain silencing (Hong et al., 2005).
Heterochromatin and transposon silencing in the animal germline
Small RNA-dependent transcriptional silencing pathways are genome surveillance mechanisms in animal germline cells. This pathway relies on the piRNAs, which direct Piwi proteins to transposon and repeat-rich RNAs for degradation (Malone and Hannon, 2009). While the piRNA pathway is largely believed to function through post-transcriptional RNAi, evidence suggests that it also silences transcription (Fig. 2C) (Pal-Bhadra et al., 2004; Brower-Toland et al., 2007; Grewal and Elgin, 2007; Watanabe et al., 2011).
In Drosophila, PIWI associates with chromatin in an RNA-dependent manner and directly binds the chromodomain protein Heterochromatin Protein 1 (HP1α) (Brower-Toland et al., 2007). Both HP1 localization and heterochromatin formation depend on the RNAi machinery, specifically the piRNA pathway proteins Piwi, Aubergine, and Spindle-E (Pal-Bhadra et al., 2004). HP1α binds to H3K9me nucleosomes and directly to PIWI through an HP1-interacting P×V×L motif (Brower-Toland et al., 2007). Recently, AGO2 and DCR2 were found on the chromatin interacting with transcriptional machinery and implicated in regulation of RNA polymerase II processivity in (Cernilogar et al., 2011).
In mice, piRNAs have been implicated in transcriptional silencing of the imprinted Rasgrf1 locus. This process requires the Piwi family proteins Mili and Miwi2, the de novo DNA methyltransferase Dnmt3b, and transcription of noncoding RNAs at the Rasgrf1 locus (Kuramochi-Miyagawa et al., 2008; Watanabe et al., 2011).
DNA elimination in Tetrahymena
In the ciliated protozoan Tetrahymena thermophila there are 2 nuclei, a somatic macronucleus, and a germline micronucleus, which performs reproductive functions (Ishuzi et al., 2011). During reproduction the micronucleus produces a new micronuclei and a new macronuclei. The developing new macronucleus undergoes programmed DNA elimination. The entire genome is restructured, losing ∼6,000 repeat and transposable elements (Ishuzi et al., 2011).
Elimination is mediated by ∼28-nt-long scan RNAs (scnRNAs), which bind the Piwi homolog Twi1p (Mochizuki et al., 2002). Scan RNAs are produced from the original micronuclear genome, including all of the sequences to be eliminated (Mochizuki and Gorovsky, 2005). Scan RNAs loaded into Twi1p are transported into the original macronucleus for selection of scnRNAs that do not share homology (Mochizuki et al., 2002; Noto et al., 2010). Scan RNAs which remain and are therefore unique to the original micronucleus enter into the developing macronucleus to direct deletion of unwanted parental sequences, which are primarily transposon repeat elements (Aronica et al., 2008).
Elimination involves docking of scnRNA–Twi1p complexes through recognition of nascent Pol II noncoding RNA transcripts. The chromodomain protein Pdd1p, an HP1 homolog, is recruited and may bind H3K9me and H3K27me chromatin marks deposited by the Ezl1p methyltransferase (Fig. 2D) (Madireddi et al., 1996; Mochizuki and Gorovsky, 2004; Liu et al., 2007; Kataoka and Machizuki, 2011). DNA elimination is a unique example of small RNA-guided genome surveillance. However, at the mechanistic level it shares striking similarities with the RdDM and RITS pathways.
AGO and other RNAi factors in cell nuclei
Small guide RNAs and RNAi machinery have been reported in the nucleus of a number of organisms (Katahira and Yoneda, 2011). In the worm C. elegans, AGO protein NRDE-3 shuttles siRNAs into the nucleus and interacts with NRDE-2. Both proteins are recruited to nascent transcripts by complementary siRNAs, resulting in silencing of gene transcription (Guang et al., 2008, 2010).
piRNA-like RNAs have been found in human cancer cells (Cheng et al., 2011) and in a panel of samples from mouse, monkey, and fruit fly (Yan et al., 2011). In human cell nuclei, an abundance of mature miRNAs have been cataloged, with numbers comparable to those in the cytoplasm (Liao et al. 2010; Jeffries et al., 2011).
In mammalian cells, the existence of nuclear RNAi pathways was first inferred from the observations that nuclear miRNAs and siRNAs could cause cleavage of RNA targets, such as 7SK small nuclear RNA (Meister et al., 2004; Robb et al., 2005). Endogenous human AGO2 has been shown to be nuclear with highly specific antibodies (Rudel et al., 2008; Tan et al., 2009; Chu et al., 2010). Fluorescence correlation and cross-correlation spectroscopy revealed AGO2 and AGO2–small RNA complexes in the nucleus (Ohrt et al., 2008) and also suggested that AGO2–small RNA complexes are predominantly loaded in the cytoplasm and imported into the nucleus (Ohrt et al., 2008). In support of AGO2 nuclear localization, importin-8 was found to affect accumulation of nuclear AGO2 (Weinmann et al., 2009).
In addition to AGO2, other RNAi factors have been reported in the nucleus of mammalian cells. Recently, we reported the presence of AGO1, 2, 3, and 4 in the nuclei of multiple human cell lines (Chu et al., 2010). In adult mouse stem cells as well as other somatic cell lines from mouse, monkey, and fruit flies, Piwi proteins have been detected at significant levels (Wu et al., 2010; Yan et al., 2011). Finally, RNAi factors like Dicer and GW182 have been observed in the nucleus, suggesting that RNAi pathways have the necessary components to function in the nucleus of various mammalian cell types (Till et al., 2007; Sinkkonen et al., 2010; Liang and Crooke, 2011).
Potential Targets for Small RNA-Bound AGO: Nuclear RNAs
Less than 2% of the human genome encodes exonic mRNA sequence, which is translated into protein, while ∼25% encodes introns (Lander et al., 2001; Venter et al., 2001). However, up to 98% of the human genome is believed to be transcribed at some point (Kapranov et al., 2002, 2007; Carninci et al., 2005; Birney et al., 2007). Nearly half of all transcripts in the human genome are unannotated, have unknown functions, and are only found in the nucleus (Cheng et al., 2005). Thus, the vast majority of RNA is noncoding and provides a rich and unappreciated pool of potential targets for nuclear RISC.
