Abstract
Staphylococcus aureus is a commensal bacterium associated with the skin and mucosal surfaces of humans and animals that can also cause chronic infection. The emergence of antibiotic-resistant strains such as methicillin-resistant S. aureus (MRSA) and strains causing chronic intramammary infections (IMI) in cows results in severe human and livestock infections. Conventional approaches to vaccine development have yielded only a few noneffective vaccines against MRSA or IMI strains, so there is a need for improved vaccine development. CD4 T lymphocytes are required for promoting gamma interferon (IFN-γ) mediated immunoglobulin isotype switching in B lymphocytes to produce high-affinity IgG antibodies and IFN-γ-mediated phagocyte activation for an effective resolution of bacterial infection. However, the lack of known CD4 T cell antigens from S. aureus has made it difficult to design effective vaccines. The goal of this study was to identify S. aureus proteins recognized by immune CD4 T cells. Using a reverse genetics approach, 43 antigens were selected from the S. aureus Newman strain. These included lipoproteins, proteases, transcription regulators, an alkaline shock protein, conserved-domain proteins, hemolysins, fibrinogen-binding protein, staphylokinase, exotoxin, enterotoxin, sortase, and protein A. Screening of expressed proteins for recall T cell responses in outbred, immune calves identified 13 proteins that share over 80% sequence identity among MRSA or IMI strains. These may be useful for inclusion in a broadly protective multiantigen vaccine against MRSA or IMI.
INTRODUCTION
Staphylococcus aureus is a Gram-positive, opportunistic pathogen associated with asymptomatic colonization of the skin and mucosal surfaces of healthy animals and humans. In cattle, S. aureus is the leading cause of mastitis, a disease affecting the mammary gland. Mastitis results in significant losses to U.S. (>$1.8 billion annually) and global dairy industries (76). S. aureus-induced mastitis is often subclinical and results in chronic bovine intramammary infections (IMI), even in animals placed under antibiotic regime (2, 25). The emergence of beta-lactam and methicillin-resistant S. aureus (MRSA) strains resistant to vancomycin and of recalcitrant staphylococcal IMI in cattle has necessitated the development of alternative therapeutic strategies (2, 25, 26, 31–33, 37, 51, 64). Furthermore, S. aureus can be transmitted from farm and companion animals to humans (15, 24, 42, 46, 80, 81). Currently, there are no commercially available vaccines available for MRSA or IMI.
Effective resolution of infection by bacterial pathogens is mediated by a combination of neutralizing antibody and inflammatory responses of activated macrophages and neutrophils. Therefore, to prevent S. aureus infection, a vaccine should be comprised of a broad repertoire of antigens that elicit IgG, driven by a CD4 Th1 microenvironment. However, decades of conventional vaccination attempts using various S. aureus antigens have yielded only a few defined proteins as vaccine candidates, of which many have proven to be ineffective (35, 69, 72). One consideration for vaccine failure is the interaction of S. aureus protein A with the Fcγ region of immunoglobulin molecule, which interferes with antibody function of opsonization and downstream phagocytosis and induction of apoptosis of marginal-zone and B-1 B cells (14). A second reason for vaccine failure has been the lack of consideration of antigens that elicit CD4 T cell responses.
To address this second problem, we identified 43 surface proteins and virulence factors from the S. aureus Newman strain by using a reverse genetics approach with a goal of identifying novel CD4 T cell antigens. A comparative proteomic and genomic analysis was performed using the S. aureus Newman strain and different bovine IMI and MRSA isolates to identify predicted surface-exposed proteins that are known virulence factors and virulence-associated factors that could be targeted by antibody and that are conserved across strains (11, 28). Holstein calves expressing different major histocompatibility complex class II (MHC-II) molecules were immunized with heat-killed S. aureus until an S. aureus-specific immune response was obtained. The 43 antigens were individually expressed by in vitro transcription and translation (IVTT), affinity-purified, and tested for immunogenicity using lymphocyte proliferation assays. Using this rapid screening method, we identified 13 novel antigens from the S. aureus Newman strain which have potential for use in a vaccine against S. aureus infection of cattle and, perhaps, humans. Until now, the dearth of knowledge of CD4 T cell antigens from S. aureus has hindered the rational design of multivalent vaccines for this pathogen, and the results of this investigation begin to bridge that critical knowledge gap.
MATERIALS AND METHODS
S. aureus.
S. aureus strain Newman was used to prepare formalin- or heat-inactivated bacterial suspensions for immunization and in vitro assays. The S. aureus Newman strain genome does not have genes encoding 11 out of 12 known superantigens (tst-1, seb, seg, sek, sei, sek, sem, sen, seo, sep, and seq), and only the sea gene is present (48). Bacteria were cultured overnight from frozen stocks in Trypticase soy broth (TSB) at 37°C with shaking (250 rpm), diluted 1:200 in fresh TSB the following morning, and then cultured to the early stationary phase of growth (optical density at 600 nm [OD600] of 2.0). To prepare inactivated suspensions, bacteria were harvested by centrifugation (8,000 × g for 10 min) and washed three times with phosphate-buffered saline (PBS), pH 7.2, to remove secreted proteins, including SEA, and resuspended in PBS. Serial 10-fold dilutions in PBS were then plated on Trypticase soy agar plates to determine the number of CFU. Bacterial suspensions were heat inactivated at 60°C for 1 h. For formalin inactivation, bacteria were resuspended in 3% formaldehyde in PBS and incubated overnight at room temperature with shaking at 60 rpm. Bacteria were then centrifuged, washed three times, and resuspended in the same volume of PBS and stored at 4°C until use. For both inactivation methods, 0.1 ml of each suspension was plated on blood agar and incubated at 37°C for up to 1 week to check for lack of growth.
Cattle.
Five 3-month-old Holstein steers (animals 30864, 30900, 30904, 30916, and 30919) with different MHC class II haplotypes were purchased from a local dairy, quarantined for 2 weeks, and immunized six times over a period of 18 weeks until a strong S. aureus-specific immune response was obtained. Immune serum collected before immunization and 2 weeks after each immunization was stored at −20°C. The animals were immunized initially by subcutaneous inoculation of 1.0 ml of 3.3 × 109 formalin-killed S. aureus bacteria emulsified in complete Freud's adjuvant (Sigma-Aldrich) and with the same number of bacteria in incomplete Freund's adjuvant 3 and 6 weeks later. Antibody responses were not detected following this regime, so animals were then immunized subcutaneously three times 3 weeks apart with 1010 heat-killed S. aureus bacteria emulsified in 2 ml of 5 mg/ml Quil-A-saponin (Sigma-Aldrich) in PBS, which was divided into two 1.0-ml subcutaneous inoculations given on each side of the neck. Antibody responses were detected after immunization number four and thereafter. Quil-A-saponin is used routinely in our lab to stimulate strong type 1 immune responses and protective immunity against Anaplasma marginale (9, 55, 56).
The bovine leukocyte antigen (BoLA) DRB3 alleles were determined by the PCR restriction fragment length polymorphism (PCR-RFLP) method as described previously (91). The nomenclature of bovine MHC class II genes can be found at the following websites: http://www.ebi.ac.uk/ipd/mhc/bola and http://www.projects.roslin.ac.uk/bola/bolahome.html. BoLA-DRB3-RFLP haplotypes for the cattle in this study are as follows: animal 30864, 12/16 (where 12/16 indicates RFLP haplotypes designated DRB3 12 and DRB3 16, each inherited from one parent); animal 30900, 23/27; animal 30904, 11/16; animal 30916, 3/24; and animal 30919, 10/22. During the course of this study, all animals maintained recall T cell responses, which enabled the development of S. aureus-specific short-term T cell lines. Two calves expressing different MHC-II haplotypes, 16/22 (animal 35287) and 8/23 (animal 583), and immunized with A. marginale outer membranes (OM) as described previously (55, 65) were used as a source of peripheral blood mononuclear cells (PBMC) for lymphocyte proliferation assays with S. aureus antigens as negative controls. Animals used in this study were in compliance with the Washington State University Institutional Animal Care and Use Committee.
Identification and cloning of ORFs encoding surface exposed antigens.
By employing a reverse genetics and comparative genomics approach using S. aureus Newman strain (GenBank accession number AP009351) and MRSA strains COL and USA300 as well as a bovine mastitis isolate RF122 (GenBank accession numbers CP000046, CP000255, and AJ938182.1, respectively), we identified 43 surface proteins and virulence factors from S. aureus Newman strain, a human isolate (4). The proteome data based on matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) in conjunction with immuno-proteomics were used to verify the surface exported proteins analyzed by in silico software (17, 82, 92). The protein class of open reading frames (ORFs) coding for known cytoplasmic functions was eliminated, whereas the other classes of proteins were selected for further analysis. A second screening step aimed at identifying putative proteins with a cellular localization spanning the inner membrane to outside the bacterium was applied. BLAST, FASTA, MOTIFS, FINDPATTERNS, and PSORT (http://psort.nibb.ac.jp), as well as the ProDom, Pfam, and Blocks databases, were used to predict features typical of surface-associated proteins such as transmembrane domains, leader peptides, homologies to known surface proteins, lipoprotein signature, outer membrane anchoring motifs, and host cell binding domains such as RGD. We also included antigens identified as anchorless cell wall proteins, deemed to be potentially immunogenic by subtractive proteome analysis (28) The ORFs selected were >80% identical over 80% of the length at nucleotide and amino acid levels across the four strains (Table 1). All 43 ORFs were amplified from the S. aureus Newman strain using freshly isolated genomic DNA (Qiagen) by high-fidelity polymerase (Pfu Ultra II Hot Start; Agilent Technologies). Subsequently, the PCR products were gel eluted (Qiagen) and cloned into the primary entry vector pENTR/DTOPO (Invitrogen) according to manufacturer's instructions. The cloning was directional, as dictated by the CACC overhang incorporated into the forward gene-specific primer. The reverse primer was designed with a FLAG sequence, which resulted in a carboxy-terminal FLAG epitope (Table 1). Positive colonies were identified by colony PCR using M13 forward and reverse primers. All positive clones were sequenced bidirectionally using M13 forward and reverse primers, and their integrity was confirmed by BLAST analysis. For making final expression constructs containing a carboxy-terminal FLAG and two amino-terminal epitopes (six-histidine [His6] and Xpress), the plasmid DNAs containing the 43 ORFs (pENTR/DTOPO) were isolated individually and recombined into the pEXP1-DEST vector using Clonase LR (Invitrogen). TOP 10 Escherichia coli cells transformed with individual plasmids were selected on carbenicillin (100 μg/ml)–Luria-Bertani (LBC) plates. The positive clones were sequenced, and their integrity was confirmed again using T7 forward and reverse primers. The gene-specific primers used to amplify individual ORFs are listed in Table 1.
