Abstract
Autophagy is an evolutionarily conserved mechanism for the degradation of cellular components, but its role in enucleation during differentiation has not been established. Tetrahymena thermophila is a unicellular eukaryote with two functionally distinct nuclei, the somatic (macro-) and the germ line (micro-) nuclei. These nuclei are produced during sexual reproduction (conjugation), which involves differentiation and selective degradation of several specific nuclei. To examine the role of autophagy in nuclear degradation, we studied the function of two ATG8 genes in Tetrahymena. Through fluorescent protein tagging, we found that both proteins are targeted to degrading nuclei at specific stages, with some enrichment on the nuclear periphery, suggesting the formation of autophagosomes surrounding these nuclei. In addition, ATG8 knockout mutant cells showed a pronounced delay in nuclear degradation without apparently preventing the completion of other developmental events. This evidence provided direct support for a critical role for autophagy in programmed nuclear degradation. The results also showed differential roles for two ATG8 genes, with ATG8-65 playing a more significant role in starvation than ATG8-2, although both are important in nuclear degradation.
INTRODUCTION
Macroautophagy (here referred to as autophagy), a conserved pathway for the recycling of cellular components, secludes surplus or damaged proteins and organelles from the cytosol by utilizing de novo-generated double membrane structures, autophagosomes, to help maintain homeostasis or deal with development and stress (62). Most cellular membrane-bound and non-membrane-bound organelles, such as the endoplasmic reticulum, Golgi apparatus, mitochondria, and ribosomes, have been reported to be eliminated by autophagy. However, for most organisms, the cell nucleus has not been known to be a target of autophagy, even in cases where enucleation occurs as a normal part of differentiation (37, 39, 55).
Tetrahymena thermophila is a binucleated unicellular organism that carries out a complex developmental process of nuclear differentiation and degradation during sexual reproduction (conjugation). Several earlier reports suggested the involvement of apoptosis- or autophagy-like processes in nuclear degradation in this species, making it a potentially excellent model by which to understand the role of autophagy in nuclear elimination (1, 2, 16–18, 33, 34, 41, 63).
Tetrahymena thermophila, like most ciliated protozoa, contains two functionally distinct nuclei in one cell: the somatic macronucleus and germ line micronucleus. The larger macronucleus is polyploid, containing around 45 times the amount of DNA present in a haploid genome (45C), and it is active in transcription during vegetative growth. It divides by amitosis, a special process of nuclear division with no chromosome condensation or spindle formation. On the other hand, the smaller micronucleus is diploid and divides by typical mitosis. After starvation and interaction of cells of different mating types, Tetrahymena undergoes sexual reproduction by conjugation; this process reveals a series of dynamic nuclear reorganization events (11, 63).
The micronucleus goes through meiosis and produces four meiotic products (haploid nuclei); one of them is selected to produce two gametic nuclei following one round of mitosis, while the other three are degraded. One gametic nucleus from each cell migrates over to the partner cell and fuses with the remaining gametic nucleus to produce a diploid zygotic nucleus. This zygotic micronucleus undergoes two rounds of mitosis to produce four diploid nuclei, of which two develop further to form the new macronuclei (new MAC). Among the other two nuclei that remain in the micronucleus state, one is eventually degraded. During this time, the parental (old) macronucleus goes through an apoptosis-like degradation process called programmed nuclear death (PND) and finally disappears, presumably absorbed by the cytoplasm (16). Therefore, a total of 10 nuclei in each mating pair (six meiotic products, two postzygotic products, and two old macronuclei) are degraded and absorbed during one round of conjugation. The PND of old MAC is a remarkable event. It has been shown that some steps of this process share features similar to apoptosis in mammalian cells, such as caspase-like activities and DNA fragmentation (16, 17, 33). However, most of the molecular details, including target selection and recognition, remain largely unknown.
PND in Tetrahymena appears similar to enucleation events in metazoans, in the sense that both processes remove unwanted nuclei from the cell while keeping the cytoplasm largely intact. Although autophagy is an evolutionarily conserved mechanism for recycling cellular organelles (32), previous studies have shown that it is not involved in the enucleation process in mammalian cells, such as erythrocytes, lens periphery cells, and megakaryocytes (35, 38, 39). In Tetrahymena, however, electron microscopic studies have revealed the presence of autophagosomes both in starved cells (43) and surrounding degenerating old MAC during conjugation (57). In addition, inhibition of the phosphatidylinositol 3-kinase (PI3K) complex, which is the initiator of autophagy, during early conjugation resulted in the accumulation of extra nuclei, including old MAC (61). Finally, by using monodansylcadaverine (MDC) staining, a recent microscopic study showed that degenerating old MACs have the features of autophagosomes (2). These studies suggest that autophagy might play a direct role in PND in Tetrahymena. To ascertain this possibility and further understand this important process, molecular analyses of the autophagy-related genes (ATG) are warranted. No studies of autophagy genes in Tetrahymena or other ciliated protozoa have been reported so far. In this study, we identified three ATG8 orthologous genes in the Tetrahymena genome and further analyzed two of them that possibly have functional roles during the conjugation stage. Through tagging with green fluorescent protein (GFP) and mCherry, we show that Atg8s are specifically associated with degrading nuclear structures. In addition, through genetic knockout studies, we show that these two genes have distinct functions in starvation and that both are critical for PND during conjugation. Our study demonstrates a clear role for autophagy in nuclear degradation and establishes a platform for further understanding of autophagy in eukaryotes.
