Abstract
Major histocompatibility complex class I chain-related gene A (MICA-129) dimorphism was investigated in 73 autoimmune diabetes patients (type 1 diabetes and latent autoimmune diabetes in adults) and 75 controls from Algeria. Only MICA-129 Val allele and MICA-129 Val/Val genotype frequencies were higher among patients than in the control group. Statistical analysis of the estimated extended HLA-DR-DQ-MICA haplotypes shown that individual effects of MICA alleles on HLA-DQ2-DR3-MICA-129 Val/Val and HLA-DQ8-DR4-MICA-129 Val/Val haplotypes were significantly higher in patients than in the control groups. These preliminary data might suggest a relevant role of MICA-129 Val/Val single nucleotide polymorphism (weak/weak binders of NKG2D receptor) in the pathogenesis of T1D and LADA.
INTRODUCTION
Type 1 diabetes (T1D) is a typical autoimmune disease that results from the destruction of insulin-producing β cells of the pancreas; the disease was once thought to be mediated exclusively by CD4+ T cells and is now recognized as one in which autoreactive CD8+ T cells play a fundamental pathogenic role (37). The etiology of T1D is complex and involves both genetic and environmental factors which play important roles (7, 11, 12).
A permissive genetic background is required for the development of the islet autoimmune process generating antibodies (Ab) against insulin (IAA), glutamic acid decarboxylase isoform 65 (GADA65-Ab), and protein tyrosine phosphatase (IA2) (6, 39). In 1986, Groop et al. showed that islet cell antibodies identify latent type 1 diabetes mellitus (T1DM) in patients aged 35 to 75 years at diagnosis (20). These subclasses of diabetes have been referred to as latent autoimmune diabetes in adults (LADA). Autoantibodies to glutamate decarboxylase (GAD65Ab) are the most sensitive and specific markers for these subgroups of diabetes (39). Autoimmunity is also assumed to be the major cause of LADA because this category of diabetes shares biochemical markers of cell-directed autoimmunity with classic type 1 diabetes. LADA is considered a mild form of type 1 diabetes. “Mild” indicates the fact that patients with LADA do not by clinical judgment need insulin from the time of diagnosis. However, within the first few years after the diagnosis of diabetes, a need for insulin treatment develops in many patients with LADA (30). However, LADA, like classic T1D, is associated with HLA class II genes (13, 24). The biological function of other genes encoded within this region generated much interest. Among these genes are the family of major histocompatibility complex class I (MHC-I) chain-related genes (MICA and MICB). MICA protein is encoded by the MICA gene present on chromosome 6p21.3 in the MHC locus, at 46.4 kb centromeric to the HLA-B gene. The MICA protein comprises a transmembrane MHC-I alpha-like chain and is not associated to the β-2-microglobulin and does not bind to peptides (15). The MICA chain is a stress-induced protein and is expressed on the basolateral membrane of intestinal epithelium cells and in epithelium-derived tumors (18, 42). The receptor of MICA chain was identified as NKG2D homodimer. MICA protein is highly polymorphic and binds to its NKG2D receptor found on the surface of natural killer (NK) and CD8 αβ T cells (8, 31).
Considerable polymorphism has been observed so far with 54 alleles of MICA and 16 alleles of MICB (5). Trinucleotide repeat (GCT) microsatellite polymorphisms, which were found on exon 5 by sequence analysis, encode the transmembrane region of the MICA protein. A change of the amino acid methionine (Met) to valine (Val) at position 129 of the (α)2-heavy chain domain is in linkage disequilibrium with other allelic changes and seems to classify MICA proteins of these alleles into strong and weak binders of NKG2D receptor, which, in turn, influences effector cell function (18). MICA molecules were proven to play prominent roles in immune processes (33). The functional relevance of this variant in NK and T-cell activation led us to hypothesize that the type of NKG2D and MICA interaction with either high-affinity (MICA-129 Met) or low-affinity (MICA-129 Val) alleles favors chronic inflammation or tumor escape in subjects genetically predisposed to these immune disorders (2, 14). The association of the HLA complex present on chromosome 6 does not alone explain the total linkage of the HLA region to T1D, leading to the hypothesis of the probable existence of additional causal genes for immune-related disorders in this region. Studies report that polymorphism on the MICA genes could be a potential candidate for association with T1D and LADA (1, 17, 29, 35, 38). In this study, we aimed to study allele polymorphism and the functionally relevant dimorphism (129 Val/Met) of the MICA gene in younger T1D and LADA patients in the Algerian population.