Table 1.
Primers designed to amplify S. aureus Newman strain ORFs for protein expression
| Gene locus no. | Function | Forward primer (5′ to 3′)a | Reverse primer (5′ to 3′)b |
|---|---|---|---|
| NWMN_0161 | Putative lipoprotein | ATGGGAAAATCAAAAATTTATATCTTG | TGAATCATCTCCAAAAATTTATG |
| NWMN_0364 | Putative lipoprotein | ATGGGAAAATTAAAATCATTAGCAG | GTGATCTTGCTCACTCTTTAATAC |
| NWMN_0389 | Enterotoxin | ATGGGAAAAATGAAAAATATTGC | TGCTTTTATAACTTTGATTTC |
| NWMN_0390 | Exotoxin 8 | ATGGGAAAAATGAGAACAATTGC | CTTATCATTAGCATTATTTTG |
| NWMN_0429 | Alanine amidase | ATGGGAGTGCAAAAAAAAGTAATTGCAGC | GTGAATATATCTATAATTATTTAC |
| NWMN_0458 | Erythritol kinase | ATGGGAATATATGAAACGGCACCAG | TCCTAATAGTCTAACTAAGTAC |
| NWMN_0601 | Putative lipoprotein | ATGGGAAAAAAATTAGTACCTTTA | TTCATGCTTCCGTGTACAGTTTC |
| NWMN_0668 | Fructose PO4 kinase | ATGGGAATTTATACAGTGACTTTC | CTCCCCATCAAGTACGCTAAT |
| NWMN_0724 | LysM domain protein | ATGGGAAAAAAATCTCTTACAGTG | GTGGATGTAATTATATTTTCCTG |
| NWMN_0734 | CofD-like proteins | ATGGGAAGACAAATAAAAGTTGTAC | TTTACGTTTATCACTTGGTACG |
| NWMN_0738 | Putative protein | ATGGGATCGAATCAAAATTACG | GTTATCATTAGCATTATTTTG |
| NWMN_0753 | Putative lipoprotein | ATGGGAAAAAAAGTAATGGGGATA | TACTTCATCTAAACCACTGTGG |
| NWMN_0791 | CBS domain protein | ATGGGAGTGATCATTGCCATAATTA | TATAGATACGTTGCGATTTTTCCG |
| NWMN_0925 | LytR domain protein | ATGGGAAATAAATTTTTAAAATAC | ATTTACAACATTTTGG |
| NWMN_1067 | FPRL1 inhibitory protein | ATGGGAAAAAAAAATATCACAAAAAC | ATACCAAGTAATCGAGTCGATTTC |
| NWMN_1069 | Fibrinogen-binding protein | ATGGGAAAAAATAAATTGATAGC | TTTAACTAATCCTTGTTTTAATAC |
| NWMN_1236 | Thermonuclease | ATGGGAAAGTCAAATAAATCGCTTG | TTTACTCCAAATATTTAATTTC |
| NWMN_1398 | Putative lipoprotein | ATGGGAAAAAAATTCATTGGATCAG | TTCATCTTTACTGTGCACCCCATATTC |
| NWMN_1607 | Thiol peroxidase | ATGGGAACTGAAATAACATTCAAAG | AATATTTTTGTATGCAGCTAAAGC |
| NWMN_1623 | Glycosyl transferase | ATGGGAACGAATCAAGACAATC | ACGCTTTTTTCGATTTAACAAATG |
| NWMN_1690 | Putative lipoprotein | ATGGGAAAATTCAAAGCTATCGTTG | TTTAACTTTAGTTTCTTCAG |
| NWMN_1701 | Serine protease SplF | ATGGGAAATAAAAATATAATCATCAAAAGTATTGCAG | TTTATCTAAATTATCTGCAATG |
| NWMN_1703 | Serine protease SplD | ATGGGAAATAAAAATATAATCATCAAAAGTATTGCG | TTTATCTAAATTATCTGCAATAAATTTC |
| NWMN_1704 | Serine protease SplC | ATGGGAAATAAAAATATAGTCATTAAAAGC | TTGTTCAATGTGCTTTTGAATAAAATC |
| NWMN_1705 | Serine protease SplB | ATGGGAAACAAAAACGTAGTCATCAAG | TTTATCTATGTTTTCTGCAATG |
| NWMN_1706 | Serine protease SplA | ATGGGAAATAAAAATGTAATGGTTAAAG | TTTTTCAATATTATTTTGAATAAATTG |
| NWMN_1726 | RNAIII-activating protein | ATGGGAAAGAAACTATATACATC | TTTCATGCTTCCGTGTACAGTTTC |
| NWMN_1733 | Foldase protein | ATGGGAAAGATGATAAACAAATTAATC | TTGGCTCATGCCGGATTGTC |
| NWMN_1876 | Complement inhibitor | ATGGGAAAAATTAGAAAATCTATAC | ATATTTACTTTTTAGTGCTTC |
| NWMN_1880 | Staphylokinase | ATGGGACTCAAAAGAGGTTTATTA | TTTCTTTTCTATAATAACCTTTG |
| NWMN_1926 | β-Hemolysin | ATGGGATATCCAAACTGGGGGC | TTTACTATAGGCTTTGATTGG |
| NWMN_1999 | Transglycosylase sceD | ATGGGAAAGAAAACATTACTCGCATC | TGCAGTAACCCAATGTCCAGC |
| NWMN_2043 | Dps family protein | ATGGGAAGTAATCAACAAGATGTTG | GCTTAAGTAAGATTTAAACATC |
| NWMN_2086 | Alkaline shock protein | ATGGGAACTGTAGATAACAATAAAG | TTGTAAACCTTGTCTTTCTTGG |
| NWMN_2270 | Putative lipoprotein | ATGGGAAAAAGATTAGTTACAGGGTTAC | TTGTTGGTAGTTTGGATCAGTAAC |
| NWMN_2318 | γ-Hemolysin A | ATGGGAATTAAAAATAAAATATTAAC | CTTAGGTGTGATGCTTTTAATTTTTAC |
| NWMN_2319 | γ-Hemolysin C | ATGGGACTTAAAAATAAAATATTAAC | ATTCTGTCCTTTCACCTTGATTTC |
| NWMN_2320 | γ-Hemolysin B | ATGGGAAAAATGAATAAATTAGTC | TTTATTGTTTTCAGTTTCTTTTG |
| NWMN_2426 | Sortase A | ATGGGAAAAAATGGACAAATCG | TTTGACTTCTGTAGCTACAAAG |
| NWMN_2469 | Antigen A | ATGGGAAAAAAGACAATTATGGCATC | GAATCCCCAAGCACCTAAACCTTG |
| NWMN_2537 | Antigen B | ATGGGAAATAAAACCAGTAAAGTTTG | TTTACTTGTTTTGTATGGTGTATG |
| NWMN_2544 | Isochorismatase | ATGGGATCTCGAAAAACGGCGCTATTAG | TAAACTACTTTTCCATGACTC |
| NWMN_2579 | Putative lipoprotein | ATGGGACATAAAAAATATTTATTGC | TCTTCATAATCCCGATTCCC |
The forward primers were designed with a CACC overhang at the 5′ end to facilitate directional cloning.
The reverse primers were designed with a TCATTTGTCGTCGTCGTCTTTATAGTC overhang to incorporate a FLAG epitope and a stop codon at the 3′ end.
Expression of antigens by IVTT and bead purification.
Approximately 600 ng of each pEXP1-DEST clone was used as a template in a 50-μl in vitro transcription and translation (IVTT) reaction volume using an RTS-100 E. coli HY kit (5-Prime) to achieve expression of cell-free recombinant protein according to the manufacturer's instructions. Three separate expression experiments were performed. The IVTT protein products were purified as previously described, with slight modifications (53, 86). Briefly, 100 μl of protein G carboxylate latex microspheres that had 526 μg of protein G/ml of beads (Polysciences, Inc.) was washed in A/G buffer (0.1 M Tris-HCl, 0.15 M NaCl, pH 7.4) three times. The washed beads were bound to 300 μg of anti-FLAG monoclonal antibody (MAb) (Sigma-Aldrich) in a 300-μl volume of A/G buffer by incubation at 4°C on a continuous rocking shaker. After 2 h the anti-FLAG MAb-coupled protein G latex microspheres were pelleted at 10,000 × g for 5 min, washed three times in A/G buffer, and resuspended in 400 μl of A/G buffer. This anti-FLAG MAb-protein G bead stock solution was diluted to a concentration of 10 μg of protein G beads/ml and bound to 10 μl of each IVTT product individually in a 200-μl volume of A/G buffer in a 1.5-ml microcentrifuge tube at 4°C for 2 h on a continuous shaker. The mixture was washed three times in A/G buffer and resuspended in 200 μl of complete RPMI medium to give stock solutions of 2 and 10 μg of protein G beads/ml for proliferation assays. Protein quantity in IVTT reaction mixtures was estimated by dot blot analysis using a MAb specific for the FLAG epitope as previously described, with slight modifications (86). A HybriDot Manifold (BRL Life Technologies, Inc.) was used to spot a prewetted nitrocellulose membrane with 1 μl of IVTT product diluted in 200 μl of PBS (pH 7.0). The membranes were washed in I-Block reagent (I-Block124; Applied Biosystems) containing 0.05% Tween 20 (Bio-Rad) (referred to here as I-Block) for 1 h, with a subsequent 1 h of incubation with either anti-FLAG MAb or anti-Xpress MAb (Invitrogen) diluted 1:10,000 in blocking reagent and extensive washing. The membranes were then incubated for 1 h with alkaline phosphatase-conjugated goat anti-mouse IgG (Applied Biosystems) diluted 1:10,000 in blocking reagent and washed. Antibody binding was detected with a Western-Star reagent system (Applied Biosystems). Dot blot densitometry was analyzed using AlphaEase FC, version 4.0.0, software (Alpha Innotech) to determine relative expression of each IVTT product. As only proteins expressing the carboxy-terminal FLAG epitope were certain to be full-length, protein concentrations were based on microgram levels of the FLAG epitope on recombinant merozoite surface antigen-1 (MSA1) from the M07 strain Babesia bovis (85), as a protein standard.