MATERIALS AND METHODS
Cell culture, starvation, and mating induction.
Wild-type T. thermophila strains CU428 (Mpr/Mpr [VII, mp-s]) and CU427 (Chx/Chx [VI, cy-s]) were obtained from Peter Bruns (Cornell University, Ithaca, NY). All Tetrahymena strains were maintained in Neff medium at room temperature (10). Cells cultured for experiments were grown in Neff medium at 30°C. For studies of starvation sensitivity, log-phase cells were washed in 10 mM Tris buffer (Tris-HCl; pH 7.4) and incubated in the same buffer at 30°C for the indicated time periods. For studies of the pairing rate and conjugation process, stationary-phase cells were starved in diluted Neff medium by directly adding 9 volumes of Tris buffer to the cell culture.
Identification of autophagy-related genes.
The identification of autophagy-related genes in Tetrahymena was based on computational comparison. Amino acid sequences of the functional domains predicted from yeast autophagy genes, including ATG1 to ATG10, ATG12 to ATG14, ATG16, and ATG18, were obtained from the Pfam database (48). Those amino acid sequences were used as probes for analysis using BLAST of proteins with a similar functional domain in the Tetrahymena genome database (TGD; http://ciliate.org/index.php/home/welcome). Full-length amino acid sequences of those genes with E values lower than 0.05 were reanalyzed against the Pfam database to verify the existence of the functional domains. Only those genes encoding proteins with confirmed functional domains were chosen as candidate genes.
Construction of gene disruption vectors.
ATG8-2 and ATG8-65 were disrupted by using pNeo4-ATG8-2-KO and pNeo4-ATG8-65-KO vectors. Approximately 1-kb sequences upstream and downstream of the DNA sequences of ATG8-2 and ATG8-65 genomic loci were cloned into each knockout vector as the 5′- and 3′-flanking sequences. The backbone of these vectors is the pNeo4 vector, which contains neoTet, a Tetrahymena codon-optimized neomycin resistance gene (gift from Mochizuki Kazufumi, Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria) (40). neoTet is under the control of the promoter of the Cd2+-inducible metallothionein 1 (MTT1) gene. Before biolistic transformation, vectors were linearized by restriction enzyme digestion to release the knockout fragment with the neoTet cassette (8). The neoTet cassette was inserted into the locus of ATG8-2 or ATG8-65 by homologous recombination to replace the entire coding sequence. Transformants were selected in SPP medium containing 120 μg/ml of paromomycin (Sigma-Aldrich, St. Louis, MO) and 1 μg/ml of CdCl2 (Sigma-Aldrich, St. Louis, MO). Germ line knockout candidates were confirmed by Southern blotting.
Construction of fluorescent protein fusion vectors.
The construction of fluorescent protein fusion vectors was carried out by following the Gateway cloning protocol (Invitrogen, Carlsbad, CA). Fluorescent protein-tagged Atg8ps were overexpressed from pVGF-Opti-EGFP-ATG8-2 and pVGF-Opti-mCherry-ATG8-65, which were recombinant products of destination vectors pVGF-Opti-EGFP-gtw and pVGF-Opti-mCherry-gtw and entry vectors pCR8-ATG8-2 and pCR8-ATG8-65. For the construction of the destination vectors, the Gateway cassette with attR sites was amplified from the pIGF-gtw vector (gift from Douglas Chalker, Washington University, St. Louis, MO) by using a primer set containing restriction sites for XhoI and ApaI. The PCR-amplified product XhoI-Gateway cassette-ApaI was used to replace the sequence between these two enzymes in the pVGF vector (gift from Aaron Turkewitz, The University of Chicago) to yield the vector pVGF-gtw. The enhanced GFP-tagged and the monomer mCherry-tagged Tetrahymena codon-optimized sequences from the pEGFP-neo4 and pmCherry-neo4 vectors (gifts from Mochizuki Kazufumi) were used to replace the monomer GFP in the pVGF-gtw to generate the destination vectors. For the construction of entry vectors, targeted regions ATG8-2 (381 bp) and ATG8-65 (369 bp) were amplified from reverse-transcribed cDNA of conjugating cells (4-hour stage) by PCR and cloned into the pCR8/GW/TOPO vector (Invitrogen, Carlsbad, CA) between attL sites. Finally, the recombination between destination vectors and entry vectors was performed by LR reaction (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol to generate the expression vectors.