MATERIALS AND METHODS
Patients and controls.
The study was carried out with blood samples taken from 73 unrelated antoimmune diabetes patients with autoimmune diabetes, positive antibodies against insulin and glutamic acid decarboxylase, and very low C peptide and insulin; the sex ratio was 1.6 (45 males/28 females). Table 1 shows the clinical characteristics of the population studied. The patient cohort was collected consecutively upon diagnosis by diabetology at the Centre Hospitalier Universitaire Mustapha, Algeria, with the diagnosis being made according to the National Diabetes Data Group and American Diabetes Association (ADA) criteria (27). We divided patients into two different groups on the basis of age at the onset of the disease: group 1 consisted of 30 juveniles with T1D, 16 to 30 years old (median age, 21.06 ± 3.69 years), and group 2 consisted of 43 LADA patients 25 to 40 years old (median age, 39.57 ± 4.68 years). All patients were on insulin therapy upon hospital discharge and presented with ketoacidosis. Case genotypes were compared to 75 healthy unrelated blood donor volunteers from the same locality; they included 45 males and 30 females with a median age of 45 years. None had a first-degree relative affected with diabetes or other autoimmune disorder. Nondiabetic control subjects with no family history of diabetes were selected from the same geographic area. Informed consent was obtained from both the patients and the healthy donors.
Table 1.
Clinical characteristics of the study population
| Parameter | Value for the group |
||
|---|---|---|---|
| T1D (n= 30) | LADA (n= 43) | Control (n= 75) | |
| Median age (yrs) | 21.06 ± 3.69 | 39.57 ± 4.68 | 30 ± 7.9 |
| Median age at diagnosis (yrs) | 18.06 ± 3.87 | 26.32 ± 1.90 | |
| Median duration of diabetes (yrs) | 2.6 ± 4.87 | 3.90 ± 3.23 | |
| Insulin dose (μU/ml) | 16.61 ± 3.55 | 13.39 ± 3.76 | 7.78 ± 1.55 |
| C peptide (ng/ml) | 0.61 ± 0.06 | 0.98 ± 0.24 | 1.57 ± 0.34 |
| Hemoglobin A1c (%) | 8.23 ± 1.26 | 8.5 ± 2.01 | 6 ± 1.006 |
| BMI (kg/m2)a | 22 ± 6.8 | 25.98 ± 5.62 | 25.98 ± 5.62 |
BMI, body mass index.
HLA-DR-DQ and MICA-129 genotyping.
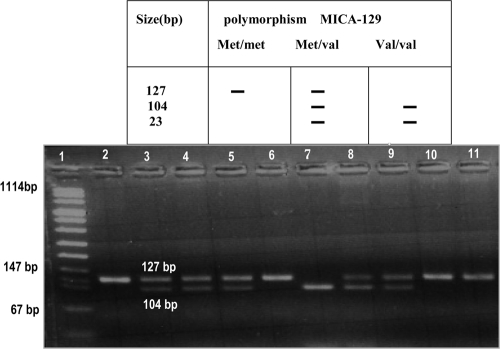
Genomic DNA was isolated from EDTA-treated peripheral blood samples using chloroform-phenol assay. HLA DR and DQ alleles were typed by means of PCR sequence-specific priming (PCR-SSP) (low resolution; Dynal, Compiègne, France), and the MICA-129 polymorphism was explored at the DNA level (A-to-G change in exon 3 at nucleotide position 454) as previously described using a nested PCR-restriction fragment length polymorphism (PCR-RFLP) procedure using an automated thermal cycler (9600; Perkin-Elmer, Cerus, CA). The MICA gene-specific amplicon was used as a template in a second round of amplification of its exon 3 with each primer (Table 2). The presence of the MICA-129 Val allele was identified by the presence of a restriction site for RsaI enzyme (Fig. 1) created by a deliberately introduced mismatch in the reverse primer (2). For clarity, the explored MICA variants are designated MICA-129 Val and MICA-129 Met.