Expression of recombinant proteins in E. coli.
NWMN_0601, NWMN_0364, NWMN_2086, and A. marginale control protein VirB9-2 were expressed in E. coli and purified for use in Western blot assays. The plasmids (pEXP1-DEST, expression vector backbone, encoding His6 and FLAG epitopes) containing these five individual ORFs were transformed, according to the manufacturers' specifications, into BL21(DE3) LysS One Shot chemically competent E. coli (Invitrogen). Individual colonies were selected and grown in LB broth supplemented with 100 μg/ml carbenicillin (LBC broth). To produce recombinant proteins, individual bacterial colonies containing each antigen of interest were expanded in LBC broth overnight, diluted 1:100 in fresh LBC broth, grown for 3 h at 37°C, and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 8 h. Bacteria were harvested after centrifugation at 4,000 × g for 20 min and resuspended in 6 M guanidine hydrochloride, 20 mM dibasic sodium phosphate, and 500 mM sodium chloride, pH 7.8, for 30 min and then sonicated four times at 350 W for 45 s each time. Proteins were extracted from the E. coli lysate using nickel chelating resin (ProBond purification system) following the manufacturer's protocol (Invitrogen). Recombinant proteins were purified using a ProBond system two consecutive times, followed by dialysis against PBS using Slide-A-Lyzer dialysis cassettes with a 10-kDa molecular-mass cutoff (Thermo Fisher Scientific). Protein concentrations were determined using a Quick Start Bradford assay, and proteins were frozen at −20°C.
Western blot assays.
Western blot analysis was performed to detect S. aureus-specific antibody responses and to determine titers against individual recombinant antigens. Briefly, 1010 heat-killed S. aureus bacteria were lysed in 200 μl of lysis buffer (130 μl of 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1.2% Triton X-100, 10 μl of 2 mM phenylmethylsulfonyl fluoride [PMSF], 60 μl of 2 mg/ml lysostaphin, 1 μl of 5 U/ml Benzonase) and incubated at 37°C for 1 h. The reaction was stopped by the addition of 20 μl of stop buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 20% SDS) and incubated for 5 min at 95°C. Protein concentrations were estimated with a Quick Start Bradford assay (Bio-Rad), and lysates were diluted to 1 μg/μl in PBS. Forty micrograms of lysate was added to sample loading buffer (10 μl of 1 M dithiothreitol [DTT] 50 μl of Nupage loading blue [Invitrogen]) and incubated at 95°C for 10 min. Next, 15 μl of the reaction mixture was loaded on a 4 to 12% Tris-HCl gradient for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Criterion gels; Bio-Rad) and electrophoresed under denaturing conditions for 1 h at 100 V. The proteins were transferred to 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membranes at 100 V for 30 min. The PVDF membranes were blocked for 1 h in 5% milk powder (Bio-Rad) and washed three times in TBST buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20). The pre- and postimmune sera from each animal were added to individual strips at 1:100 and 1:1,000 dilutions and incubated for 6 h and then washed with TBST buffer. Alkaline phosphatase-conjugated mouse anti-bovine IgG (1:5,000) was used as a secondary antibody, and blots were developed in a substrate solution containing nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP; Bio-Rad) for 1 to 5 min.
To determine the titers of S. aureus-specific antibodies against recombinant NWMN_0601, NWMN_0364, NWMN_2086, and A. marginale VirB9-2 as a control, proteins were boiled in SDS sample buffer and loaded at 1 μg/lane on 4 to 20% SDS-PAGE precast Tris-HCl polyacrylamide Ready Gels (Bio-Rad). One gel was stained with Coomassie brilliant blue, and the second was transferred to a nitrocellulose membrane for immunoblot analysis using anti-FLAG MAb (86). Immunoblotting was also used to determine IgG1 and IgG2 antibody titers specific for S. aureus using pre- and postimmune sera. Serum was collected before immunization and 2 weeks after each immunization and stored at −20°C until the assays were performed simultaneously on all samples. After adsorption with E. coli lysate, the serum was serially diluted up to 1:4,000 in I-Block containing 5% bovine serum albumin (BSA) and 0.5% Tween 20 (New England BioLabs) as described previously, with the following modifications (34). Individual recombinant proteins were boiled in sample buffer, and 1 μg protein/lane was electrophoresed and transferred to nitrocellulose membranes. Individual membrane strips were incubated with 1:200 dilutions of mouse anti-bovine IgG1 and mouse anti-bovine IgG2 (both from Serotec). Preimmune serum was used as a negative control. Serum from an A. marginale-infected calf (04B92) with known VirB9-2-specific IgG1 and IgG2 titers was used as a positive control to standardize the immunoblots (34, 55).
Proliferation assays.
Freshly harvested PBMC obtained before immunization and 2 weeks after the last immunization were assayed in triplicate using 2.5 × 105 viable cells/well in complete RPMI 1640 medium (Gibco) as previously described, with minor modifications (1, 34). Cells were stimulated in 96-well round-bottomed plates in a volume of 100 μl/well with 5 × 103 to 5 × 106 heat-killed S. aureus cells. T cell growth factor (TCGF) diluted 1:10 in complete RPMI 1640 medium served as a positive control, and complete RPMI medium served as a negative control. Cells were cultured for 6 days at 37°C in a 5% CO2 incubator, radiolabeled for 18 h with 0.25 μCi of [3H]thymidine (Dupont), and harvested onto glass filters; radionucleotide incorporation was measured with a beta counter.
For T cell proliferation assays using individual IVTT products, 2-week CD4 T cell lines were established as previously described, with slight modifications (53, 86). We have previously shown that the use of T cell-enriched lines enables a more sensitive detection of responses to specific proteins (53). Briefly, PBMC were isolated from S. aureus-immunized steers, plated at 4 × 106 cells/well in 24-well flat-bottom plates for 7 days with 5 × 104 heat-killed S. aureus bacteria/well, and incubated at 37°C with 5% CO2 in air. On day 7, the cells were harvested and recultured for 1 week at 0.7 × 106 cells/well with 2 × 106 cells/well of freshly harvested, irradiated (3,000 rads) autologous PBMC as a source of antigen presenting cells (APC) without antigen (resting). The cells were harvested after 7 days, washed repeatedly in complete RPMI medium, and tested for a proliferative response to individual affinity-purified S. aureus antigens (bead-bound IVTT products). Cells were cultured in triplicate wells of 96-well U-bottom plates at 3 × 104 cells/well with 2 × 105 APC and incubated at 37°C in 5% CO2 in air with medium or various concentrations of antigen. Positive controls consisted of 5 × 104 or 5 × 106 heat-killed S. aureus bacteria/well and 10% TCGF. The negative-control antigens included a no-DNA control IVTT reaction mixture incubated with beads and IVTT-expressed A. marginale VirB9-2 bound to beads (53). The IVTT antigen-beads were tested at a final concentration of 1 and 5 μg of protein G/ml in 100-μl triplicate cultures. After 3 days, the cells were radiolabeled and harvested.
For an additional negative control to rule out nonspecific proliferation to the IVTT-expressed proteins, 2-week T cell lines were derived from two A. marginale outer membrane (OM)-vaccinated cattle as described previously (65). PBMC were cultured for 1 week with 1 μg/ml A. marginale OM and rested for 1 week and then tested as described above in proliferation assays.
In some assays, the 2-week cell lines were depleted of CD4+ cells, CD8+ cells, and/or γδ T cells and cultured in 4-day proliferation assays. Depletion was accomplished by incubating cells with MAb, obtained from the Washington State Monoclonal Antibody Center, specific for bovine CD4 (ILA-11, mouse IgG2a), CD8 (7C2B, mouse IgG2a), or γδ T cells (GB21A, mouse IgG2b), at 15 μg of MAb per 107 cells, rotating at 4°C for 30 min. Cells were repeatedly washed with compete RPMI medium and magnetic cell sorting (MACS) bead buffer (Miltenyi) containing 5% bovine serum albumin (BSA). MAb-labeled cells were incubated on ice for 20 min with 80 μl of goat anti-mouse IgG magnetic microbeads (Miltenyi) per 107 cells. MAb- and bead-labeled cells were depleted by filtration through an LD magnetic column (Miltenyi) under a strong magnetic field, and washed with 3 ml of MACS buffer containing BSA. Depletion was determined by flow cytometry after cells were stained with MAbs ILA-11 (CD4), 7C2B (CD8), and GB21A (γδ) or with isotype-matched negative control MAbs COLIS205B (IgG2a) and COLIS169A (IgG2b) and a 1:10,000 dilution of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories). The cells were assayed for proliferation to antigen as described above for cell lines and PBMC.
Proliferation was expressed as either mean counts per minute (cpm) ± 1 standard deviation (SD) for triplicate cultures or as a stimulation index (SI), determined as the mean cpm of cells cultured with S. aureus antigen divided by the mean cpm of cells cultured with negative-control antigen.
Detection of IFN-γ in supernatants of T cell cultures.
S. aureus-specific 2-week cell lines were cultured for 3 days with APC (2 × 106 cells per ml) and either 1 × 106 heat-killed S. aureus bacteria per well or 1 and 5 μg of protein G per ml of bead-bound IVTT proteins, and supernatants were tested for gamma interferon (IFN-γ) using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Bovigam; CSL Ltd.,) according to the manufacturer's protocol. The IFN-γ activity in culture supernatants, diluted 1:2, was determined by comparison with a standard curve obtained with a recombinant bovine IFN-γ as described previously (49). The results are presented as units of IFN-γ per ml of supernatant.
Statistical analyses.
T cell proliferation and IFN-γ secretion to S. aureus antigens were compared with negative-control antigens VirB9-2 beads and no-DNA IVTT beads using a Student's one-tailed t test or a one-way analysis of variance (ANOVA) with Dunnett's posttest for multiple comparisons (P < 0.05), as indicated (see Table 2 and Fig. 3 and 4). In addition, proliferative responses to IVTT-expressed antigens were considered to be biologically relevant if the SI was ≥2.0 and if the mean cpm was ≥1,000.
Table 2.