Long noncoding RNAs
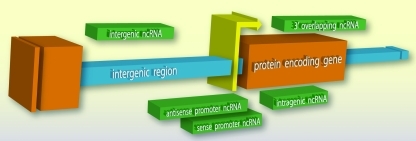
Transcriptome studies have identified numerous RNAs that are not protein coding, with many falling into an important class >200 nts known as long ncRNA (lncRNA) (Kapranov et al., 2002, 2007; Bertone et al., 2004; Birney et al., 2007). Long ncRNAs can share traits with coding genes, such as defined promoters and chromatin marks at their genomic loci and splicing (Guttman et al., 2009). Long ncRNAs are autonomous sequence domains or elements driven by their own promoters, which can be cryptic and difficult to define (Guttman et al., 2009). They are produced from various locations and may be sense or antisense with respect to coding genes. The prominent types are intergenic, intragenic, or gene-overlapping (Fig. 3) (Dinger et al., 2009; Faghihi and Wahlestedt, 2009).
FIG. 3.
Typical locations of long noncoding RNA (lncRNA) expression. Various sense and antisense ncRNA transcripts (green) can arise from regions between (intergenic; blue) or within (intragenic; orange) the open reading frames of protein coding genes in the genome, as well as overlap gene promoters (yellow) or gene termini.
Long ncRNA-mediated transcriptional regulation
The exact function of lncRNAs has been debated, stemming partly from technical concerns surrounding detection methods, RNA abundance, and artifacts from complementary DNA library preparations (van Bakel et al., 2010; Clark et al., 2011). A greater concern is that some transcripts, especially low abundance or rapidly degraded ncRNAs, may be transcriptional background or “noise” and have no significant function (Dinger et al., 2009). Nonetheless, a number of lncRNAs have been functionally defined and many more are being actively investigated (Khalil et al., 2009; Ponting et al., 2009; Guttman et al., 2011).
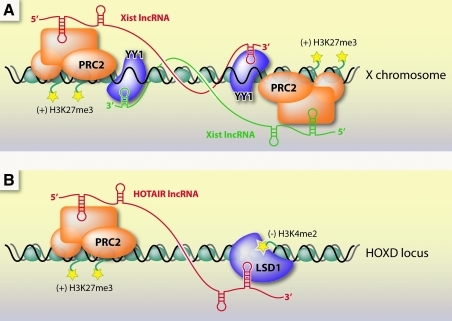
Regulatory lncRNAs might act in cis to regulate their local environment or in trans at distant regions of the genome. Xist and HOTAIR are two well-studied lncRNAs that serve as models of function. Both associate with chromatin to silence transcription of large genomic domains in a developmentally regulated manner. Xist acts in cis to coat the entire X chromosome. HOTAIR, by contrast, acts in trans; it is expressed from chromosome 12 but associates with the HOXD locus on chromosome 2 (Rinn et al., 2007; LEE, 2009).
The interactions of Xist and HOTAIR with chromatin are mediated by RNA-protein interactions and binding of chromatin-modifying enzymes (Fig. 4). Both Xist and HOTAIR recruit a histone methyltransferase complex called polycomb repressive complex 2 (PRC2) to the chromatin (Rinn et al., 2007). In addition, Xist has been found to bind a unique adaptor protein called YY1, which has separate RNA and DNA binding domains suggesting it is a key factor in tethering Xist to the chromatin (Jeon and Lee, 2011). In addition to PRC2, HOTAIR also binds a histone demethylase enzyme called LSD1 (Tsai et al., 2010).
FIG. 4.
Transcriptional silencing mechanisms by long noncoding RNAs Xist and HOTAIR. (A) Xist RNA is expressed from the X chromosome and functions in cis to silence the X chromosome. Xist coats the X chromosome locally, interacts with the chromatin through the adaptor complex YY1, and recruits the repressive polycomb group complex PRC2 to deposit histone H3 lysine 27 trimethyl (H3K27me3) marks on nucleosomes. (B) The HOTAIR lncRNA is expressed from the HOXC locus and functions in trans to silence expression from the HOXD locus. HOTAIR binds PRC2 at its 5′ end and LSD1 near its 3′ end. PRC2 deposits the repressive H3K27me3 mark, while LSD1 can erase H3K4me2 marks, both necessary for maintaining silencing at the HOXD locus.
Other lncRNAs, like Kcnq1ot1 and ANRIL, have been found to interact with PRC2 as well as other chromatin modifying complexes (Pandey et al., 2008; Yap et al., 2010). A genome-wide study found that ∼20% of human intergenic lncRNAs are bound by PRC2 (Khalil et al., 2009). The detailed mechanism of transcriptional control by XIST, HOTAIR, and other functional lncRNAs is not known. It is likely, however, that they regulate transcription at the chromatin level through protein bridges to chromosomal DNA (Khalil et al., 2009).
Long non-coding RNA and disease
Some lncRNAs have been linked to disease. HOTAIR and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) are overexpressed in various cancers (Gupta et al., 2010; Kogo et al., 2011; Lai et al., 2011; Luo et al., 2006; Yang et al., 2011). The lncRNA ANRIL is a factor in coronary artery disease risk (Broadbent et al., 2008; Schaefer et al., 2009) and the DGCR5 lncRNA has been implicated in the cause and progression of DiGeorge Syndrome (Qureshi et al., 2010). The correlation between lncRNAs and disease is expected to grow significantly as lncRNA functions become better characterized (Wapinski and Chang, 2011).