RT-PCR.
RNA preparation and quantitative PCR experiments were performed as previously described (14). In brief, total RNA samples were isolated from vegetative, starved, and conjugating cells at 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 h after cell mixing by using an RNA isolation kit (Roche Applied Science, Indianapolis, IN). Transcriptor reverse transcriptase (RT; Roche Applied Science, Indianapolis, IN) was used to synthesize the first-strand cDNAs by using an anchored oligo(dT)18 primer. Primer sets for ATG8-2, 5′-ATAGAATAGAAAGTGCATCTCC-3′ and 5′-TCTATTTATTGGTTCAGTCG-3′, and ATG8-65, 5′-AATATCAGGCTGTATGAAGA-3′ and 5′-ATACATACACCTACAAAAGAGC-3′, were used for quantitative PCR experiments. Relative quantifications of ATG8-2 and ATG8-65 mRNA expression levels were normalized using α-tubulin mRNA expression as an internal control. Vegetative cells in this study refer to growing cells. They were usually diluted from stocks 100-fold in SPP or Neff medium and cultured for 14 to 16 h at 30°C. Vegetative cells that were starved in Tris buffer for 14 to 16 h at 30°C are referred to as starved cells.
Southern blotting.
Genomic DNA for Southern blotting was isolated from wild-type or ATG8 knockout strains and digested with PacI and HindIII. Digested DNA samples were analyzed by electrophoresis in a 0.8% agarose gel. DNA was transferred and adhered to a nylon membrane (Immobilon-NY+; INYC00010; Millipore) by UV cross-linking. The membrane was hybridized with probes overnight at 42°C in digoxigenin (DIG) Easy Hyb buffer (Roche Applied Science, Indianapolis, IN). Digoxigenin-labeled probes were made by using DIG-High Prime (Roche Applied Science, Indianapolis, IN) labeling reaction mixtures with PCR products amplified from genomic DNA. After hybridization, the membrane was washed several times in 2 to 0.5× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS. Hybridized DNA was detected by using anti-DIG–allophycocyanin (AP), Fab fragment (Roche Applied Science, Indianapolis, IN), and disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD; Roche Applied Science, Indianapolis, IN) as AP substrate. The membrane was then exposed to X-ray film for several minutes to several hours.
Fluorescent dye labeling of living cells.
Acidic organelles in mating cells were labeled using 1 μl of 200-fold-diluted reagent of the Lyso-ID Red lysosomal detection kit (Enzo Life Sciences, Farmingdale, NY) in 500-μl samples. A nuclear counterstain was performed with Hoechst 33342 (4 μg/ml; Sigma-Aldrich, St. Louis, MO). Living cells in 10 mM Tris buffer were stained with these fluorescent compounds at 30°C for at least 30 min. After incubation, cells were treated with NiCl2 (1 mM; Sigma-Aldrich, St. Louis, MO) for an additional 10 min and then immediately analyzed by fluorescence microscopy.
Conjugation progression and DNA damage detection.
For monitoring conjugation progression, mating cells at the stages of conjugation at 2, 4, 6, 8, 10, 12, 14, 16, 18, 24, 48, and 72 h were fixed in 2% paraformaldehyde. After fixation, cells were washed twice with phosphate-buffered saline (PBS) buffer. Cellular nuclei were then stained with 40 ng/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO) for evaluation of nuclear states. For DNA damage detection, a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed using the In Situ Cell Death Detection kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. In brief, paraformaldehyde-fixed cells were attached to glass slides coated with poly-l-lysine (Sigma, St. Louis, MO). Cells were permeabilized with PBS buffer containing 0.3% Triton X-100 at 30°C for 30 min. After washing out the detergent with PBS buffer, permeabilized cells on slides were incubated with TUNEL reaction mixture at 37°C for 4 h. After the reaction, cellular nuclei were visualized by DAPI staining, and the results of TUNEL staining were examined by fluorescence microscopy.
RESULTS
Autophagy-related genes in Tetrahymena.
To determine whether the autophagy machinery is conserved in Tetrahymena, we searched for autophagy-related genes (ATG) in the Tetrahymena genome database TGD (http://ciliate.org/index.php/home/welcome). Since the autophagy pathway has been well-characterized in Saccharomyces cerevisiae (29), amino acid sequences from the functional domains of each yeast autophagy protein were used to probe the database (see Materials and Methods). The analysis showed that most genes of the core autophagy machinery could be found in the Tetrahymena genome, including ATG1s, ATG3, ATG4s, ATG5, ATG6, ATG7, ATG8s, ATG10, ATG16, and ATG18 (see Table S1 in the supplemental material). This finding suggests that Tetrahymena, like other eukaryotes, contains the autophagy machinery.