Table 2.
List of MICA primer sequences, their positions, and the sizes of the amplified products
| PCR type and primer name | Primer sequence | Location (position [nt])a | Product size (bp) |
|---|---|---|---|
| MICA-specific PCR | |||
| MICA-FG | 5′-CGTTCTTGTCCCTTTGCCCGTGTGC-3′ | Intron 1 (6823–6847) | 2,201 |
| MICA-RF | 5′-GATGCTGCCCCCATTCCCTTCCCAAA-3′ | Intron 5 (8999–9023) | |
| MICA nested PCR | |||
| MICA-FM | 5′-GGGTCTGTGAGATCCATGA-3′ | Exon 3 (350–368) | 127 |
| MICA-RMb | 5′-TGAGCTCTGGAGGACTGGGGTA-3′ | Exon 3 (455–476) |
Positions are based on GenBank accession number X92841. nt, nucleotide.
The exon 3 reverse primer has a deliberately introduced mismatch to create an RsaI restriction site.
Fig 1.
PCR-RFLP amplification for the studied MICA-129 gene polymorphisms. Lane 1, DNA size marker; lane 2, product of PCR 2; lanes 3, 4, 5, 8, and 9, samples from patients with heterozygous Met/Val alleles; lanes 6, 10, and 11, samples from patients with homozygous Met/Met alleles; lane 7, sample from patient with homozygous Val/Val alleles.
Statistical analysis.
Statistical analysis was performed using the Compare 2 software, version 1.02 (Chicago, IL). Allele frequencies were calculated from the observed number of genotypes. The significance of differences in the allele frequencies between each of three groups was determinate by Fisher's exact test. The chi-square testing (with Yates' correction or Fisher exact test for a low number of cases) was used to assess the differences in frequencies for each risk factor. MICA-129 gene polymorphisms will be designated here as the MICA-129 Val or Met allele or their allelic combination into genotype. The P values were corrected (Pc) by multiplying the P value by the number of tested alleles at each studied locus. Findings were considered statistically significant at Pc values of less than 0.05. The odds ratio (OR) was calculated for each risk factor and is given with its 95% confidence interval (CI).
RESULTS
MICA amino acid 129 polymorphism.
We observed that the MICA-129 Val allele was significantly more frequent in young T1D and LADA patients than in the controls (for T1D patients, 78.33% versus 65.33% for the controls, Pc of 0.034, OR of 1.92, and 95% CI of 0.916 to 4.214; for LADA patients, 82.56% versus 65.33% for the controls, Pc of 2.2 × 10−3, OR of 2.51, and 95% CI of 1.266 to 5.18), whereas MICA-129 Met allele was significantly more frequent in control than in patient groups (for T1D, 34.67% versus 21.67%, Pc of 0.034, OR of 0.52, and 95% CI of 0.237 to 1.092; for LADA, 34.67% versus 17.44%, Pc of 2.2 × 10−3, OR of 0.40, and 95% CI of 0.193 to 0.79) (Table 3). The analysis of the distribution of the different genotypes (MICA-129 Met/Met, Met/Val, and Val/Val) between the three groups revealed that the MICA-129 Val/Val genotype is significantly associated with juvenile T1D and LADA rather than with controls (for T1D, 60.00% versus 41.33%, Pc of 0.043, OR of 2.13, and 95% CI of 0.827 to 5.571; for LADA, 67.44% versus 41.33%, Pc of 3.3 × 10−3, OR of 2.94, and 95% CI of 1.254 to 7.02). Furthermore, we did not find statistical differences in the distribution of MICA-129 Val allele and MICA-129 Val/Val genotype between the young T1D and the LADA patients (Pc values of 0.334 and 0.342, respectively). However, we noticed that polymorphism of the MICA-129 Val allele has more risks for the LADA than the juvenile T1D patients (OR of 2.51 and 1.92, respectively). We found that the MICA-129 Val allele, which was more frequent in the juvenile T1D and LADA groups than in the control group (78.33% and 82.56% for T1D and LADA, respectively, versus 65.33% for the control), may be the risk factor for T1D and LADA; genotype frequencies were 36.67% and 30.23% for T1D and LADA, respectively, versus 48% for the control for the heterozygous MICA-129 Met/Val, 3.33% and 2.33%, respectively, versus 10.67% for the homozygous MICA-129 Met/Met, and 60.00% and 67.44%, respectively, versus 41.33% for the homozygous MICA-129 Val/Val.