Alternative splicing and disease
On average, pre-mRNA transcript is overwhelmingly intronic by a factor of 25:1 and therefore largely noncoding (Venter et al., 2001). Pre-mRNA is made as a continuous transcript, and exons must be spliced together to produce a mature mRNA. When alternate exons are used, products of diverse function and expression are generated (Ben-Dov et al., 2008; Keren et al., 2010). Approximately 95% of multi-exonic genes are estimated to undergo alternative splicing in humans (Pan et al., 2008).
Alternative splicing provides an opportunity for targeting and regulation with small RNAs. In the diseases neurofibromatosis and ataxia-telangiecstasia, about 50% of the mutations in the NF1 and ATM genes affect splicing, respectively (Pagani and Baralle, 2004). The symptoms of spinal muscular atrophy and Duchenne's muscular dystrophy might be alleviated if splicing of the respective genes, SMN2 and dystrophin, can be redirected (Lorson et al., 2010; Lu et al., 2010). Correlations between disease and alternative splicing exist and will continue to be discovered with the increasing application of high-throughput sequencing technologies (Ben-Dov et al., 2008; Pan et al., 2008).
Controlling Transcription and Splicing with Small RNAs Inside Mammalian Nuclei
Suppressing transcription with promoter targeted small RNAs
In 2004, Morris and co-workers reported that siRNAs complementary to the EF1A promoter could silence expression when introduced into human cells. Inhibition depended on histone deacetylase and DNA methyltransferase activities (Morris et al., 2004). Subsequently, other reports appeared describing the silencing of various genes, including E-cadherin, RASSF1, TGFβ receptor II, progesterone receptor, major vault protein, androgen receptor, cyclooxygenase-2, CDH1, and c-myc (Castanotto et al., 2005; Janowski et al., 2005; Ting et al., 2005; Kim et al., 2007; Napoli et al., 2009; Green and Weinberg, 2011).
The mechanism of transcriptional regulation by small RNAs is under investigation. The laboratories examining mammalian small RNA-mediated modulation of transcription employ different targets, some near the transcription start site and some hundreds of bases distant (MORRIS, 2009a; Green and Weinberg, 2011). Several studies implicate lncRNAs that overlap gene promoters as critical recognition points for promoter-targeted small RNAs (Han et al., 2007; Gonzalez et al., 2008; Schwartz et al., 2008; MORRIS, 2009b). These lncRNAs may be sense or antisense with respect to the affected coding mRNA.
All reports indicate a role for either AGO1 or AGO2 (Janowski et al., 2006; Kim et al., 2006; Chu et al., 2010). In most studies, chromatin binding or modifying proteins, like HP1 and DNMT3, or RNA-binding proteins, like heterogeneous nuclear RNP K, are also implicated (Janowski et al., 2006; Kim et al., 2006; Schwartz et al., 2008; MORRIS, 2009a; Green and Weinberg, 2011). Although reports differ on the extent and necessity of chromatin modification (COREY, 2005; Green and Weinberg, 2011), the current model of small RNA-directed transcriptional silencing in mammals has a number of similarities with natural pathways like RdDM and RITS (Fig. 5A).
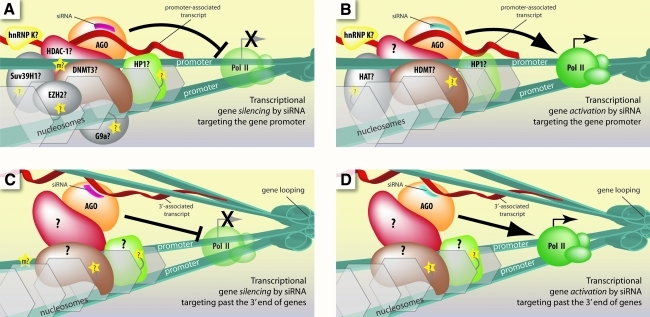
FIG. 5.
Current models for the molecular mechanism of transcriptional control by small RNAs that target gene promoters and regions past the 3′ end of genes in mammalian cells. (A, B) Promoter-targeted siRNAs guide AGO protein to promoter-associated transcripts. A number of chromatin modification and remodeling enzymes have been implicated in this process and are recruited to either silence or activate gene expression. (C, D) siRNAs targeted to regions past the 3′-ends of genes can guide AGO to noncoding transcripts overlapping those regions. Gene looping, which brings the 3′-end into close proximity to the promoter, enables recruitment of factors that can remodel or modify the promoter chromatin to modulate RNA polymerase II activity, either repressing or activating transcription.
Activating transcription with promoter targeted small RNAs
In 2006 Li and colleagues reported that duplex RNAs complementary to the promoter regions of E-cadherin, p21, or VEGF increased transcription of the respective gene (Li et al., 2006). AGO2 protein was required for activation and H3K9me was reduced. They have gone on to activate a number of other genes in different mammalian cell types, including p53, PAR4, WT1, NKX3-1, cyclin B1, and CXCR4 (Huang et al., 2010).
Our laboratory observed that small RNAs targeting the promoter of progesterone receptor can increase expression by up to 10-fold in MCF7 breast cancer cells (Janowski et al., 2007). Small RNAs complementary to the promoter of LDL receptor were also shown to activate expression (Matsui et al., 2010). The mechanism of transcriptional gene activation with small RNAs in mammalian cells shares common features with transcriptional silencing, such as involvement of AGO2 protein and promoter-associated lncRNAs (Fig. 5B) (Janowski et al., 2007; Schwartz et al., 2008; Chu et al., 2010).
Targeting past the 3′ end of genes with small RNAs
Noncoding RNAs often overlap the 3′ termini of mRNA. Small RNAs targeting lncRNAs downstream from the 3′ end of progesterone receptor and BRCA1 can either activate or silence transcription, depending on the basal level of expression in the cell (Younger and Corey, 2011b; Yue et al., 2010). The mechanism requires AGO2 and gene looping brings the 3′ ends into close proximity with promoter regions, allowing transcriptional control across tens of thousands of bases (Fig. 5C, D).
Are nuclear small RNAs natural regulators of gene expression?