Summary of antigens that induced significant T cell proliferation in three experiments using 2-week cell lines from S. aureus-immunized calves
| Expt no. | Proliferative response of T cell lines to the indicated antigen(s) by animala |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 30864 (12/16)b |
30900 (23/27) |
30904 (11/16) |
30916 (3/24) |
30919 (10/22) |
||||||
| Antigen | SI | Antigen | SI | Antigen | SI | Antigen | SI | Antigen | SI | |
| 1 | NWMN_0364 | 25.7c | NWMN_2086 | 9.6c | NWNM_0601 | 38.2c | NWMN_0601 | 2.3c | NWMN_2086 | 8.3c |
| NWMN_0601 | 24.4c | NWMN_0364 | 2.4 | NWMN_1999 | 4.3 | |||||
| 2 | NWMN_0364 | 46.7c | NWMN_2086 | 17.2c | NWMN_0601 | 6.7c | NWMN_0601 | 5.3c | NWMN_2086 | 9.3c |
| NWMN_0601 | 20.1c | NWMN_0601 | 3.0d | NWMN_0429 | 4.4 | NWMN_2086 | 2.2d | NWMN_2469 | 2.6d | |
| NWMN_0161 | 25.3 | NWMN_2270 | 2.3d | |||||||
| NWMN_0390 | 10.4 | NWMN_2318 | 7.8d | |||||||
| NWMN_2319 | 7.2d | |||||||||
| NWMN_2320 | 2.2d | |||||||||
| NWMN_2469 | 9.6d | |||||||||
| 3 | NWMN_0364 | 38.9c | NWNM_2086 | 4.2c | NWMN_0601 | 2.7c | NWMN_0601 | 18.9c | NWMN_2086 | 6.0c |
| NWMN_0601 | 19.3c | NWMN_0601 | 2.4d | NWMN_1733 | 2.7 | NWMN_2086 | 12.5d | NWMN_2469 | 13.7d | |
| NWMN_2086 | 9.4 | NWNM_2320 | 4.7 | NWMN_2270 | 12.8d | NWMN_2320 | 5.3 | |||
| NWMN_2318 | 29.3d | |||||||||
| NWMN_2319 | 24.0d | |||||||||
| NWMN_2320 | 16.2d | |||||||||
| NWMN_2469 | 34.5d | |||||||||
Different batches of IVTT-expressed proteins were tested in each experiment. Antigens were determined to induce significant responses based on the following criteria: mean cpm of >1,000, SI of >2.0, and P of <0.05 compared to medium, beads, and VirB9 beads. Significant proliferation was determined using ANOVA with Dunnett's posttest for multiple comparisons (P < 0.05). Assays were performed using 43 antigens tested in triplicate from three expression experiments. SI was determined as the mean cpm in response to 1 or 5 μg/ml S. aureus antigen/mean cpm in response to Virb9 beads, whichever was higher.
MHC class II haplotypes were determined by RFLP analysis of exon II from the DRB3 locus. For example, 12/16 refers to the RFLP haplotypes designated DRB3 12 and DRB3 16, each inherited from one parent.
Antigens that induced significant proliferation in three independent assays for each animal.
Antigens that induced significant proliferation in two independent assays for each animal.
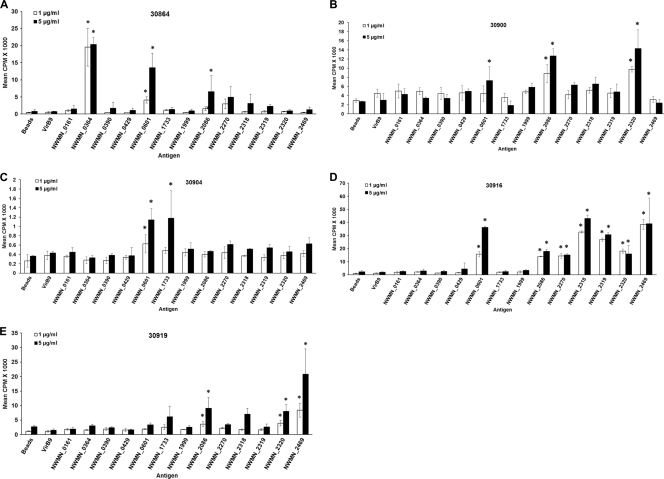
Fig 3.
Antigens that induced significant proliferation for one or more calves. Proliferation assays were performed with 2-week T cell lines from the immunized calves using 1 and 5 μg/ml protein-G bead-bound IVTT-expressed S. aureus proteins. Negative-control antigens include IVTT-expressed A. marginale VirB9-2 beads and no-DNA control beads. Data are presented from one screening assay for each animal. Responses were considered significant (indicated by asterisks) if mean the cpm was >1,000, the SI compared with negative-control antigens was >2.0, and P was <0.05 compared to negative-control antigens and medium (using ANOVA with Dunnett's posttest for multiple comparisons).
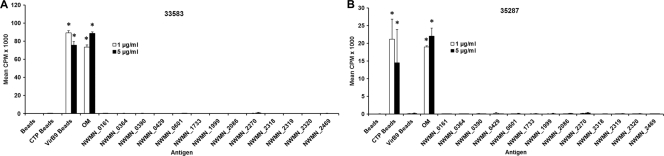
Fig 4.
S. aureus antigens do not induce significant proliferation of 2-week T cell lines from A. marginale-immunized cattle. S. aureus proteins were tested on 2-week T cell lines from cattle immunized with A. marginale OM: animal 35287 that expresses the DRB3 RFLP haplotype 16/22 (A) and animal 33583 that expresses the DRB3 RFLP haplotype 8/23 (B). A. marginale OM and IVTT-expressed VirB9-1 (CTP) and VirB9-2 (VirBa) bound to protein G beads were used as positive controls. Responses were considered significant (indicated by asterisks) if the mean cpm was >1,000, the SI compared with negative-control antigen was >2.0, and P was <0.05 compared to negative-control antigens and medium (using ANOVA with Dunnett's posttest for multiple comparisons).
RESULTS
Antibody and PBMC responses from calves immunized with S. aureus Newman strain.
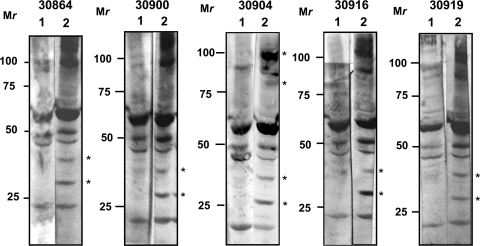
The current study was designed to screen and test the immunogenicity of selected S. aureus proteins that could be included in a potential vaccine in cattle that might translate to use in humans. Five age-matched Holstein steers with different MHC class II haplotypes were immunized with S. aureus until S. aureus-specific immune responses were obtained. Responses were first observed after the fourth immunization. Western blot analysis of S. aureus whole-cell lysates showed the presence of bands specific for S. aureus in all five immune sera obtained 2 weeks after the fourth immunization (Fig. 1). Bands observed in postimmunization sera that were not visible in preimmunization sera include bands at the approximate molecular masses of 80 and 100 kDa (animal 30900) and 30 and 40 kDa (all animals). In addition, the ∼100-kDa band identified by preimmunization sera was more intense with postimmunization sera (animals 30864, 30900, 30916, and 30919). The presence of the same bands identified by both pre- and postimmunization sera indicates that the cattle may have been exposed to S. aureus or other bacteria that share antigenic epitopes with S. aureus before the animals were immunized. The identities of these proteins were not determined.
Fig 1.
Immunoblotting to detect S. aureus-specific antibody responses. Lysate (5 μg/lane) from heat-killed S. aureus was loaded and probed with preimmune (lane 1) and postimmune (lane 2) bovine serum obtained after the fourth immunization and diluted 1:100 or 1:1,000. Alkaline phosphatase-conjugated mouse anti-bovine IgG (1:5,000) was used as a secondary antibody, and blots were developed in a substrate solution containing NBT/BCIP for 1 to 5 min. Asterisks indicate unique bands which appeared after the fourth immunization. Mr, relative molecular weight in thousands.
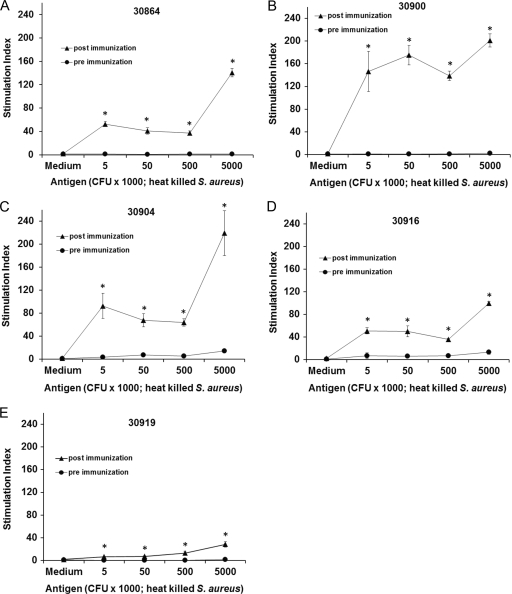
Lymphocyte responses from all five calves were determined by proliferation assays using PBMC obtained before and 2 weeks after the last immunization and heat-killed S. aureus as antigen. The postimmunization PBMC had a significant proliferative response compared to preimmunization PBMC (Fig. 2A to E). Furthermore, the postimmunization PBMC from all calves responded in a concentration-dependent manner to heat-killed S. aureus. As we typically observe for outbred cattle (53, 54), there is considerable variation in the levels of immune response between animals.
Fig 2.
Proliferation assays on pre- and postimmune PBMC to confirm S. aureus-specific T cell responses. Heat-killed S. aureus cells were used as the antigen at the indicated concentrations. TCGF (interleukin-2) was used as a positive control. The assay was performed with the indicated PBMC obtained before immunization and 2 weeks after the last immunization. Responses are presented as the SI compared to the negative control (RPMI medium) ± 1 SD for triplicate cultures. Responses that were significantly greater than medium are indicated with asterisks where P is <0.05 using a one-tailed Student's t test.
Identification of S. aureus antigens that induced significant T cell and IgG responses.