Table 3.
MICA-129 allele and genotype frequencies among T1D juvenile, LADA, and control groups
| MICA-129 polymorphism | Distribution analysis by groupa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| T1D |
LADA |
Frequency in the controls (no. [%]) | |||||||
| Frequency (no. [%]) | Pc | χ2 | OR (95% CI) | Frequency (no. [%]) | Pc | χ2 | OR (95% CI) | ||
| Alleles | |||||||||
| MICA-129 Val | 47 (78.33) | 0.034 | 2.8 | 1.92 (0.916–4.214) | 71 (82.56) | 2.2 × 10−3 | 7.152 | 2.51 (1.266–5.18) | 98 (65.33) |
| MICA-129 Met | 13 (21.67) | 0.034 | 2.8 | 0.52 (0.237–1.092) | 15 (17.44) | 2.2 × 10−3 | 7.152 | 0.40 (0.193–0.79) | 52 (34.67) |
| Genotypes | |||||||||
| MICA-129 Val/Val | 18 (60.00) | 0.043 | 2.297 | 2.13 (0.827–5.571) | 29 (67.44) | 3.3 × 10−3 | 6.44 | 2.94 (1.254–7.02) | 31 (41.33) |
| MICA-129 Met/Val | 11 (36.67) | 0.151 | 0.7 | 0.63 (0.236–1.619) | 13 (30.23) | 0.031 | 2.859 | 0.47 (0.194–1.106 | 36 (48.00) |
| MICA-129 Met/Met | 1 (3.33) | 0.153 | 0.68 | 0.29 (0.006–2.345) | 1 (2.33) | 0.062 | 1.645 | 0.20 (0.004–1.594) | 8 (10.67) |
Groups consist of 30 T1D and 43 LADA patients and 75 controls. OR, odds ratio; 95% CI, 95% confidence interval. Boldface indicates significant differences in the allele and genotype frequencies between each of the three groups (Pc < 0.05).
To illustrate these results, the prevalence of the estimated extended DR-DQ-MICA-129 haplotypes was analyzed (Table 4). The frequencies of the haplotypes HLA-DQ2/DR3-MICA-129 Val/Val and Q8/DR4-MICA-129 Val/Val were significantly increased in juvenile T1D and LADA patients relative to levels in the controls: for HLA-DQ2/DR3-MICA-129 Val/Val, 53.33% for T1D versus 9.33% for the control (OR, 11; Pc, 1.3 10−6; 95% CI, 3.448 to 37.291) and 48.84% for LADA versus 9.33% (Pc, 9.1 10−7; OR, 9.27; 95% CI, 3.194 to 28.845); for DQ8/DR4-MICA-129 Val/Val, 30.00% for T1D versus 5.33% for the control (Pc, 5.2 10−4; OR, 7.61; 95% CI, 1.848 to 36.4) and 34.88% for LADA versus 5.33% (OR, 9.51; Pc, 2.0 10−5; 95% CI, 2.665 to 41.896). These haplotypes confer significant susceptibility to T1D.
Table 4.