The observation of robust transcriptional silencing and activation has led to searches for natural small RNAs and pathways. Many miRNAs have strong complementarity to promoter regions or sequences beyond 3′ gene termini (Younger et al., 2009; Younger and Corey, 2011a, 2011b). Synthetic miRNAs based on computationally predicted matches to promoter sequences have been shown to inhibit and activate gene transcription (Klase et al., 2007; Kim et al., 2008; Place et al., 2008; Tan et al., 2009; Majid et al., 2010; Younger and Corey, 2011a). However, definitive characterization of endogenous small RNA transcriptional activators or repressors remains a primary goal for the field.
Designing and using promoter-targeted small RNAs
Accomplishing gene modulation and then demonstrating that it is not an off-target effect can be time consuming. Thus, it is important that the target gene be chosen for compelling reasons, such as for an important research or therapeutic goal.
Preliminary work includes having a firm understanding of the transcription start site. For well-characterized genes, a database search may be sufficient, but poorly characterized genes will require determination by 5′-rapid amplification of cDNA ends (RACE). Noncoding transcription across the targeted promoter site is also critical and should be experimentally validated by methods like reverse transcriptase-PCR and RACE.
Once a promising target region is identified (upstream from the +1 site of mRNA transcription and overlapped by ncRNA), several small RNAs should be designed following basic siRNA design rules. If gene activation is desired, a low basal level of expression will make detection of increased expression simpler. Once transcriptional activation or silencing is observed, multiple mismatched or scrambled siRNAs must be tested to help minimize the chance that modulation is an of off-target effect. In support of specificity, RNA immunoprecipitation can be performed to test AGO recruitment to the ncRNA (Schwartz and Corey, 2011). Finally, chromatin immunoprecipitation of RNA Polymerase II or nuclear run-on assays can provide evidence for gene modulation at the transcriptional level.
Controlling alternative splicing with small RNAs
Alternative splicing allows cells to make different proteins from one mRNA (Keren et al., 2010). In some cases, promoting or inhibiting a particular splice event can lead to production of a therapeutically valuable protein isoform.
Precedence for modulating splicing with small RNAs comes from synthetic therapeutic approaches and natural small RNAs. Antisense oligonucleotides that target pre-mRNAs have successfully modulated splicing in cell lines, animals, and clinical trials (Aartsma-Rus and van Ommen, 2007; Bauman et al., 2009). Small RNAs derived from processed small nucleolar RNAs of the imprinted HBII-52 genomic region have been shown to affect splicing of serotonin 5-HT2C receptor and the U1 small nuclear RNP naturally modulates splicing by binding to a 9-bp sequence at 5′ splice sites (Khanna and Stamm, 2010).
Kornblihtt and co-workers have reported small RNAs that affect the splicing of fibronectin 1 (FN1) pre-mRNA (Allo et al., 2009). They targeted siRNAs to intron 33 of FN1 and found greater inclusion of the neighboring exon. This effect was dependent on AGO1 and antisense transcription. It also affected elongation by RNA polymerase, caused changes in H3K9 and H3K27 methylation and involved HP1, suggesting a coupling of transcription and splicing regulation.
Recently, our laboratory observed RNA-mediated alteration of splicing by a different mechanism (Liu et al., 2011). We targeted introns and exons of an engineered beta-globin gene, dystrophin, and SMN2 genes with fully complementary and central mismatch-containing siRNAs and observed exon exclusion (Fig. 6). AGO2 was required to redirect splicing with small RNAs, but not AGO1. No changes in H3K9 or H3K27 methylation were observed. Knockdown of HP1α or inhibition of histone deacetylation or DNA methylation had no effect on splicing modulation. These studies show that splice site alteration can be achieved with small RNAs, perhaps using more than one mechanism.
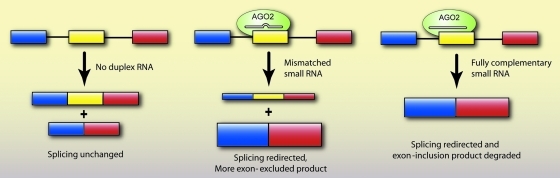
FIG. 6.
Small RNAs of full or partial complementarity can redirect splicing in human cells. siRNAs loaded into AGO2 force exon exclusion or inclusion by targeting intronic or exonic sequences involved in splicing. siRNAs containing central mismatches can target exons without causing degradation, thereby expanding the potential target sequence for redirecting splicing. On the other hand, fully complementary siRNAs can redirect splicing and also help remove an unwanted mRNA species that contains a specific exon by causing post-transcriptional degradation.
Is AGO a slicer in the nucleus?
When fully complementary small RNAs guide slicer-competent AGO proteins to target RNAs, the expected outcome has traditionally been site-specific cleavage. However, the siRNA-mediated alternative splicing that we have observed could not occur if small RNAs are causing degradation of the pre-mRNA target (Liu et al., 2011). AGO2 binding to nuclear transcripts has been detected using primer sets that are on either side of the predicted cleavage site, which would be impossible if the noncoding transcript were cut (Chu et al., 2010). In addition, when we modulate progesterone receptor transcription with small RNAs, levels of promoter-associated lncRNA are unchanged (Schwartz et al., 2008; Yue et al., 2010).
Hirose and colleagues have compared gene silencing of nuclear targets by duplex RNAs and antisense oligonucleotides that function through a mechanism involving RNAse H. Consistent with our observations, they observe that antisense oligonucleotides silence nuclear RNA targets, while duplex RNAs are inactive (Ideue et al., 2009).
By contrast, other reports suggest that addition of small RNAs can cause degradation of noncoding nuclear transcripts. One early study reported siRNA-mediated reduction of 7SK snRNA (Robb et al., 2005). A more recent study has used short hairpin RNAs, which are processed into siRNAs inside cells, to interrogate the function of 226 lncRNAs in mouse embryonic stem cells (Guttman et al., 2011). Expression was successfully reduced by an average of 75% for 147 lncRNAs. Other examples of knocking down specific lncRNAs with small RNAs have been reported (Huarte et al., 2010; Lai et al., 2011). It is possible that AGO2 may be a slicer in some contexts but not others, and this issue will need to be resolved for future investigation of small RNA and AGO mechanisms in mammalian nuclei. At a minimum, researchers who use duplex RNAs to reduce expression of noncoding transcripts in the nucleus should ensure the experiments include proper controls before concluding that silencing is an “on-target” effect.