The 43 antigen-encoding ORFs were expressed by IVTT, and their expression was confirmed by dot blot analysis using carboxy-terminal FLAG and amino-terminal anti-Xpress MAbs. The amount of protein expressed by IVTT varied between ∼16.0 and 36.5 μg/ml, compared with recombinant B. bovis MSA1 (expressing a FLAG epitope) by dot blot densitometry. All 43 antigens from S. aureus Newman strain were tested with 2-week T cell lines from calves immunized with heat-killed S. aureus. Before testing individual antigens, we compared the proliferative response of 2-week T cell lines and freshly obtained PBMC to heat-killed S. aureus cells. As observed previously with A. marginale proteins (53), the T cell lines had a higher SI to S. aureus than fresh PBMC (data not shown). Therefore, 2-week T cell lines were used to assay all 43 IVTT-expressed proteins. The T cells were cultured with IVTT-expressed proteins bound to protein G beads via anti-FLAG MAb and tested at 1 and 5 μg/ml protein G beads (based on concentration of protein G on the beads). The amount of FLAG-specific MAb bound to protein G beads ranged between 90 and 105 μg/ml. This was determined by comparing the concentrations of FLAG-specific MAb in the supernatants before and after binding.
The 43 proteins were expressed by IVTT three separate times, but they did not express uniformly each time. During the first round of expression and screening, antigens NWMN_2043 and NWMN_2579 could be detected only when 3 μl, instead of 1 μl, of the IVTT mixture was blotted onto the membranes. However, all of the proteins were comparably expressed during the second round of screening. During the third round of expression and screening, NWMN_1607, NWMN_1726, NWMN_1733, NWMN_1926, NWMN_1999, NWMN_2043, and NWMN_2086 were expressed at <16 μg/ml, but two of these bead-bound proteins (NWMN_1733 and NWMN_2086) were sufficient to elicit T cell proliferation in one or more calves (Table 2). Figure 3 provides examples of assays in which S. aureus proteins induced significant proliferation of 2-week T cell lines from one or more calves (30864, 30900, 30904, 30916, and 30919) in one or more of the assays. Negative controls included A. marginale VirB9-2 beads (expressed in the same way using IVTT) and no-DNA control beads. In contrast, the S. aureus proteins did not induce significant T cell proliferation of 2-week cell lines derived from A. marginale OM-immunized cattle 33583 and 35287 (Fig. 4A and B). These cell lines did respond to A. marginale OM and responded variably to type 4 secretion system proteins VirB9-1 (CTP) and VirB9-2 (VirBa) expressed by IVTT and bound to protein G beads, as we have shown previously (53, 65, 86).
The cumulative results of three rounds of expression and screening identified 13 T cell antigens, two of which induce serum antibodies in humans (NWMN_2318 [γ-hemolysin A] and NWMN_2469 [antigen A]) (11–13). However, the remaining 11 proteins (NWMN_0161 [putative lipoprotein], NWMN_0364 [putative lipoprotein], NWMN_0390 [exotoxin 8], NWMN_0429 [alanine amidase], NWMN_0601 [putative lipoprotein], NWMN_1733 [foldase protein], NWMN_1999 [transglycosylase], NWMN_2086 [alkaline shock protein], NWMN_2270 [putative lipoprotein], NWMN_2319 [γ-hemolysin C], and NWMN_2320 [γ-hemolysin B]) were not previously identified as antigenic or used as a vaccine (Table 2). The 13 T cell antigens include four putative lipoproteins, three hemolysins, various enzymes, and an alkaline shock protein. Three antigens were designated the “best” in these experiments (NWMN_0601, NWMN_0364, and NWMN_2086) because they induced significant T cell proliferation in three independent assays in at least one animal and stimulated responses from two or more animals that expressed different MHC class II haplotypes (Table 2). Antigens NWMN_0601 and NWMN_2086 each stimulated T cell responses from four cattle, and antigen NWMN_0364 stimulated responses from two cattle. Furthermore, only two cattle shared one MHC class II haplotype; animals 30864 (12/16) and 30904 (11/16) have the DRB3 RFLP haplotype 16. Thus, the finding that these three antigens stimulate recall responses from two to four animals with completely different haplotypes and the fact that these haplotypes are common among North American Holstein cattle (65, 77) suggest that these are good candidates for inclusion in a multiantigen vaccine that would be broadly protective in Holstein cattle. Two of these antigens are putative lipoproteins, and one is the alkaline shock protein.
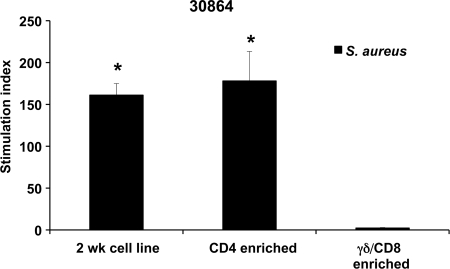
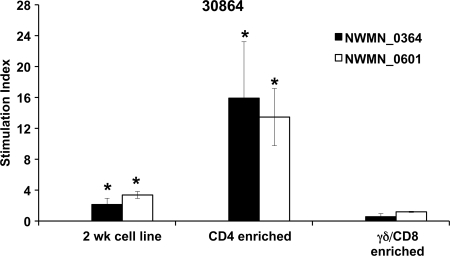
Antigen-specific lymphocytes in 2-week T cell lines were further characterized by depleting CD4+ cells or γδ and CD8+ cells and repeating proliferation assays. Flow cytometric analysis indicated that the 2-week T cell lines from all five steers contained 21 to 46% CD4+ cells, 31 to 36% γδ T cells, and 7 to 11% CD8+ cells. Following enrichment, CD4+ T cells were increased to 94 to 96% of total T cells, with only 4 to 6% γδ and CD8 T cells remaining. Similarly, upon enrichment for γδ and CD8+ cells, these increased to 95 to 96% of total T cells, while CD4+ T cells fell to 4 to 5%. CD4-enriched and γδ- and CD8-enriched cell populations were then tested for proliferation to S. aureus and two IVTT-expressed proteins. The CD4-enriched 2-week cell lines from all three animals showed significant proliferation against S. aureus compared to γδ- and CD8-enriched T cells in this assay (representative data are shown in Fig. 5). Furthermore, when tested against IVTT-expressed proteins from the third round of expression, CD4-enriched T cells from animal 30919 showed significant responses to antigens NWMN_2086, NWMN_1999, NWMN_2318, and NWMN_2469; those from animal 30904 responded significantly to NWMN_1733, and those from animal 30864 responded significantly to NWMN_0364 and NWMN_0601 (data from 30864 are shown in Fig. 6). The γδ T cell- and CD8-enriched populations had insignificant proliferation to heat-killed S. aureus and all of the proteins tested. This indicates that the antigen-specific lymphocytes obtained following S. aureus immunization were CD4 T cells. Enriching for CD4+ T cells increased the response of the cell lines to the individual proteins compared with untreated T cell lines, just as we have shown previously that T cell lines are more sensitive than PBMC to detect IVTT-expressed proteins (53).
Fig 5.
The cells responding to S. aureus immunization are predominantly CD4 T cells. Two-week cell lines were undepleted or depleted of CD4+ cells or CD8+ cells and γδ T cells. Heat-killed S. aureus (5 × 106 bacteria/well) was used as the antigen. Responses are presented as the SI compared to medium ± 1 SD for triplicate cultures. Significant responses of each cell population are indicated with asterisks, where P is <0.05 using a one-tailed Student's t test.
Fig 6.
Response by enriched CD4 T cells to two S. aureus proteins. Two-week cell lines from animal 30865 were depleted of CD4+ cells or CD8+ cells and γδ T cells. Antigens NWMN_0364 and NWMN_0601 were used at 1 μg/ml in proliferation assays with undepleted 2-week cell lines, CD4-enriched cells, and γδ- and CD8+-enriched cells. Responses are presented as the SI compared to medium ± 1 SD for triplicate cultures. Significant responses of each cell population are indicated with asterisks, where P is <0.05 using a one-tailed Student's t test.
Secretion of IFN-γ is a hallmark of the Th1-type response, and IFN-γ induces opsonizing antibodies and activates macrophages to kill intracellular bacteria and therefore likely plays an important role in immunity to S. aureus (5, 30, 38, 62, 70). The 13 IVTT-expressed S. aureus proteins shown to stimulate T cell proliferation were therefore tested, in comparison with medium and negative-control VirB9-2-beads, for inducing secretion of IFN-γ using 2-week T cell lines. Eight of the 13 proteins induced IFN-γ secretion in T cell lines from one or more calves (Table 3). The three best antigens, NWMN_0601, NWMN_0364, and NWMN_2086, induced high levels of IFN-γ in culture, ranging from 206 to 429 U/ml. Antigens NWMN_1733, NWMN_2270, NWMN_2318, NWMN_2319, and NWMN_2469 induced generally lower levels of IFN-γ, ranging from 44 to 219 U/ml. Animal 30919 had a high background response to the negative-control VirB9 beads so none of the responses to S. aureus proteins were significant.
Table 3.