MICA alleles and HLA-DR-DQ haplotypes in relation to T1D juvenile, LADA, and control groups
| HLA-DR-DQ haplotype | MICA-129 genotype | Distribution analysis by groupa |
||||||
|---|---|---|---|---|---|---|---|---|
| LADA (n = 43) |
Juveniles (n = 30) |
Frequency (no. [%]) in the controls (n = 75) | ||||||
| Frequency (no. [%]) | Pc | OR (95% CI) | Frequency (no. [%]) | Pc | OR (95% CI) | |||
| DR3 DQ2 | Met/Met | 1 (2.33) | 0.303 | 1.76 (0.022–140.14) | 1 (3.33) | 0.201 | 2.55 (0.031–202.950) | 1 (1.33) |
| Met/Val | 10 (23.26) | 0.118 | 1.76 (0.600–5.09) | 11 (36.67) | 0.007 | 3.37 (1.117–10.013) | 11 (14.67) | |
| Val/Val | 21 (48.84) | 9.1 × 10−7 | 9.27 (3.194–28.84) | 16 (53.33) | 1.3 × 10−6 | 11 (3.448–37.291) | 7 (9.33) | |
| DR4 DQ8 | Met/Met | 1 (2.33) | 0.303 | 0.00 (0.015–18.30) | 0 (0) | 0.0 | 0(0.0) | |
| Met/Val | 5 (11.63) | 0.167 | 1.83 (0.395–8.51) | 5 (16.67) | 0.057 | 2.8 (0.584–13.155) | 5 (6.67) | |
| Val/Val | 15 (34.88) | 2.0 × 10−5 | 9.51 (2.665–41.89) | 9 (30.00) | 5.2 × 10−4 | 7.61 (1.848–36.4) | 4 (5.33) | |
| DR15/DQ6 | Met/Met | 0 (0.0) | 0.156 | 0.0 (0.000–72.56) | 0 (0.0) | 0.249 | 0.0 (0.000–6.089) | 3 (4) |
| Met/Val | 1 (2.33) | 0.041 | 0.17 (0.004–1.35) | 0 (0.0) | 0.027 | 0.0 (0.000–1.207) | 9 (12) | |
| Val/Val | 6 (13.95) | 0.105 | 0.51 (0.153–1.52) | 1 (3.33) | 5.4 × 10−3 | 0.11 (0.003–0.772) | 18 (24) | |
| Other | Met/Met | 0 (0) | 0.097 | 0.0 (0.000–1.54) | 0 (0.0) | 0.174 | 0.0 (0.000–3.795) | 4 (5.33) |
| Met/Val | 6 (13.95) | 0.341 | 0.77 (0.222–2.42) | 1 (3.45) | 0.032 | 0.16 (0.004–1.210) | 13 (17.33) | |
| Val/Val | 10 (23.26) | 0.021 | 2.94 (0.908–9.90) | 4 (13.33) | 0.239 | 1.49 (0.294–6.450) | 7(9.33) | |
OR, odds ratio; CI, confidence interval; Pc, corrected P value. Boldface indicates significant differences in the allele and genotype frequencies between each of the three groups (Pc < 0.05).
DISCUSSION
In previous studies, some authors have hypothesized that MICA could engage both adaptive and innate immunity (32). In addition, MICA alleles may also be associated with susceptibility or resistance to developing autoimmunity (10). Recently, several authors have shown that the MICA-129 polymorphism was associated with several pathologies (9, 14, 23). In contrast, it has been demonstrated that a change of methionine to valine at position 129 of the α2-heavy chain domain classifies MICA alleles into strong (MICA-129 Met) or weak (MICA-129 Val) binders of NKG2D receptor (33). However, the role of MICA-129 dimorphism in T1D susceptibility has not been investigated yet. This shortcoming has been addressed in this study by investigating whether MICA-129 dimorphism was associated with susceptibility to/or resistance against developing T1D. We found that the MICA-129 Val allele, which was more frequent in the juvenile T1D and LADA than in the control group, may be a risk factor for autoimmune diabetes.
That the MICA-129 Met/Val heterozygote and MICA-129 Met/Met homozygote (strong NKG2D binders)were significantly more prevalent in the control group than in the patient groups may suggest a beneficial role in healthy individuals and may be associated with protection against the autoimmune diabetes. It can be hypothesized that the presence of MICA-129 Met/Met and MICA-129 Met/Val genotypes may modify NK and CD8 T lymphocyte activation to mount an adequate immune response, thereby allowing an immune response to viral infections including enterovirus, adenovirus, coxsackie B virus, cytomegalovirus, and hepatitis C virus (26).