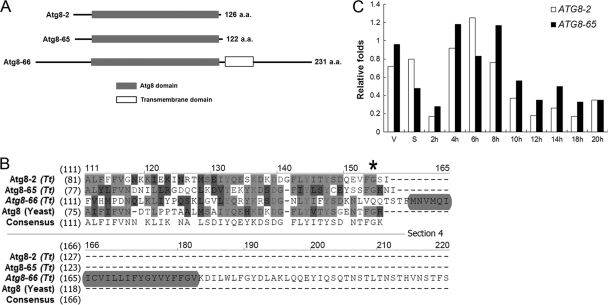
In order to understand the process of autophagy in Tetrahymena, especially in nuclear reorganization during sexual reproduction, we first focused on the autophagy marker protein Atg8p. Atg8p is known to be a ubiquitin-like protein in several eukaryotes and is required for autophagosome formation (60). Interestingly, Tetrahymena, like humans, has more than one ATG8 gene in its genome: TTHERM_00037460 (ATG8-2), TTHERM_00522490 (ATG8-65), and TTHERM_00526360 (ATG8-66). The conserved yeast Atg8p functional domain can be found in all three Tetrahymena Atg8ps, but a predicted transmembrane domain is located at the carboxy terminus of Atg8-66p (Fig. 1A). Atg4p is the key posttranslational processing enzyme that exposes the carboxyl-terminal glycine of Atg8p for conjugation to phosphatidylethanolamine (PE), and this carboxyl-terminal processing is also critical for Atg8p functions (31). We thus checked for the presence of the corresponding glycine residue in each Tetrahymena Atg8p by comparisons with yeast Atg8p. The glycine residue was found in Atg8-2p and Atg8-65p but not in Atg8-66p (Fig. 1B). These findings suggested that Atg8-2p and Atg8-65p, but not Atg8-66p, could be the functional homologues of Atg8ps in Tetrahymena; hence, we focused our studies on these two genes.
Fig 1.
Characterization of Atg8ps in Tetrahymena. (A) Schematic representation of Atg8 proteins. Black solid lines show the full lengths of the Atg8 proteins. (B) Multiple-sequence alignment of the C-terminal region of Tetrahymena Atg8 proteins. The asterisk indicates the position of the conserved C-terminal glycine residue. Light gray, conservative or identical amino acids; dark gray, similar amino acids; light gray rectangle, the transmembrane domain of theAtg8-66 protein. (C) Quantitative RT-PCR findings for ATG8 mRNA expression levels. Total RNA samples isolated from vegetative (V), starved (S), and conjugating cells (2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 h after postmixing) were used as templates. Quantification was normalized by using α-tubulin mRNA.
In order to determine the gene expression pattern at different stages of the Tetrahymena life cycle, RT-PCR analyses of ATG8-2 and ATG8-65 mRNA expression were carried out. The results showed that both genes were expressed during vegetative growth, starvation, and conjugation. The expression levels of both genes were higher at the 4- to 8-h stage of conjugation than during starvation (Fig. 1C). Since this stage coincides with the time of degradation of meiotic products and the beginning of PND (Fig. 2A), the data support the possibility that these two genes play a role in nuclear degradation.
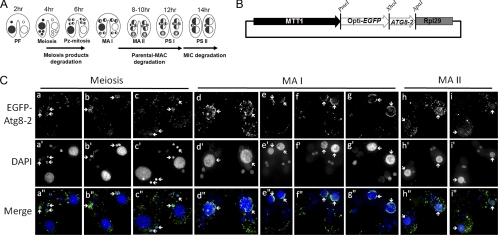
Fig 2.
EGFP–Atg8-2p locates to degenerating nuclei during conjugation. (A) Developmental process and nuclear degradation events of Tetrahymena during conjugation. PF, pair formation; Pz-mitosis, postzygotic mitosis; MA I, macronuclear anlagen I; MA II, macronuclear anlagen II; PS I, pair separation I; PS II, pair separation II. The indicated times (hours of conjugation) show the approximate time period after mixing different mating type cells. (B) Schematic representation of the employed EGFP–ATG8-2 overexpression vector. MTT1, the promoter of the Cd2+-inducible metallothionein (MTT1) gene; opti-EGFP, Tetrahymena codon-optimized enhanced GFP; ATG8-2, Tetrahymena ATG8-2 gene; RPL29, terminator of the 60S ribosomal protein gene L29. (C) Localization of EGFP–Atg8-2ps in mating cells at different conjugation stages. EGFP–Atg8-2-overexpressing mating cells were fixed in 2% paraformaldehyde at 2-hour intervals. Cellular nuclei were stained with DAPI (40 ng/ml), and subsequently, fixed cells were classified into different stages according to cellular and nuclear morphologies. Fluorescent images were taken with a DeltaVision deconvolution microscope. Arrows, location of EGFP–Atg8-2–positive nuclei.