IFN-γ induced by S. aureus proteins
| Culture condition | Amt of antigen (μg/ml) | IFN-γ secretion (U/ml) by 2-week cell lines from the indicated animala |
||||
|---|---|---|---|---|---|---|
| 30864 | 30900 | 30904 | 30916 | 30919 | ||
| Medium | 1.0 | 40.0 | 1.0 | 1.0 | 6.4 | |
| S. aureus cells | 429.6 | 298.0 | 596.2 | 111.5 | 294.3 | |
| TCGF | 415.1 | 235.0 | 171.1 | 126.1 | 239.8 | |
| IVTT-expressed protein | ||||||
| VirB9 | 1 | 2.0 | 33.0 | 7.0 | 12.3 | 190.4 |
| 5 | 8.1 | 60.0 | 24.0 | 21.8 | 251.2 | |
| NWNM_0161 | 1 | 16.1 | ND | 126.5 | ND | ND |
| 5 | 28.5 | ND | 29.8 | ND | ND | |
| NWMN_0364 | 1 | 325.5* | ND | ND | ND | 225.0 |
| 5 | 401.1* | ND | ND | ND | 219.4 | |
| NWMN_0390 | 1 | 22.5 | ND | ND | ND | ND |
| 5 | 30.7 | ND | ND | ND | ||
| NWMN_0429 | 1 | 29.5 | 33.4 | 0.6 | ND | ND |
| 5 | 16.1 | 33.0 | 1.0 | ND | ND | |
| NWMN_0601 | 1 | 381.9* | 25.1 | 19.8 | 39.1 | ND |
| 5 | 429.2* | 43.2 | 11.3 | 90.3* | ND | |
| NWMN_0724 | 1 | 2.08 | ND | 34.6 | ND | ND |
| 5 | 17.4 | ND | 15.1 | ND | ND | |
| NWMN_1733 | 1 | 1 | ND | 114.5* | ND | 139.8 |
| 5 | 17.68 | ND | 80.0* | ND | 196.5 | |
| NWMN_1880 | 1 | ND | ND | ND | 12.4 | ND |
| 5 | ND | ND | ND | 24.6 | ND | |
| NWMN_1999 | 1 | ND | ND | ND | ND | 168.4 |
| 5 | ND | ND | ND | ND | 211.1 | |
| NWMN_2086 | 1 | 17.1 | 206.3* | ND | 13.4 | 179.8 |
| 5 | 49.7 | 198.0* | ND | 40.4 | 211.2 | |
| NWMN_2270 | 1 | 67.7* | 55.4 | ND | 11.7 | ND |
| 5 | 99.9 | 36.4 | ND | 25.6 | ND | |
| NWMN_2318 | 1 | ND | 66.4 | ND | 27.5 | 258.8 |
| 5 | ND | 87.4 | ND | 53.1* | 273.7 | |
| NWMN_2319 | 1 | 36.8 | 34.1 | ND | 26.3 | ND |
| 5 | 73.5* | 26.4* | ND | 44.4* | ND | |
| NWMN_2320 | 1 | 22.8 | 87.1 | ND | 10.9 | 220.1 |
| 5 | 45.3 | 108.2 | ND | 20.3 | 213.5 | |
| NWMN_2469 | 1 | 39.8 | ND | ND | 145.9* | 212.8 |
| 5 | 63.8 | ND | ND | 219.4* | 206.5 | |
| NWMN_2579 | 1 | 22.8 | 41.8 | ND | 11.4 | ND |
| 5 | 45.3 | 44.6 | ND | 60.7 | ND | |
Data are presented as the mean U/ml of IFN-γ secreted by 2-week T cell lines cultured in triplicate with 1 × 106 heat-killed S. aureus bacteria or the indicated antigens. Boldface numbers indicate significant (P < 0.05) IFN-γ secretion with respect to medium and bead-bound VirB9. Asterisks (*) indicate antigens that induced significant proliferation. ND, not determined.
IgG titers specific for the three best T cell antigens in S. aureus-vaccinated calves were determined using affinity-purified recombinant proteins. Animal 04B92 immunized with A. marginale OM had a titer (both IgG1 and IgG2) of at least 4,000 against VirB9-2, and this serum served as a positive control (54). Recombinant S. aureus proteins NWMN_0601, NWMN_0364, and NWMN_2086 were expressed in E. coli and purified, and their expression was confirmed by immunoblotting with an anti-FLAG MAb (data not shown). All five animals had positive antibody responses to all three recombinant proteins. IgG1 titers were at least 10 times higher than IgG2 titers in most of the animals for all three antigens (Table 4).
Table 4.
Titers of IgG1 and IgG2 to recombinant proteins in cattle immunized with S. aureus
| Animal |
S. aureus-specific IgG titer to the indicated antigena |
|||||
|---|---|---|---|---|---|---|
| rNWMN_0364 |
rNWMN_0601 |
rNWMN_2086 |
||||
| IgG1 | IgG2 | IgG1 | IgG2 | IgG1 | IgG2 | |
| 30864 | 4,000 | 400 | 4,000 | 400 | 4,000 | 400 |
| 30900 | 4,000 | 400 | 4,000 | 400 | 4,000 | <400b |
| 30904 | 4,000 | 400 | 4,000 | 400 | 4,000 | <400 |
| 30916 | 4,000 | 400 | 4,000 | 400 | 4,000 | <400 |
| 30919 | 4,000 | 400 | 4,000 | 400 | 4,000 | <400 |
Serum obtained 2 weeks after the last immunization were adsorbed with E. coli lysate, diluted 1:400 to 1:4,000, and tested for reactivity to recombinant protein (indicated by the “r” prefix) by immunoblotting. The titer is defined as the reciprocal of the highest serum dilution giving a positive signal.
No specific bands were detected at a 1:400 dilution, and no other dilutions were tested.
DISCUSSION
Identifying candidate vaccine antigens for S. aureus has been problematic due to its complex and plastic genetic machinery, and protection using a single-component vaccine has been elusive to date (52). Identification of potential vaccine antigens using patient infection serum is a common strategy (19, 94) although the usefulness of this strategy for S. aureus has not been proven (16). The failure of bivalent capsular polysaccharide type 5 (CP5) and CP8 conjugate vaccine, StaphVAX, in human clinical trials can be partly explained by the fact that S. aureus switches to an acapsular phenotype and can still be lethal (18, 63, 66). Various vaccination trials have shown that partial protection against S. aureus challenge in mice can be achieved by using S. aureus surface carbohydrates such as poly-N-acetylglucosamine or poly-N-succinyl glucosamine or individual surface proteins such as clumping factor A or B (ClfA or ClfB, respectively), iron-regulated surface determinant B (IsdB), truncated fibronectin-binding protein (FnBP), live attenuated S. aureus, or a combination of FnBP and ClfA (3, 29, 44, 57, 60, 67, 73–75, 84, 89). Other vaccines that are currently under different stages of clinical trials by various pharmaceutical companies include CP5 and/or CP8, combined with alpha toxin and Panton-Valentine leukocidin (PVL), whole-cell bacteria, and proteins such as IsdB, siderophore receptors, porins, secreted proteins, and enterotoxin B (16). However, none of these antigens has been tested for induction of CD4 T cell responses in animals or humans (16). These approaches raise serious concerns regarding immune protection and safety since an IsdB-based vaccine (V710) was abruptly terminated during a recent phase II/III clinical trial in humans (61).
Although extensive immunization trials in mice have revealed protective S. aureus antigens (3, 20–23, 36, 39, 41, 44, 57, 60, 67, 74, 75, 78, 79, 84, 89), these have not translated to a vaccine for cattle or humans. One promising approach is the use of a mutated SpA protein (KKAA) which has amino acid substitutions incorporated into each of the five Ig-binding domains of protein A. This protein A mutant can no longer bind to IgG and interfere with antibody function and does not induce B cell apoptosis (43). Immunization of mice with this protein elicited antibody that neutralized the native protein A molecule and provided some protection in mice against virulent MRSA (43). However, there are limitations with the mouse model. S. aureus is highly adapted to humans, with virulence factors more potent in humans than in mice (87, 90, 96, 98). Additionally, inbred mice are not optimal animal models for identifying antigens from S. aureus because of MHC differences. Cattle are evolutionarily more closely related to humans than are mice, are outbred, and have polymorphic MHC-II alleles similar to those of humans. Although the class II DP, DO, DM, DQ, and DR genes are generally conserved among mice, humans, and cattle, there are some important differences (45). In mice the class II genes have undergone contraction, and about half of the strains lack functional I-E (DR orthologue) class II molecules, whereas cattle and humans have the same number of functional DR and DQ genes, and both sets of molecules present antigen to CD4 T cells (27, 50, 88). Therefore, cattle with different haplotypes would be more likely than mice to identify a wide range of S. aureus antigens that would be relevant for vaccine design. Furthermore, it is very likely that antigens discovered here could be useful for vaccine development against human MRSA. Both S. aureus γ-hemolysin A and antigen A, identified by T cells in this study (Table 2), induced serum antibody responses in humans (11–13), and recognition of the same proteins by cattle and human T cells has been shown for other bacterial pathogens, such as mycobacterial ESAT-6 and Ag85 (93).
One significant omission from the mouse and human studies is the paucity of known S. aureus CD4 T cell antigens, making it difficult to design efficacious multivalent vaccines against human and animal infections. To address this problem, we used cattle as a large outbred animal model to screen S. aureus antigens, selected using reverse genetics, to identify those antigens that elicited recall CD4 T cell responses. The S. aureus Newman strain was chosen because it displays robust virulence properties in animal models and has been extensively analyzed for its molecular traits of staphylococcal pathogenesis (4). Furthermore, this strain also shares high homology (>80% amino acid identity) within the 43 full-length antigen-encoding ORFs found in IMI and MRSA strains, which will increase the potential for stimulating cross-reactive immune responses for these strains. Notably, genes encoding staphylokinase (sak, NWMN_1880) and staphylococcal complement inhibitor (scin, NWMN_1876) were absent in S. aureus RF122 and COL strains. Analysis of 58 S. aureus genomes from 14 different clonal complex lineages revealed that 24 surface proteins involved in adhesion and 13 secreted proteins implicated in immune evasion are located on the stable core genome (28, 58). Eight of these 37 surface-exposed or secreted proteins that had ORFs of ≤1.2 kb (an optimal size) were included for expression by IVTT. These are β-hemolysin (NWMN_ 1926), fibrinogen-binding protein (NWMN_1069), staphylokinase (NWMN_1880), exotoxin (NWMN_0390), enterotoxin (NWMN_0389), sortase (NWMN_2426), antigen A (NWMN_2469), and antigen B (NWMN_2537) (28, 58). We also identified antigens through our analysis which included eight surface-exposed lipoproteins (NWMN_0161, NWMN_0364, NWMN_0601, NWMN_0753, NWMN_1398, NWMN_1609, NWMN_1690, and NWMN_2579), five proteins encoded by the serine protease operon splFDCBA located in the pathogenicity island (NWMN_1701, NWMN_1703, NWMN_ 1704, NWMN_1705, and NWMN_1706), global transcription regulator RNAIII activating protein (NWMN_1726), and an alkaline shock protein (NWMN_2086). The complete list of 43 antigens included in the initial screening to determine T cell immunogenicity is summarized in Table 1.
Over one-fourth of the candidate antigens were immunogenic for one or more cattle, and specificity was shown by lack of stimulation of T cells from control A. marginale-immune cattle that shared MHC class II alleles with the S. aureus-immune cattle. Furthermore, eight of the S. aureus antigens (NWMN_0364, NWMN_0601, NWMN_1733, NWMN_2086, NWMN_2318, NWMN_2319, NWMN_2270, and NWMN_2469) induced IFN-γ secretion, the signature Th1 cytokine. Five antigens (NWMN_0161, NWMN_0390, NWMN_0429, NWMN_1999, and NWMN_2320) that failed to induce T cell proliferation also failed to induce IFN-γ in this assay. In cattle, IFN-γ produced by antigen-activated T cells can correlate with T cell proliferation (8).