Studies performed in different populations showed an association of different MICA alleles with type 1 diabetes (1, 29, 36, 38). In addition, several studies demonstrated the association of MICA alleles with the age of the patient presenting the clinical onset of T1D (16, 21) or an association with LADA (16, 31). In our study carried out on the Algerian population, the MICA-129 Val allele was found to be associated with adult-onset T1D (<25 years old; OR of 1.92) and with LADA (>25 years old; OR of 2.51). We also found that a combination of the MICA-129 Val/Val genotype and DR3-DQ2 or DR4-DQ8 conferred an increased risk for adult-onset T1D (OR of 11 or 6.45, respectively) and for LADA (OR of 9.27 or 9.51, respectively).
In addition, we provided new evidence of the significant association between the MICA-129 Val allele and adult-onset T1D. This association was also observed in a patient with LADA, suggesting a common genetic background for LADA and adult-onset T1D. We have indirectly confirmed the autoimmune origin of LADA based on our findings of the association between LADA and MICA-129 gene polymorphisms. These clinical and autoimmune markers do vary in the different age categories, supporting the conclusion that MICA contributes to the disease irrespective of the autoimmune markers and residual β cell function. These findings and interpretations were in line with other pathological contexts (3, 9, 14, 23, 25, 29, 34, 41).
In the context of autoimmune disease, alleles at the MICA locus can be defined as strong or weak binders to the NKG2D receptor. The strong NKG2D binding alleles share methionine at position 129 with high affinity (MICA-129 Met), whereas weak binding alleles have valine at this position with low affinity (MICA-129 Val). The significance of high/low affinity for the NKG2D receptor in terms of immune activation is not completely understood yet. However, in this study and in the context of T1D, it can be hypothesized that the presence of the weak engagement of NKG2D receptors by the weak binder MICA-129 Val allele implies both pathways (14). The ultimate consequence of such weak interaction could be the impairment of natural killer (NK) or cytotoxic T lymphocyte cell activation and costimulation, possibly skewing the Th1 pathway toward Th2, with consequent B cell activation and Ab production. The weak interaction could lead to overexpression of NKG2D receptors as previously shown in autoimmune diseases (19, 22, 28). This situation could favor the binding of other ligands such as UL-16 and UL-16 binding proteins (ULBPs) (4). Thus, in a microenvironment highly enriched in interleukin-15 (IL-15), repeated T and NK stimulation favors an autoimmune-like situation, breaking down the self-tolerance with consequent auto-Ab production. The presence of autoimmunity against pancreatic β cells in the T1D and LADA group was assessed by detection of autoantibodies in the peripheral blood. It has been shown that autoantibody production has been detected up to 5 years prior to the development of hyperglycemic events, and this indicates that autoantibody production precedes the clinical manifestation of T1D (40). Molecular mimicry is by far one of the most well studied processes associated with viral triggering mechanisms and T1D disease induction. Viruses produce proteins similar to those of the host, and certain viral components share a distinct homology with identified β-cell antigens targeted in an autoimmune response (26).
In conclusion, our findings highlight that the MICA-129 gene polymorphism is a functionally relevant gene associated with T1D and LADA and seems to play a potential role in the development of T1D in the patient population in central Algeria. This study might provide a better understanding of the implications of MICA genetic variation in T1D immunology, thus helping to design future detection and monitoring assays. We are planning additional work in order to determine the conditional extended transmission disequilibrium between MICA-129 and HLA class I.
ACKNOWLEDGMENTS
We acknowledge the invaluable contributions of the patients and their families.
This work was supported by the Laboratory of Immunogenetics and Immunopathology, Algeria.