Atg8ps are localized in and around degrading macro- and micronuclei.
To obtain more direct evidence, the localizations of amino-terminal fluorescence-tagged Atg8-2p and Atg8-65p were analyzed during conjugation. Fluorescent-tagged Atg8ps (i.e., EGFP–Atg8-2p and mCherry–Atg8-65p) were expressed under the control of the MTT1 promoter, which was induced by the addition of cadmium (Fig. 2B and 3A) (50). We focused on the stages between late macronuclear anlagen I (MA I) and pair separation I (PS I), when PND of the pa-MAC occurred (Fig. 2A), and captured images by using a DeltaVision deconvolution fluorescence microscope to follow EGFP–Atg8-2p in cells fixed at different mating time points. As shown in Fig. 2C (panels d to i, arrows), EGFP–Atg8-2p appeared in degenerating pa-MAC but not in newly developing micronuclei (MIC) or macronuclei. Interestingly, the GFP signal in the peripheral areas of the pa-MAC appeared to be stronger than that inside the nucleus, suggesting that the degenerating nucleus might be enclosed by autophagic structures. Furthermore, by carefully reviewing mating cells at the MA I stage, we found GFP-positive structures either partially or almost completely surrounding pa-MAC (Fig. 2C, panels d to g, arrows). Three-dimensional (3D) reconstructions of the z-stack images of GFP fluorescence obtained using Imaris 6.4 (Bitplane) revealed clear spatial distributions of those GFP structures that were in the process of enveloping the nucleus (see Videos S2 and S3 in the supplemental material). This pattern agrees with the formation process of autophagic structures and is unlikely generated by random aggregations or other causes. It is noteworthy that the pa-MAC at this stage still appeared as normal nuclei in morphology and were without pronounced PND features (33). These results indicated that the Tetrahymena autophagic marker protein Atg8-2p could mark the pa-MAC at the beginning of the PND process.
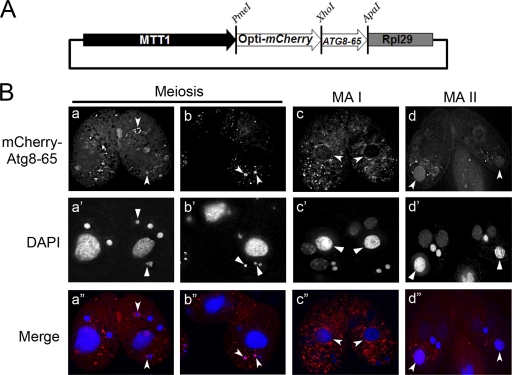
Fig 3.
mCherry–Atg8-65p is localized to degenerating nuclei during conjugation. (A) Schematic representation of the mCherry–ATG8-65 overexpression vector. (B) Localization of mCherry–Atg8-65ps in mating cells at different conjugation stages. The sample processing was the same as for samples shown in Fig. 2C, and images were taken with a DeltaVision deconvolution microscope.
Another PND process in ciliates is the elimination of three of the four meiotic products. To address whether autophagy also participates in this process, we examined cells at the stage of meiosis. We found that EGFP–Atg8-2p was associated with micronuclei and appeared to be enriched at the nuclear periphery (Fig. 2C, panels a to c, arrowheads; see also the 3D reconstruction Video S1 in the supplemental material). It is not possible to morphologically distinguish degrading micronuclei from those destined to produce the gametic nuclei. However, it is known that degrading nuclei tend to migrate to the posterior portion of the cell, while gametic nuclei remain in the anterior portion. To better correlate Atg8-2p with nuclear degradation, we calculated the percentage of GFP-marked MIC-sized nuclei at either the anterior or posterior area. The results showed that an average of 89.5 ± 2.0% (mean ± standard deviation) of posterior and 31.6 ± 18.9% of anterior MIC-sized nuclei (in a total of 312 pairing cells from at least three independent experiments) were marked with GFP. Taking into account that some nuclei may have been in early stages before degradation markers appeared, this result indicated that most degrading meiotic products were surrounded by EGFP–Atg8-2ps (Fig. 2C, panels a to c).
To determine whether Atg8-65p, like Atg8-2p, is also associated with degenerating meiotic products and pa-MAC, we tagged Atg-65p with mCherry and examined its distribution in mating cells. Although the background fluorescence was increased, we found that mCherry–Atg8-65ps was also associated with meiotic products and degrading pa-MAC (Fig. 3B). In addition, the fluorescence patterns, such as the strong peripheral distribution, were similar to those of EGFP–Atg8-2ps (Fig. 3B, panels a and b [meiotic products] and c and d [pa-MAC]).
During starvation, these fluorescent proteins form distinctive vesicular structures consistent with the appearance of autophagosomes. Their associations with Lyso-ID staining signals are also consistent with their relationship with lysosomes (see Fig. S2 and Videos S4 and S5 in the supplemental material).