Five of the 13 T-cell antigens we identified are lipoproteins. The three best antigens were an alkaline shock protein (NWMN_2086) and two lipoproteins (NWMN_0364 and NWMN_0601). Although these have not been previously described as immunogenic, their immunogenicity is not surprising since they are secreted and are involved in a variety of extracellular functions. The alkaline shock protein imparts pH tolerance during bacterial infection by maintaining an optimal pH (7.0 to 7.5) during the initial phase of staphylococcal colonization and probably functions as an accessory virulence factor during infection (47). Lipoproteins are an integral part of bacterial peripheral membrane and perform diverse functions, including adhesion, antibiotic resistance and prevention of phage superinfection, transportation, and secretion of enzymes and redox and sensory processes, and they serve as important virulence factors (40). Lipoproteins can also function as pathogen-associated molecular patterns, activating Toll-like receptor 2 (TLR2) signaling cascades, which can lead to increased pathology (10, 83). Putative lipoprotein, NWMN_0601 (mntC) belongs to manganese transport regulator operon, mntABCD (71) and plays a critical role in maintaining Mn2+ homeostasis during infection. Therefore, mntC may serve as a component of a new multivalent vaccine against S. aureus along with NWMN_2086 and NWMN_0364.
Previously identified antigens staphylokinase (NWMN_1880) (12), immunodominant antigen A (NWMN_2469) (11, 28), and transglycosylase sceD (NWMN_ 1999) (28) were also immunogenic in our T cell assays, but staphylokinase failed to induce IFN-γ. However, of the previously reported vaccine antigens that we tested, including fibrinogen-binding protein (NWMN_1069), β-hemolysin (NWMN_1926), staphylococcal complement inhibitor (NWMN_1876), and sortase (NWMN_2426), none was immunogenic in our T cell assays, which may explain their failure to afford protective immunity.
The predominant T cell population responding in proliferation assays is CD4 T lymphocytes. Targeting CD4 T cell antigens is important because CD4 T cells are required for T cell-dependent immunoglobulin isotype switching to produce high-affinity, opsonizing IgG antibodies and for IFN-γ-mediated phagocyte activation (7, 56, 95), effectors important for protective immunity against intracellular bacterial pathogens (5, 30, 38, 62, 70). In mice it was shown that a Th1 response is also important for inducing optimal IgG for opsonization and complement-mediated lysis against S. aureus (30). Thus, in the context of appropriate adjuvants, antigens that stimulate IFN-γ and IgG production would be good choices for vaccine development. The three best T cell antigens were also recognized by IgG1 from all five immune animals, and two of the three antigens were recognized by IgG2 although IgG1 titers were 10 times higher than IgG2. The consequence of this response regarding protection against S. aureus in cattle immunized with killed bacteria is unknown. However, higher titers of IgG1 and IgG2 could be induced using recombinant proteins administered with adjuvant such as CpG oligodeoxynucleotide or RIBI adjuvant (a mixture of monophosphoryl lipid A, trehalose dimycolate, and cell wall skeleton), which have been shown to stimulate these isotypes in cattle (68, 97). Nevertheless, both IgG1 and IgG2 have been shown to be opsonins and to activate macrophage-mediated phagocytosis in humans and cattle (6, 59).
In summary, we identified 13 S. aureus antigens that induce CD4 T cell responses in immunized cattle. Eight antigens, including the top three antigens that elicit IgG and IFN-γ responses, are good candidates for testing in vaccination-challenge trials. These antigens may prove useful as broadly cross-protective components of multivalent vaccine against IMI in cattle and MRSA in humans.
ACKNOWLEDGMENTS
We thank Emma Karel, Kathleen White, and Shelley Whidbee for assistance with the cattle and cell culture and Paul Beare and Robert Heinzen, NIAID, NIH Rocky Mountain Laboratories, for helping with the IVTT expression system.
This research was funded through a grant provided by Sanofi Pasteur, France.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Abbott JR, Palmer GH, Howard CJ, Hope JC, Brown WC. 2004. Anaplasma marginale major surface protein 2 CD4 T cell epitopes are evenly distributed in conserved and hypervariable regions (HVR), whereas linear B-cell epitopes are predominantly located in the HVR. Infect. Immun. 72:7360–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almeida RA, Matthews KR, Cifrian E, Guidry AJ, Oliver SP. 1996. Staphylococcus aureus invasion of bovine mammary epithelial cells. J. Dairy Sci. 79:1021–1026 [DOI] [PubMed] [Google Scholar]
- 3. Arrecubieta C, et al. 2008. Vaccination with clumping factor A and fibronectin binding protein A to prevent Staphylococcus aureus infection of an aortic patch in mice. J. Infect. Dis. 198:571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belardelli F. 1995. Role of interferons and other cytokines in the regulation of the immune response. APMIS 103:161–179 [DOI] [PubMed] [Google Scholar]
- 6. Bergius RG, et al. 1993. Phagocytosis of Staphylococcus aureus and Haemophilus influenzae type B opsonized with polyclonal human IgG1 and IgG2 antibodies. Functional hFc gamma RIIa polymorphism to IgG2. J. Immunol. 151:1463–1472 [PubMed] [Google Scholar]
- 7. Bradley LM, Dalton DK, Croft M. 1996. A direct role for IFN-γ in regulation of Th1 cell development. J. Immunol. 157:1350–1358 [PubMed] [Google Scholar]
- 8. Brown WC, et al. 2001. Highly conserved regions of the immunodominant major surface protein 2 of the genogroup II ehrlichial pathogen Anaplasma marginale are rich in naturally derived CD4 T lymphocyte epitopes that elicit strong recall responses. J. Immunol. 166:1114–1124 [DOI] [PubMed] [Google Scholar]
- 9. Brown WC, et al. 1998. CD4+ T lymphocyte and IgG2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 103:13831–13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke SRK, et al. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J. Infect. Dis. 193:1098–1108 [DOI] [PubMed] [Google Scholar]
- 12. Collen D, Stockx L, Lacroix H, Suy R, Vanderschueren S. 1997. Recombinant staphylokinase variants with altered immunoreactivity. IV: Identification of variants with reduced antibody induction but intact potency. Circulation 95:463–472 [DOI] [PubMed] [Google Scholar]
- 13. Croze M, et al. 2009. Serum antibodies against Panton-Valentine leukocidin in a normal population and during Staphylococcus aureus infection. Clin. Microbiol. Infect. 15:144–148 [DOI] [PubMed] [Google Scholar]
- 14. Daum RS, Spellberg B. 2012. Progress toward a Staphylococcal aureus vaccine. Clin. Infect. Dis. 54:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Neeling AJ, et al. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 16. Deresinski S, Herrera V. 2010. Immunotherapies for Staphylococcus aureus: current challenges and future prospects. Infect. Control Hosp. Epidemiol. 31(Suppl 1:S45–S47 [DOI] [PubMed] [Google Scholar]
- 17. Dreisbach A, et al. 2010. Profiling the surfacome of Staphylococcus aureus. Proteomics 10:3082–3096 [DOI] [PubMed] [Google Scholar]
- 18. Esen N, Wagoner G, Philips N. 2010. Evaluation of capsular and acapsular strains of S. aureus in an experimental brain abscess model. J. Neuroimmunol. 218:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Etz H, et al. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 99:6573–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fattom A, et al. 1990. Synthesis and immunologic properties in mice of vaccines composed of Staphylococcus aureus type 5 and type 8 capsular polysaccharides conjugated to Pseudomonas aeruginosa exotoxin A. Infect. Immun. 58:2367–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fattom A, et al. 1993. Laboratory and clinical evaluation of conjugate vaccines composed of Staphylococcus aureus type 5 and type 8 capsular polysaccharides bound to Pseudomonas aeruginosa recombinant exoprotein A. Infect. Immun. 61:1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fattom AI, Horwith G, Fuller S, Propst M, Naso R. 2004. Development of StaphVAX, a polysaccharide conjugate vaccine against S. aureus infection: from the lab bench to phase III clinical trials. Vaccine 22:880–887 [DOI] [PubMed] [Google Scholar]
- 23. Flock JI. 1999. Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol. Med. Today 5:532–537 [DOI] [PubMed] [Google Scholar]
- 24. Flynn N, Cohen SH. 2008. The continuing saga of MRSA. J. Infect. Dis. 197:1217–1219 [DOI] [PubMed] [Google Scholar]
- 25. Fournier B, Philpott DJ. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 18:521–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frank AL, Marcinak JF, Mangat PD, Schreckenberger PC. 1999. Increase in community-acquired methicillin-resistant Staphylococcus aureus in children. Clin. Infect. Dis. 29:935–936 [DOI] [PubMed] [Google Scholar]
- 27. Glass EJ, Oliver RA, Russell GC. 2000. Duplicated DQ haplotypes increase the complexity of restriction element usage in cattle. J. Immunol. 165:134–138 [DOI] [PubMed] [Google Scholar]
- 28. Glowalla E, Tosetti B, Krönke M, Krut O. 2009. Proteomics-based identification of anchorless cell wall proteins as vaccine candidates against Staphylococcus aureus. Infect. Immun. 77:2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gómez MI, García VE, Gherardi MM, Cerquetti MC, Sordelli DO. 1998. Intramammary immunization with live-attenuated Staphylococcus aureus protects mice from experimental mastitis. FEMS Immunol. Med. Microbiol. 20:21–27 [DOI] [PubMed] [Google Scholar]
- 30. Gómez MI, Sordelli DO, Buzzola FR, García VE. 2002. Induction of cell-mediated immunity to Staphylococcus aureus in the mouse mammary gland by local immunization with a live attenuated mutant. Infect. Immun. 70:4254–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorak EJ, Yamada SM, Brown JD. 1999. Community-acquired methicillin-resistant Staphylococcus aureus in hospitalized adults and children without known risk factors. Clin. Infect. Dis. 29:797–800 [DOI] [PubMed] [Google Scholar]
- 32. Gorwitz RJ. 2008. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and update. Pediatr. Infect. Dis. J. 27:925–926 [DOI] [PubMed] [Google Scholar]
- 33. Gorwitz RJ, et al. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J. Infect. Dis. 197:1226–1234 [DOI] [PubMed] [Google Scholar]
- 34. Han S, et al. 2010. Anaplasma marginale infection with persistent high-load bacteremia induces a dysfunctional memory CD4 T lymphocyte response but sustained high IgG titers. Clin. Vaccine Immunol. 17:1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harro C, et al. 2010. Safety and immunogenicity of a novel Staphylococcus aureus vaccine: results from a first-in-human dose-ranging study. Clin. Vaccine Immunol. 17:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harro JM, et al. 2010. Vaccine development in Staphylococcus aureus: taking the biofilm phenotype into consideration. FEMS Immunol. Med. Microbiol. 59:306–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hensinger RN. 2006. Impending danger: community-acquired methicillin-resistant Staphylococcus aureus. J. Pediatr. Orthop. 26:701–702 [DOI] [PubMed] [Google Scholar]