Footnotes
Published ahead of print 8 February 2012
REFERENCES
- 1. Alizadeh BZ, et al. 2007. MICA marks additional risk factors for Type 1 diabetes on extended HLA haplotypes: an association and meta-analysis. Mol. Immunol. 44: 2806– 2812 [DOI] [PubMed] [Google Scholar]
References
- 2. Amroun H, et al. 2005. Early-onset ankylosing spondylitis is associated with a functional MICA polymorphism. Hum. Immunol. 66: 1057– 1061 [DOI] [PubMed] [Google Scholar]
- 3. Aquino-Galvez AM, et al. 2009. MICA polymorphisms and decreased expression of the MICA receptor NKG2D contribute to idiopathic pulmonary fibrosis susceptibility. Hum. Genet. 125: 639– 648 [DOI] [PubMed] [Google Scholar]
- 4. Bahram S, et al. 2005. MIC and other NKG2D ligands: from none to too many. Curr. Opin. Immunol. 17: 505– 509 [DOI] [PubMed] [Google Scholar]
- 5. Baham S, et al. 1994. A second lineage of mammalian major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. U. S. A. 91: 6259– 6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonifacio E, et al. 1990. Quantification of islet cell antibodies and prediction of insulin dependent diabetes. Lancet 335: 147– 149 [DOI] [PubMed] [Google Scholar]
- 7. Borchers AT, Uibo R, Gershwin ME. 2010. The geoepidemiology of type 1 diabetes. Autoimmun. Rev. 9: A355– A365 [DOI] [PubMed] [Google Scholar]
- 8. Borrego F, et al. 2002. Structure and function of major histocompatibility complex (MHC) class I specific receptors expressed on human natural killer (NK) cells. Mol. Immunol. 38: 637– 660 [DOI] [PubMed] [Google Scholar]
- 9. Boukouaci W. 2009. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood 114: 5216– 5224 [DOI] [PubMed] [Google Scholar]
- 10. Caillat-Zucman S. 2006. How NKG2D ligands trigger autoimmunity? Hum. Immunol. 67: 204– 207 [DOI] [PubMed] [Google Scholar]
- 11. Concannon P, et al. 2009. Genetics of type 1A diabetes. N. Engl. J. Med. 360: 1646– 1654 [DOI] [PubMed] [Google Scholar]
- 12. Csorba TR, et al. 2010. Autoimmunity and the pathogenesis of type 1 diabetes. Crit. Rev. Clin. Lab. Sci. 47: 51– 71 [DOI] [PubMed] [Google Scholar]
- 13. Cucca F, et al. 1993. Combinations of specific DRB1, DQA1, and DQB1 haplotypes are associated with insulin-dependent diabetes mellitus in Sardinia. Hum. Immunol. 37: 85– 94 [DOI] [PubMed] [Google Scholar]
- 14. Douik H, et al. 2009. Association of MICA-129 polymorphism with nasopharyngeal cancer risk in a Tunisian population. Hum. Immunol. 70: 45– 48 [DOI] [PubMed] [Google Scholar]
- 15. Frigoul A, Lefranc MP. 2005. MICA: standardized IMGT allele nomenclature, polymorphisms and diseases. Recent Res. Devel. Hum. Genet. 3: 95– 145 [Google Scholar]
- 16. Gambelunghe G, et al. 2001. Two distinct MICA gene markers discriminate major autoimmune diabetes types. J. Clin. Endocrinol. Metab. 86: 3754– 3760 [DOI] [PubMed] [Google Scholar]
- 17. Gambelunghe G, et al. 2007. MICA gene polymorphism: the pathogenesis of type 1 diabetes. Ann. N. Y. Acad. Sci. 1110: 92– 98 [DOI] [PubMed] [Google Scholar]
- 18. Groh V, et al. 1996. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Nati. Acad. Sci., U. S. A. 93: 12445– 12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groh V, et al. 2003. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 100: 9452– 9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Groop L, et al. 1986. Islet cell antibodies identify latent type 1 diabetes in patients aged 35–75 years at diagnosis. Diabetes 35: 237– 241 [DOI] [PubMed] [Google Scholar]
- 21. Gupta ML, et al. 2003. Association between the transmembrane region polymorphism of MHC class I chain related gene-A and type 1 diabetes mellitus in Sweden. Hum. Immunol. 64: 553– 561 [DOI] [PubMed] [Google Scholar]
- 22. Hue S, et al. 2004. Direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21: 367– 377 [DOI] [PubMed] [Google Scholar]