Conclusions
Gene-specific control of transcription or splicing has opened up new therapeutic opportunities. Oligonucleotides have previously been shown to function in the nucleus by targeting pre-mRNA or nuclear-localized RNAs. The presence and demonstrated function of AGO protein and other RNAi factors in the nucleus of mammalian cells indicates a new and exciting therapeutic option. Small RNAs entering the nucleus can, in complex with AGO, recognize ncRNAs or pre-mRNAs and affect nuclear processes like transcription or splicing.
Nuclear pathways that use AGO–RNA complexes to control gene expression in diverse organisms from plants to yeast to man suggests an endogenous function in mammalian cells. A surprising degree of similarity among these pathways, including lncRNAs as targets and related protein factors like chromatin modifiers and remodelers, indicates the conservation of nuclear RNAi mechanisms. The robustness with which synthetic RNAs control gene expression or splicing in mammalian cells supports this conclusion. However, the significance of nuclear AGO is largely unknown. As we learn more about the function of ncRNAs and roles for nuclear AGO, it is likely that we will better understand the potential for small RNAs to act as natural regulators of gene expression in the nucleus. This understanding will in turn lead to recognition of novel intervention points for therapy.
Acknowledgments
This work was supported by the National Institutes of Health (GM77253 and GM73042 to DRC and 1F32HD060377 to KTG) and the Robert A. Welch Foundation (I-1244 to DRC).
Author Disclosure Statement
No competing financial interests exist.
References
- ALLO M. BUGGIANO V. FEDEDA J.P. PETRILLO E. SCHOR I. DE LA MATA M. AGIRRE E. PLASS M. EYRAS E. ELELA S.A., et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- ARAVIN A. GAIDATZIS D. PFEFFER S. LAGOS-QUINTANA M. LANDGRAF P. IOVINO N. MORRIS P. BROWNSTEIN M.J. KURAMOCHI-MIYAGAWA S. NAKANO T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- ARONICA L. BEDNENKO J. NOTO T. DESOUZA L.V. SIU K.W. LOIDL J. PEARLMAN R.E. GOROVOSKY M.A. MOCHIZUKI K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AARTSMA-RUS A. VAN OMMEN G.J. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTEL D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- BARTEL D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUMEN J. JEARAWIRIYAPAISARN N. KOLE R. Therapeutic potential of splice switching oligonucleotides. Oligonucleotides. 2009;19:1–13. doi: 10.1089/oli.2008.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEN-DOV C. HARTMANN B. LUNDGREN J. VALCARCEL J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- BERTONE P. STOLC V. ROYCE T.E. ROZOWSKY J.S. URBAN A.E. ZHU X. RINN J.L. TONGPRASIT W. SAMANTA M. WEISSMAN S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- BIRNEY E. STAMATOYANNOPOULOS J.A. DUTTA A. GUIGO R. GINGERAS T.R. MARGULIES E.H. WENG Z. SNYDER M. DERMITZAKIS E.T. THURMAN R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOHMERT K. CAMUS I. BELLINI C. BOUCHEZ D. CABOCHE M. BENNING C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROADBENT H.M. PEDEN J.F. LORKOWSKI S. GOEL A. ONGEN H. GREEN F. CLARKE R. COLLINS R. FRANZOSI M.G. TOGNONI G., et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- BROWER-TOLAND B. FINDLEY S.D. JIANG L. LIU L. YIN H. DUS M. ZHOU P. ELGIN S.C. LIN H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUHLER M. VERDEL A. MOAZED D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- CARMELL M.A. XUAN Z. ZHANG M.Q. HANNON G.J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- CARNINCI P. KASUKAWA T. KATAYAMA S. GOUGH J. FRITH M.C. MAEDA N. OYAMA R. RAVASI T. LENHARD B. WELLS C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- CASTANOTTO D. TOMMASI S. LI M. LI H. YANOW S. PFEIFER G.P. ROSSI J.J. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol. Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- CERNILOGAR F.M. ONORATI M.C. KOTHE G.O. BURROUGHS A.M. PARSI K. M. BREILING A. SARDO F.L. SAXENA A. MIYOSHI K. SIOMI H., et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENG J. GUO J.M. XIAO B.X. MIAO Y. JIANG Z. ZHOU H. LI Q.N. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- CHENG J. KAPRANOV P. DRENKOW J. DIKE S. BRUBAKER S. PATEL S. LONG J. STERN D. TAMMANA H. HELT G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- CHU Y. YUE X. YOUNGER S.T. JANOWSKI B.A. COREY D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK M.B. AMARAL P.P. SCHLESINGER F.J. DINGER M.E. TAFT R.J. RINN J.L. PONTING C.P. STADLER P.F. MORRIS K.V. MORILLON A., et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COREY D.R. Regulating mammalian transcription with RNA. Trends Biochem. Sci. 2005;30:655–658. doi: 10.1016/j.tibs.2005.09.007. [DOI] [PubMed] [Google Scholar]
- DENLI A.M. TOPS B.B. PLASTERK R.H. KETTING R.F. HANNON G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- DINGER M.E. AMARAL P.P. MERCER T.R. MATTICK J.S. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief. Func. Genomic. Proteomic. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- ELBASHIR S.M. HARBORTH J. LENDECKEL W. YALCIN A. WEBER K. TUSCHL T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- EULALIO A. BEHM-ANSMANT I. IZAURRALDE E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Bio. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- FAGARD M. BOUTET S. MOREL J.B. BELLINI C. VAUCHERET H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAGHIHI M.A. WAHLESTEDT C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Bio. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANK F. SONENBERG N. NAGAR B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- GAO Z. LIU H.L. DAXINGER L. PONTES O. HE X. QIAN W. LIN H. XIE M. LORKOVIC Z.J. ZHANG S., et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILDIYAL M. ZAMORE P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALEZ S. PISANO D.G. SERRANO M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- GREEN V.A. WEINBERG M.S. Small RNA-induced transcriptional gene regulation in mammals mechanisms, therapeutic applications, and scope within the genome. Prog. Mol. Biol. Transl. Sci. 2011;102:11–46. doi: 10.1016/B978-0-12-415795-8.00005-2. [DOI] [PubMed] [Google Scholar]