- 38. Hsieh CS, et al. 1993. Development of TH1 CD4 T cells through IL-12 produced by Listeria-induced macrophages. Science 260:547–549 [DOI] [PubMed] [Google Scholar]
- 39. Hume EBH, Dajcs JJ, Moreau JM, O'Callaghan RJ. 2000. Immunization with alpha-toxin toxoid protects the cornea against tissue damage during experimental Staphylococcus aureus keratitis. Infect. Immun. 68:6052–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. 2009. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 17:13–21 [DOI] [PubMed] [Google Scholar]
- 41. Josefsson E, Hartford O, O'Brien L, Patti JM, Foster T. 2001. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184:1572–1580 [DOI] [PubMed] [Google Scholar]
- 42. Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 43. Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O. 2010. Nontoxic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J. Exp. Med. 207:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuklin N, et al. 2006. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74:2215–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumanovics A, Tada T, Lindahl KF. 2003. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 21:629–657 [DOI] [PubMed] [Google Scholar]
- 46. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 9:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuroda M, Ohta T, Hayashi H. 1995. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 207:978–984 [DOI] [PubMed] [Google Scholar]
- 48. Kusch K, et al. 2011. The influence of SaeRE and σ(B) on the expression of superantigens in different Staphylcoccus aureus isolates. Int. J. Med. Microbiol. 301:488–499 [DOI] [PubMed] [Google Scholar]
- 49. Lahmers KK, Norimine J, Abrahamsen MS, Palmer GH, Brown WC. 2005. The CD4 T cell immunodominant Anaplasma marginale major surface protein 2 stimulates γδ T cell clones that express unique T cell receptors. J. Leukoc. Biol. 77:199–208 [DOI] [PubMed] [Google Scholar]
- 50. Lewin HA, Russell GC, Glass EJ. 1999. Comparative organization and function of the major histocompatibility complex of domesticated cattle. Immunol. Rev. 167:145–158 [DOI] [PubMed] [Google Scholar]
- 51. Liu C, et al. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52:285–292 [DOI] [PubMed] [Google Scholar]
- 52. Liu G. Y. 2009. Molecular pathogenesis of Staphylococcus aureus infection. Pediatr. Res. 65:71R–77R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lopez JE, et al. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 332:129–141 [DOI] [PubMed] [Google Scholar]
- 54. Lopez JE, et al. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins. Infect. Immun. 75:2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez JE, et al. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macatonia SE, Hsieh CS, Murphy KM, O'Garra A. 1993. Dendritic cells and macrophages are required for Th1 development of CD4 T cells from αβ TCR transgenic mice: IL-12 substitution for macrophages to stimulate IFN-γ production is IFN-γ-dependent. Int. Immunol. 5:1119–1128 [DOI] [PubMed] [Google Scholar]
- 57. Maira-Litran T, Kropec A, Goldmann DA, Pier GB. 2005. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-β-(1-6)-glucosamine. Infect. Immun. 73:6752–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCarthy AJ, Lindsay JA. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 10:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McGuire TC, Musoke AJ, Kurtti T. 1979. Functional properties of bovine IgG1 and IgG2: interaction with complement, macrophages, neutrophils and skin. Immunology 38:249–256 [PMC free article] [PubMed] [Google Scholar]
- 60. McKenney D, et al. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:523–1527 [DOI] [PubMed] [Google Scholar]
- 61. Merck 2011. Update on phase II/III clinical trial of investigational Staphylococcus aureus vaccine, V710 Merck, Whitehouse Station, NJ: http://www.merck.com/newsroom/news-release-archive/research-and-development/2011_0411.html [Google Scholar]
- 62. Mills KH, Barnard A, Watkins J, Redhead K. 1993. Cell-mediated immunity to Bordetella pertussis: role of Th1 cells in bacterial clearance in a murine respiratory infection model. Infect. Immun. 61:399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Montgomery CP, et al. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570 [DOI] [PubMed] [Google Scholar]
- 64. Moran GJ, Mount J. 2003. Update on emerging infections: news from the Centers for Disease Control and Prevention. Ann. Emerg. Med. 41:148–151 [DOI] [PubMed] [Google Scholar]
- 65. Morse K, et al. 2012. Association and evidence for linked recognition of type IV secretion system proteins VirB9-1, VirB9-2, and VirB10 in Anaplasma marginale. Infect. Immun. 80:215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nabi Biopharmaceuticals 2005. Key facts about StaphVAX. Nabi Biopharmaceuticals, Boca Raton, FL: http://www.nabi.com/images/factsheets/fsStaphVAX.pdf [Google Scholar]
- 67. Nanra JS, et al. 2009. Heterogeneous in vivo expression of clumping factor A and capsular polysaccharide by Staphylococcus aureus: implications for vaccine design. Vaccine 27:3276–3280 [DOI] [PubMed] [Google Scholar]
- 68. Norimine J, et al. 2003. Stimulation of T-helper cell gamma interferon and immunoglobulin G responses specific for Babesia bovis rhoptry-associated protein 1 (RAP-1) or a RAP-1 protein lacking the carboxy-terminal repeat region is insufficient to provide protective immunity against virulent B. bovis challenge. Infect. Immun. 71:5021–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pereira UP, Oliviera DGS, Mesquita LR, Costa GM, Pereira LJ. 2011. Efficacy of Staphylococcus aureus vaccines for bovine mastitis: a systematic review. Vet. Microbiol. 148:117–124 [DOI] [PubMed] [Google Scholar]
- 70. Pie S, Truffa-Bachi P, Pla M, Nauciel C. 1997. Th1 response in Salmonella typhimurium-infected mice with a high or low rate of bacterial clearance. Infect. Immun. 65:4509–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454–1468 [DOI] [PubMed] [Google Scholar]
- 72. Rappuoli R. 2000. Reverse vaccinology. Curr. Opin. Microbiol. 3:445–450 [DOI] [PubMed] [Google Scholar]
- 73. Rennermalm A, et al. 2001. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine 19:3376–3383 [DOI] [PubMed] [Google Scholar]
- 74. Schaffer AC, Lee JC. 2008. Vaccination and passive immunisation against Staphylococcus aureus. Int. J. Antimicrob. Agents 32:S71–S78 [DOI] [PubMed] [Google Scholar]
- 75. Schaffer AC, et al. 2006. Immunization with Staphylococcus aureus clumping factor B, a major determinant in nasal carriage, reduces nasal colonization in a murine model. Infect. Immun. 74:2145–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schroeder JW. 2010. Mastitis control programs: bovine mastitis and milking management. Document AS-1129 North Dakota State University Extension Service, Fargo, ND: http://www.ag.ndsu.edu/pubs/ansci/dairy/as1129.pdf [Google Scholar]
- 77. Sharif S, et al. 1998. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim. Genet. 29:185–193 [DOI] [PubMed] [Google Scholar]
- 78. Shinefield HR. 2006. Use of a conjugate polysaccharide vaccine in the prevention of invasive staphylococcal disease: is an additional vaccine needed or possible? Vaccine 2:65–69 [DOI] [PubMed] [Google Scholar]
- 79. Shinefield H, et al. 2002. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 346:491–496 [DOI] [PubMed] [Google Scholar]
- 80. Smith TC, et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 Is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Soavi L, et al. 2010. Methicillin-resistant Staphylococcus aureus ST398, Italy. Emerg. Infect. Dis. 2:346–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Solis N, Larsen MR, Cordwell SJ. 2010. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false positive control. Proteomics 10:2037–2049 [DOI] [PubMed] [Google Scholar]
- 83. Stoll H, Dengjel J, Nerz C, Götz F. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stranger-Jones YK, Bae T, Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103:16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Suarez CE, et al. 2000. Characterization of allelic variation in the Babesia bovis merozoite surface antigen 1 (MSA-1) locus and identification of a cross-reactive inhibition-sensitive MSA-1 epitope. Infect. Immun. 68:6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sutten EL, et al. 2010. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect. Immun. 78:1314–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thomsen I, Dudney H, Creech CB. 2010. Searching for the holy grail of a staphylococcal vaccine. Hum. Vaccin. 6:1068–1070 [DOI] [PubMed] [Google Scholar]
- 88. Thorsby E, Berle E, Nousiainen H. 1982. HLA-D region molecules restrict proliferative T-cell responses to antigen. Immunol. Rev. 66:39–56 [DOI] [PubMed] [Google Scholar]
- 89. Tuchscherr LP, Buzzola FR, Alvarez LP, Lee JC, Sordelli DO. 2008. Antibodies to capsular polysaccharide and clumping factor A prevent mastitis and the emergence of unencapsulated and small-colony variants of Staphylococcus aureus in mice. Infect. Immun. 76:5738–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. van Belkum A, et al. 2009. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 199:1820–1826 [DOI] [PubMed] [Google Scholar]
- 91. van Eijk MJ, Stewart-Haynes JA, Lewin HA. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23:483–496 [DOI] [PubMed] [Google Scholar]
- 92. Vytvytska O, et al. 2002. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2:580–590 [DOI] [PubMed] [Google Scholar]
- 93. Waters WR, et al. 2011. Tuberculosis immunity: opportunities from studies with cattle. Clin. Dev. Immunol. doi:10.1155/2011/768542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Weichhart T, et al. 2003. Functional selection of vaccine candidate peptides from Staphylococcus aureus whole-genome expression libraries in vitro. Infect. Immun. 71:4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wenner CA, Guler ML, Macatonia SE, O'Garra A, Murphy KM. 1996. Roles of IFN-γ and IFN-α in IL-12-induced T helper cell-1 development. J. Immunol. 156:1442–1447 [PubMed] [Google Scholar]
- 96. Wertheim HF, et al. 2008. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 5:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Y, et al. 2003. CpG ODN 2006 and IL-12 are comparable for priming Th1 lymphocyte and IgG responses in cattle immunized with a rickettsial outer membrane protein in alum. Vaccine 21:3307–3318 [DOI] [PubMed] [Google Scholar]
- 98. Ziebandt AK, et al. 2004. The influence of agr and σB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4:3034–3047 [DOI] [PubMed] [Google Scholar]