No acidification occurred before EGFP–Atg8-2p completely labeled the periphery of degenerating nuclei.
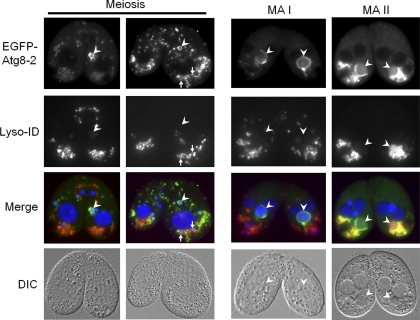
After observing the EGFP–Atg8-2p–positive structure, we were curious about its temporal formation relative to other events, such as chromatin condensation, nuclear compaction, and acidification of degrading nuclei. Chromatin condensation was assessed based on an increase in Hoechst staining intensity, and nuclear compaction was measured by decreases in nuclear size (33). Nuclear acidification was determined by staining with an acidic organelle indicator, Lyso-ID Red dye (15). We thus measured these parameters in cells at the MA I and MA II stages (Fig. 4), and we found that the mean size of the pa-MAC with a peripheral EGFP signal but without Lyso-ID Red dye staining was 6.62 μm ± 1.02 μm (n = 45; data from three individual experiments). In contrast, the average size of the pa-MAC with a peripheral EGFP signal and Lyso-ID Red dye staining was 5.17 μm ± 0.42 μm (n = 54; data from three individual experiments). In the same batch of the sample, the size of the pa-MAC in nonmating cells or mating pairs at earlier stages was 10.7 μm ± 1.63 μm (n = 53; data from three individual experiments). The size differences among these three types of nuclei were highly significant according to Student's t test (P < 0.005). In addition, we observed relatively more intense Hoechst staining in the pa-MAC of cells at the MA I stage that showed EGFP signal but no Lyso-ID Red dye staining than in cells at an earlier stage. This result showed that the pa-MAC was associated with the Atg8-2p-positive autophagic structure before acidification occurred, about the time when chromatin condensation and nuclear compaction begin to occur, indicating that autophagic events happen before lysosome fusion.
Fig 4.
EGFP–Atg8-2p labeling of the parental macronucleus occurs before nuclear acidification and condensation. The images show localization of EGFP–Atg8-2ps in mating cells at different conjugation stages, based on Lyso-ID Red dye staining. EGFP–Atg8-2p expression was induced when starved cells of different mating types were mixed. Live pairing cells were stained with Lyso-ID Red dye for acidified structures and Hoechst for DNA at conjugation, from 4 to 9 hours. Images were taken by using a conventional fluorescence microscope with differential interference contrast (DIC). Arrows, meiotic products with EGFP and Lyso-ID Red dye staining; arrowheads, nuclei with EGFP staining.
ATG8-65 but not ATG8-2 is critical for surviving starvation.
Although autophagy has been studied in several protozoan model organisms (6, 9, 36), little is known about autophagy, especially the functions of ATG genes, in ciliated protozoans. To facilitate this understanding, we used gene knockout approaches to create Tetrahymena strains defective in autophagy genes. We independently deleted ATG8-2 and ATG8-65 from the genome by homologous recombination (40). As shown in Fig. 5A and B, Southern blot analysis confirmed the correct replacements of the ATG8 genomic loci with the inserted Escherichia coli neomycin resistance gene cassette (neo). Two complete knockout strains (created by removing all gene copies in both the MIC and MAC genomes; CKO–ATG8-2 and CKO–ATG8-65) were successfully generated and further characterized for their deficiencies.
Fig 5.