- GREGORY R.I. YAN K.P. AMUTHAN G. CHENDRIMADA T. DORATOTAJ B. COOCH N. SHIEKHATTAR R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- GREWAL S.I. ELGIN S.C. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUANG S. BOCHNER A.F. BURKHART K.B. BURTON N. PAVELEC D.M. KENNEDY S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUANG S. BOCHNER A.F. PAVELEC D.M. BURKHART K.B. HARDING S. LACHOWIEC J. KENNEDY S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNAWARDANE L.S. SAITO K. NISHIDA K.M. MIYOSHI K. KAWAMURA Y. NAGAMI T. SIOMI H. SIOMI M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- GUO H. INGOLIA N.T. WEISSMAN J.S. BARTEL D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUPTA R.A. SHAH N. WANG K.C. KIM J. HORLINGS H.M. WONG D.J. TSAI M.C. HUNG T. ARGANI P. RINN J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTTMAN M. AMIT I. GARBER M. FRENCH C. LIN M.F. FELDSER D. HUARTE M. ZUK O. CAREY B.W. CASSADY J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTTMAN M. DONAGHEY J. CAREY B.W. GARBER M. GRENIER J.K. MUNSON G. YOUNG G. LUCAS A.B. ACH R. BRUHN L., et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN J. KIM D. MORRIS K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE X.J. HSU Y.F. ZHU S. LIU H.L. PONTES O. ZHU J. CUI X. WANG C.S. ZHU J.K. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009a;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE X.J. HSU Y.F. ZHU S. WIERZBICKI A.T. PONTES O. PIKAARD C.S. LIU H.L. WANG C.S. JIN H. ZHU J.K. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009b;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOCK J. MEISTER G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOCK J. WEINMANN L. ENDER C. RUDEL S. KREMMER E. RAABE M. URLAUB H. MEISTER G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO reports. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG E.J. VILLEN J. GERACE E.L. GYGI S.P. MOAZED D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- HORWICH M.D. LI C. MATRANGA C. VAGIN V. FARLEY G. WANG P. ZAMORE P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- HUANG V. QIN Y. WANG J. WANG X. PLACE R.F. LIN G. LUE T.F. LI L.C. RNAa is conserved in mammalian cells. PloS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUARTE M. GUTTMAN M. FELDSER D. GARBER M. KOZIOL M.J. KENZELMANN-BROZ D. KHALIL A.M. ZUK O. AMIT I. RABANI M., et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IDEUE T. HINO K. KITAO S. YOKOI T. HIROSE T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIDA T. NAKAYAMA J. MOAZED D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISHIZU H. NAGAO A. SIOMI H. Gatekeepers for Piwi-piRNA complexes to enter the nucleus. Curr. Opin. Genet. Dev. 2011;21:484–490. doi: 10.1016/j.gde.2011.05.001. [DOI] [PubMed] [Google Scholar]
- JANOWSKI B.A. HUFFMAN K.E. SCHWARTZ J.C. RAM R. HARDY D. SHAMES D.S. MINNA J.D. COREY D.R. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- JANOWSKI B.A. HUFFMAN K.E. SCHWARTZ J.C. RAM R. NORDSELL R. SHAMES D.S. MINNA J.D. COREY D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- JANOWSKI B.A. YOUNGER S.T. HARDY D.B. RAM R. HUFFMAN K.E. COREY D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- JEFFRIES C.D. FRIED H.M. PERKINS D.O. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011;17:675–686. doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEON Y. LEE J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANNO T. BUCHER E. DAXINGER L. HUETTEL B. BOHMDORFER G. GREGOR W. KREIL D.P. MATZKE M. MATZKE A.J. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- KANNO T. METTE M.F. KREIL D.P. AUFSATZ W. MATZKE M. MATZKE A.J. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- KAPRANOV P. CAWLEY S.E. DRENKOW J. BEKIRANOV S. STRAUSBERG R.L. FODOR S.P. GINGERAS T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- KAPRANOV P. CHENG J. DIKE S. NIX D.A. DUTTAGUPTA R. WILLINGHAM A.T. STADLER P.F. HERTEL J. HACKERMULLER J. HOFACKER I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- KATAHIRA J. YONEDA Y. Nucleocytoplasmic Transport of MicroRNAs and Related Small RNAs. Traffic. 2011;12:1468–1474. doi: 10.1111/j.1600-0854.2011.01211.x. [DOI] [PubMed] [Google Scholar]
- KATAOKA K. MOCHIZUKI K. Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Adv. Exp. Med. Biol. 2011;722:156–173. doi: 10.1007/978-1-4614-0332-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEREN H. LEV-MAOR G. AST G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- KHALIL A.M. GUTTMAN M. HUARTE M. GARBER M. RAJ A. RIVEA MORALES D. THOMAS K. PRESSER A. BERNSTEIN B.E. VAN OUDENAARDEN A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHANNA A. STAMM S. Regulation of alternative splicing by short non-coding nuclear RNAs. RNA Biol. 2010;7:480–485. doi: 10.4161/rna.7.4.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM D.H. SAETROM P. SNOVE O. ROSSI J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM D.H. VILLENEUVE L.M. MORRIS K.V. ROSSI J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- KIM J.W. ZHANG Y.H. ZERN M.A. ROSSI J.J. WU J. Short hairpin RNA causes the methylation of transforming growth factor-beta receptor II promoter and silencing of the target gene in rat hepatic stellate cells. Biochem. Biophys. Res. Commun. 2007;359:292–297. doi: 10.1016/j.bbrc.2007.