- 23. Lopez-Hernandez R, et al. 2010. Association analysis of MICA gene polymorphism and MICA-129 dimorphism with inflammatory bowel disease susceptibility in a Spanish population. Hum. Immunol. 71: 512– 514 [DOI] [PubMed] [Google Scholar]
- 24. Ludvigsson JU. 1986. HLA-DR3 is associated with a more slowly progressive form of type 1 (insulin-dependent) diabetes. Diabetologia 29: 207– 221 [DOI] [PubMed] [Google Scholar]
- 25. Mizuki N, et al. 1997. Triplet repeat polymorphism in the transmembrane region of the MICA gene: a strong association of six GCT repetitions with Behçet disease. Proc. Natl. Acad. Sci. U. S. A. 94: 1298– 1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morran MP, et al. 2008. Immunology and genetics of type 1 diabetes. Mt. Sinai J. Med. 75: 314– 327 [DOI] [PubMed] [Google Scholar]
- 27. National Diabetes Data Group 1979. Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes 28: 1039– 1042 [DOI] [PubMed] [Google Scholar]
- 28. Ogasawara K, et al. 2004. NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity 20: 757– 767 [DOI] [PubMed] [Google Scholar]
- 29. Park Y, et al. 2001. MICA polymorphism is associated with type 1 diabetes in the Korean population. Diabetes Care 24: 33– 38 [DOI] [PubMed] [Google Scholar]
- 30. Radtke MA, et al. 2009. Heterogeneity of patients with latent autoimmune diabetes in adults: linkage to autoimmunity is apparent only in those with perceived need for insulin treatment. Diabetes Care 32: 245– 250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanjeevi CB. 2006. Genes influencing innate and acquired immunity in type 1 diabetes and latent autoimmune diabetes in adults. Ann. N. Y. Acad. Sci. 1079: 67– 80 [DOI] [PubMed] [Google Scholar]
- 32. Stastny P. 2006. Introduction: MICA/MICB in innate immunity, adaptative immunity, autoimmunity, cancer, and in the immune response to transplants. Hum. Immunol. 67: 141– 144 [DOI] [PubMed] [Google Scholar]
- 33. Steinle A, et al. 2001. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics 53: 279. [DOI] [PubMed] [Google Scholar]
- 34. Tonnerre P, et al. 2010. MICA gene polymorphism in kidney allografts and possible impact of functionally relevant variants. Transplant. Proc. 42: 4318– 4321 [DOI] [PubMed] [Google Scholar]
- 35. Torn C, et al. 2003. Heterozygosity for MICA5.0/MICA5.1 and HLA-DR3-DQ2/DR4-DQ8 are independent genetic risk factors for latent autoimmune diabetes in adults. Hum. Immunol. 64: 902– 909 [DOI] [PubMed] [Google Scholar]
- 36. Triolo TM, et al. 2009. Homozygosity of the polymorphism MICA5.1 identifies extreme risk of progression to overt adrenal insufficiency among 21-hydroxylase antibody-positive patients with type 1 diabetes J. Clin. Endocrinol. Metab. 94: 1210– 1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsai S, et al. 2008. CD8+ T cells in type 1 diabetes. Adv. Immunol. 100: 79– 124 [DOI] [PubMed] [Google Scholar]
- 38. Van Autreve JE, et al. 2006. MICA is associated with type 1 diabetes in the Belgian population, independent of HLA-DQ. Hum. Immunol. 67: 94– 101 [DOI] [PubMed] [Google Scholar]
- 39. van Deutekom AW, et al. 2008. The islet autoantibody titres: their clinical relevance in latent autoimmune diabetes in adults (LADA) and the classification of diabetes mellitus. Diabet. Med. 25: 117– 125 [DOI] [PubMed] [Google Scholar]
- 40. von Herrath M, et al. 2007. Type 1 diabetes as a relapsing-remitting disease? Nat. Rev. Immunol. 7: 988– 994 [DOI] [PubMed] [Google Scholar]
- 41. Zhao J, et al. 2011. Functional MICA-129 Polymorphism and serum levels of soluble MICA are correlated with ulcerative colitis in Chinese patients. J. Gastroenterol. Hepatol. 26: 593– 598 [DOI] [PubMed] [Google Scholar]
- 42. Zwirner NW, et al. 1998. MICA, a new polymorphic HLA related antigen, is expressed mainly by keratinocytes, endothelial cells, and monocytes. Immunogenetics 47: 139– 148 [DOI] [PubMed] [Google Scholar]