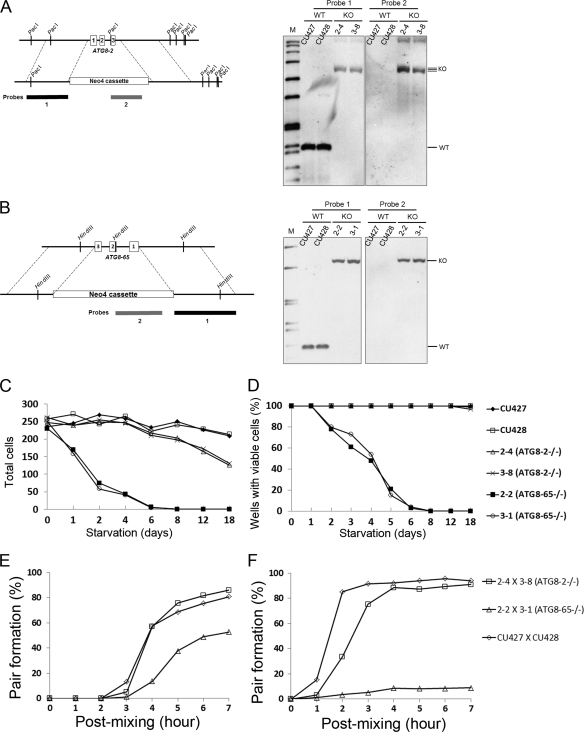
The ATG8-65 homozygous knockout was sensitive to starvation. (A, left) Schematic showing the ATG8-2 genomic locus before (upper) or after (lower) homologous recombination with the neo4 knockout cassette. Numbered boxes represent three exons. Probe 1 (1,093 bp) was used to detect the insertion accuracy of the knockout cassette. Probe 2 (0.7 kb) was used to ensure the existence of the neo knockout cassette. (Right) Southern analysis of PacI-digested genomic DNA isolated from ATG8-2−/− (2-4 and 3-8) or wild-type (CU427 and CU428) strains. KO (knockout) bands, 4.5 kb; WT (wild-type) bands, 1.5 kb; M (marker), DIG labeling marker. (B, left) Schematic showing the ATG8-65 genomic locus before (upper) and after (lower) knockout cassette replacement. The representation of numbered boxes and functions of probe 1 (1,055 bp) and probe 2 (0.7 kb) are the same as for panel A. (Right) Southern analysis of HindIII-digested genomic DNA from ATG8-65−/− (2-2 and 3-1) or WT strains. KO bands, 3.1 kb; WT bands, 1.1 kb; M, DIG labeling marker. (C) Remaining cells in ATG8 knockout or wild-type starvation pools. Cells starved in 10 mM Tris buffer for 0, 1, 2, 4, 6, 8, 12, and 18 days were sampled (200 μl) from starvation pools and fixed in 2% paraformaldehyde. Total cell numbers represent the averages of three independent samples taken from the same starvation pool. (D) Percentages of wells with viable cells in a 96-well plate seeded with starved ATG8 knockout or wild-type cells. A 25-μl aliquot of starved cells (containing an average of 30 input cells at the beginning of starvation) was sampled from the same starvation pools and at the same time point as shown in panel C and transferred to 96-well plates with rich SPP medium (50 μl) for incubation at 30°C. Three days later, wells were examined for the presence of viable cells. Percentages represent averages from three 96-well plates. Panels C and D share the same labeling as that shown at the right side of panel D. (E and F) Mating ability of ATG8 knockout and wild-type starved cells. Vegetative cells grown in Neff medium were directly diluted with Tris buffer to generate a 10-fold-diluted Neff solution for starvation. Cells of different mating types were either mixed immediately after dilution (E) or later after overnight prestarvation (F). Cells were fixed at 0, 1, 2, 3, 4, 5, 6, and 7 h postmixing to calculate the pairing rate. Panels E and F share the same labeling as shown at the right side of panel F. Percentages represent the average values of three independent samples from two individual experiments.
We measured the cell number and the viability of CKO-ATG8 cells under starvation conditions. To minimize the opportunity of feeding on cell debris, we starved cells in 10 mM Tris buffer at a very low cell density (2 cells per μl, as opposed to the conventionally used starvation cell density of ∼200 cells per μl). After starvation, a sample of cells was fixed and counted under a microscope. Total cell numbers of starved CKO–ATG8-65 strains decreased to near zero at day 6 of starvation (Fig. 5C, open circles and closed squares); by contrast, total cell numbers of CKO–ATG8-2 strains (crosses and open triangles) were similar to those of the control normal strains (closed diamonds and open squares). To further test the ability of these cells to grow and divide, we also transferred cells at the same starvation time points to growth medium in 96-well plates, with each well containing an average of 30 input cells at the onset of starvation. Cells in about 90% of wells in plates of both control and CKO–ATG8-2 strains grew, even after 18 days of starvation (Fig. 5D), indicating that at least one cell per well survived prolonged starvation. However, none of the cells in plates containing a CKO–ATG8-65 strain grew after 6 days of starvation. These results further supported the higher sensitivity of CKO–ATG8-65 cells to starvation. Since the number of CKO–ATG8-65 cells that were able to grow appeared to be significantly lower than those visible under the microscope, this strain appeared to have lost its ability to divide long before it lysed.
Since Tetrahymena needs low nutrient conditions to induce conjugation (3), we were interested in finding out whether the different sensitivities to starvation affected their abilities to form pairs. CKO–ATG8-65 cells were unable to form pairs when starved under conventional starvation conditions (10 mM Tris buffer). To improve mating, we modified the conditions by using a greatly diluted medium (10-fold-diluted Neff medium [see below]). ATG8 knockout or control strains (CU427 and CU428) were grown in Neff medium to saturation and then diluted 10-fold with 10 mM Tris buffer. In this mild starvation solution, different mating types were either mixed immediately (Fig. 5E) or after 16 h (Fig. 5F). To monitor pair formation, cells were mixed and counted under the microscope at 1-h intervals. As shown in Fig. 5E and F, rates of pair formation of CKO–ATG8-2 and wild-type cells were similar, with ∼90% of cells finally forming pairs with or without prestarvation. However, CKO–ATG8-65 cells had much lower pairing rates under both conditions, with close to 9% and 50% of cells forming pairs with and without prestarvation, respectively. Taken together, these results showed that ATG8-65 and ATG8-2 genes play distinctive roles in starvation. ATG8-65 appears to be crucial for cells to form stable pairs in the beginning of the conjugation process as well as for cell survival under prolonged starvation. ATG8-2 has a relatively minor role in these processes.