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLASE Z. KALE P. WINOGRAD R. GUPTA M.V. HEYDARIAN M. BERRO R. MCCAFFREY T. KASHANCHI F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOGO R. SHIMAMURA T. MIMORI K. KAWAHARA K. IMOTO S. SUDO T. TANAKA F. SHIBATA K. SUZUKI A. KOMUNE S., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- KULKARNI M. OZGUR S. STOECKLIN G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- KURAMOCHI-MIYAGAWA S. WATANABE T. GOTOH K. TOTOKI Y. TOYODA A. IKAWA M. ASADA N. KOJIMA K. YAMAGUCHI Y. IJIRI T.W., et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI M.C. YANG Z. ZHOU L. ZHU Q.Q. XIE H.Y. ZHANG F. WU L.M. CHEN L.M. ZHENG S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2011 doi: 10.1007/s12032-011-0004-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- LANDER E.S. LINTON L.M. BIRREN B. NUSBAUM C. ZODY M.C. BALDWIN J. DEVON K. DEWAR K. DOYLE M. FITZHUGH W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- LANDTHALER M. GAIDATZIS D. ROTHBALLER A. CHEN P.Y. SOLL S.J. DINIC L. OJO T. HAFNER M. ZAVOLAN M. TUSCHL T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAW J.A. AUSIN I. JOHNSON L.M. VASHISHT A.A. ZHU J.K. WOHLSCHLEGEL J.A. JACOBSEN S.E. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE J.T. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE Y. AHN C. HAN J. CHOI H. KIM J. YIM J. LEE J. PROVOST P. RADMARK O. KIM S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- LI L.C. OKINO S.T. ZHAO H. POOKOT D. PLACE R.F. URAKAMI S. ENOKIDA H. DAHIYA R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIANG X.H. CROOKE S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAO J.Y. MA L.M. GUO Y.H. ZHANG Y.C. ZHOU H. SHAO P. CHEN Y.Q. QU L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PloS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINGEL A. SIMON B. IZAURRALDE E. SATTLER M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- LIU J. CARMELL M.A. RIVAS F.V. MARSDEN C.G. THOMSON J.M. SONG J.J. HAMMOND S.M. JOSHUA-TOR L. HANNON G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- LIU J. HU J. COREY D.R. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucl Acids Res. 2011 doi: 10.1093/nar/gkr780. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y. TAVERNA S.D. MURATORE T.L. SHABANOWITZ J. HUNT D.F. ALLIS C.D. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y. YE X. JIANG F. LIANG C. CHEN D. PENG J. KINCH L.N. GRISHIN N.V. LIU Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LORSON C.L. RINDT H. SHABABI M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum. Mol. Genet. 2010;19:R111–R118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU Q-L. YOKOTA T. TAKEDA S. GARCIA L. MUNTONI F. PARTRIDGE T. The status of exon skipping as a therapeutic approach to Duchenne muscular dystrophy. Mol. Ther. 2010;19:9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO J.H. REN B. KERYANOV S. TSENG G.C. RAO U.N. MONGA S.P. STROM S. DEMETRIS A.J. NALESNIK M. YU Y.P, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA J.B. YE K. PATEL D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA J.B. YUAN Y.R. MEISTER G. PEI Y. TUSCHL T. PATEL D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACRAE I.J. MA E. ZHOU M. ROBINSON C.V. DOUDNA J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U. S. A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADIREDDI M.T. COYNE R.S. SMOTHERS J.F. MICKEY K.M. YAO M.C. ALLIS C.D. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- MAJID S. DAR A.A. SAINI S. YAMAMURA S. HIRATA H. TANAKA Y. DENG G. DAHIYA R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALONE C.D. HANNON G.J. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUI M. SAKURAI F. ELBASHIR S. FOSTER D.J. MANOHARAN M. COREY D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010;17:1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATZKE M. KANNO T. DAXINGER L. HUETTEL B. MATZKE A.J. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- MEISTER G. LANDTHALER M. PATKANIOWSKA A. DORSETT Y. TENG G. TUSCHL T. Human Argonaute2 mediates cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- MOAZED D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOCHIZUKI K. FINE N.A. FUJISAWA T. GOROVSKY M.A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- MOCHIZUKI K. GOROVSKY M.A. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot. Cell. 2004;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOCHIZUKI K. GOROVSKY M.A. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRIS K.V. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009a;19:299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRIS K.V. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009b;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRIS K.V. CHAN S.W. JACOBSEN S.E. LOONEY D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- MOTAMEDI M.R. VERDEL A. COMENARES S.U. GERBER S.A. GYGI S.P. MOAZED D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- NAKAYAMA J. RICE J.C. STRAHL B.D. ALLIS C.D. GREWAL S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- NAPOLI S. PASTORI C. MAGISTRI M. CARBONE G.M. CATAPANO C.V. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOTO T. KURTH H.M. KATAOKA K. ARONICA L. DESOUZA L.V. SIU K.W. PEARLMAN R.E. GOROVSKY M.A. MOCHIZUKI K. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 2010;140:692–703. doi: 10.1016/j.cell.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OHRT T. MUTZE J. STAROSKE W. WEINMANN L. HOCK J. CRELL K. MEISTER G. SCHWILLE P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucl. Acids. Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAGANI F. BARALLE F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- PAL-BHADRA M. LEIBOVITCH B.A. GANDHI S.G. RAO M. BHADRA U. BIRCHLER J.A. ELGIN S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- PAN Q. SHAI O. LEE L.J. FREY B.J. BLENCOWE B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- PANDEY R.R. MONDAL T. MOHAMMAD F. ENROTH S. REDRUP L. KOMOROWSKI J. NAGANO T. MANCINI-DINARDO D. KANDURI C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- PARKER J.S. ROE S.M. BARFORD D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLACE R.F. LI L.C. POOKOT D. NOONAN E.J. DAHIYA R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PONTING C.P. OLIVER P.L. REIK W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- QURESHI I.A. MATTICK J.S. MEHLER M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAND T.A. GINALSKI K. GRISHIN N.V. WANG X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REINHART B.J. BARTEL D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- RINN J.L. KERTESZ M. WANG J.K. SQUAZZO S.L. XU X. BRUGMANN S.A. GOODNOUGH L.H. HELMS J.A. FARNHAM P.J. SEGAL E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBB G.B. BROWN K.M. KHURANA J. RANA T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- RUDEL S. FLATLEY A. WEINMANN L. KREMMER E. MEISTER G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADIAIE M. IIDA T. URANO T. NAKAYAMA J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASAKI T. SHIOHAMA A. MINOSHIMA S. SHIMIZU N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- SAXENA S. JONSSON Z.O. DUTTA A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- SCHAEFER A.S. RICHTER G.M. GROESSNER-SCHREIBER B. NOACK B. NOTHNAGEL M. EL MOKHTARI N.E. LOOS B.G. JEPSEN S. SCHREIBER S. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5:e1000378. doi: 10.1371/journal.pgen.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWARTZ J.C. COREY D.R. Practical considerations for analyzing antigene RNAs (agRNAs): RNA immunoprecipitation of argonaute protein. Methods Mol. Biol. 2011;764:301–315. doi: 10.1007/978-1-61779-188-8_20. [DOI] [PubMed] [Google Scholar]
- SCHWARTZ J.C. YOUNGER S.T. NGUYEN N.B. HARDY D.B. MONIA B.P. COREY D.R. JANOWSKI B.A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINKKONEN L. HUGENSCHMIDT T. FILIPOWICZ W. SVOBODA P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PloS ONE. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIOMI M.C. SATO K. PEZIC D. ARAVIN A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- SONG J.J. SMITH S.K. HANNON G.J. JOSHUA-TOR L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- TAKIMOTO K. WAKIYAMA M. YOKOYAMA S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN G.S. GARCHOW B.G. LIU X. YEUNG J. MORRIS J.P.T. CUELLAR T.L. MCMANUS M.T. KIRIAKIDOU M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009a;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN Y. ZHANG B. WU T. SKOGERBO G. ZHU X. GUO X. HE S. CHEN R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009b;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TILL S. LEJEUNE E. THERMANN R. BORTFELD M. HOTHORN M. ENDERLE D. HEINRICH C. HENTZE M.W. LADURNER A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- TING A.H. SCHUEBEL K.E. HERMAN J.G. BAYLIN S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOLIA N.H. JOSHUA-TOR L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- TSAI M.C. MANOR O. WAN Y. MOSAMMAPARAST N. WANG J.K. LAN F. SHI Y. SEGAL E. CHANG H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN BAKEL H. NISLOW C. BLENCOWE B.J. HUGHES T.R. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENTER J.C. ADAMS M.D. MYERS E.W. LI P.W. MURAL R.J. SUTTON G.G. SMITH H.O. YANDELL M. EVANS C.A. HOLT R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- VERDEL A. JIA S. GERBER S. SUGIYAMA T. GYGI S. GREWAL S.I. MOAZED D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLPE T.A. KIDNER C. HALL I.M. TENG G. GREWAL S.I. MARTIENSSEN R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- WANG Y. JURANEK S. LI H. SHENG G. TUSCHL T. PATEL D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAPINSKI O. CHANG H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- WASSENEGGER M. HEIMES S. RIEDEL L. SANGER H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- WATANABE T. TOMIZAWA S. MITSUYA K. TOTOKI Y. YAMAMOTO Y. KURAMOCHI-MIYAGAWA S. IIDA N. HOKI Y. MURPHY P.J. TOYODA A., et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEINMANN L. HOCK J. IVACEVIC T. OHRT T. MUTZE J. SCHWILLE P. KREMMER E. BENES V. URLAUB H. MEISTER G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- WIERZBICKI A.T. HAAG J.R. PIKAARD C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIERZBICKI A.T. REAM T.S. HAAG J.R. PIKAARD C.S. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU Q. MA Q. SHEHADEH L.A. WILSON A. XIA L. YU H. WEBSTER K.A. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2010;396:915–920. doi: 10.1016/j.bbrc.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN K.S. YAN S. FAROOQ A. HAN A. ZENG L. ZHOU M.M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- YAN Z. HU H.Y. JIANG X. MAIERHOFER V. NEB E. HE L. HU Y. HU H. LI N. CHEN W., et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Z. ZHOU L. WU L.M. LAI M.C. XIE H.Y. ZHANG F. ZHENG S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- YAP K.L. LI S. MUNOZ-CABELLO A.M. RAGUZ S. ZENG L. MUJTABA S. GIL J. WALSH M.J. ZHOU M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YE X. HUANG N. LIU Y. PAROO Z. HUERTA C. LI P. CHEN S. LIU Q. ZHANG H. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNGER S.T. COREY D.R. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011a;39:5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNGER S.T. COREY D.R. Transcriptional regulation by miRNA mimics that target sequences downstream of gene termini. Mol. Biosys. 2011b;7:2383–2388. doi: 10.1039/c1mb05090g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNGER S.T. PERTSEMLIDIS A. COREY D.R. Predicting potential miRNA target sites within gene promoters. Bioorg. Med. Chem. Lett. 2009;19:3791–3794. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU B. YANG Z. LI J. MINAKHINA S. YANG M. PADGETT R.W. STEWARD R. CHEN X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUE X. SCHWARTZ J.C. CHU Y. YOUNGER S.T. GAGNON K.T. ELBASHIR S. JANOWSKI B.A. COREY D.R. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAMORE P.D. TUSCHL T. SHARP P.A. BARTEL D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- ZHANG X. ROSSI J.J. Phylogenetic comparison of small RNA-triggered transcriptional gene silencing. J. Biol. Chem. 2011;286:29443–29448. doi: 10.1074/jbc.R111.276378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZILBERMAN D. CAO X. JACOBSEN S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]