ATG8-2 and ATG8-65 are important for nuclear degradation.
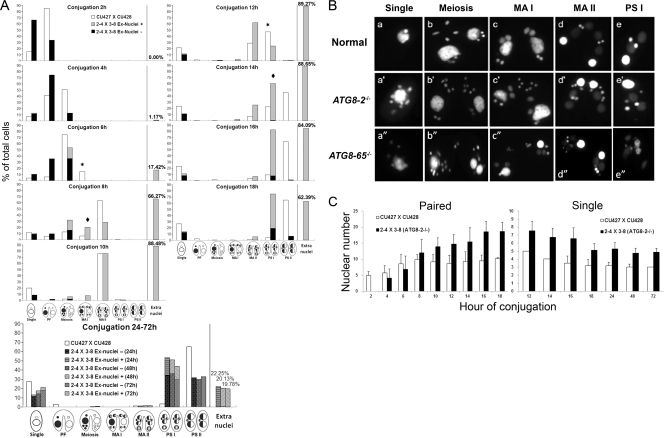
If nuclear degeneration involves autophagy, one would expect that mutations in ATG8s might delay or block this process. To test this idea, we mated CKO–ATG8-2 cells and looked for any nuclear degradation defects during conjugation. After overnight starvation in the 10-fold-diluted Neff medium, CKO–ATG8-2 cells of different mating types were mixed to induce conjugation, collected at the indicated time points, and examined by microscopy after fixation and staining. CKO–ATG8-2 cells showed an approximately 2-hour delay in mating progression, compared with control strains (CU427 and CU428), and contained a notable population of cells with extra nuclei after conjugation for 6 h (Fig. 6A), which was approximately the stage when the degradation of meiotic products occurred in normal cells (Fig. 6A, conjugation at 4 h, and 2A). Remarkably, those extra nuclei still remained in a large proportion of cells, even at 72 h (Fig. 6A, conjugation at 24 to 72 h), while conjugation was largely completed within 20 h for normal cells. Nonetheless, small proportions of CKO–ATG8-2 cells with normal nuclear configurations for the PS I and PS II stages could be observed after 24 h (Fig. 6A, conjugation at 24 to 72 h), indicating that those meiotic products were being degraded, albeit slowly. These results thus indicated that ATG8-2 deletion caused a severe delay in nuclear degradation in Tetrahymena.
Fig 6.
ATG8 complete knockout cells carried the extra nuclei phenotype caused by delayed nuclear degradation. (A) Mating progression of ATG8-2 knockout or wild-type mating cells. Mating cells at 2, 4, 6, 8, 10, 12, 14, 24, 48, and 72 h after conjugation were fixed in 2% paraformaldehyde and treated with DAPI (40 ng/ml) for staining of nuclei. Cells were classified into different stages according to cellular and nuclear morphologies. The compositions of cells with regard to different developmental stages at each examined conjugation time point are shown. Percentages represent the average values of three independent samples. White bars, wild type; black bars, mutant with normal nuclear number; gray bars, mutant with extra nuclei; asterisks, wild type; rhombuses, ATG8-2 knockout. Time intervals indicate the time period of new MAC development. (B) Nuclear morphology of wild-type and ATG8 knockout mating cells with aberrant nuclear configurations at different stages. Images were taken with a conventional fluorescence microscope. Single, single cells, including nonmating cells and pairs that have separated. (C) Nuclear numbers of pairing cells or single separated progeny sampled at the indicated time points. Percentages represent the average values of three independent samples from two individual experiments.
ATG8-65 also appeared to be important for nuclear degradation. Unlike CKO–ATG8-2 cells, CKO–ATG8-65 mutants have severe defects when starved and cannot mate under traditional conditions. To minimize the starvation effect, we mixed CKO–ATG8-65 cells in the diluted growth medium without prestarvation. As shown in Fig. 6B, panels a″ to e″, CKO–ATG8-65 mating cells were able to mate, although at low efficiency, under these conditions and showed the same extra nuclei phenotype as CKO–ATG8-2. These results strongly suggested that both Atg8-2p and Atg8-65p play important roles in the nuclear degradation of Tetrahymena.
By comparing the nuclear morphology of these mutant mating cells with those of normal cells, it appeared that the extra micronuclei in CKO–TtATG8-2 cells are likely the meiotic products (Fig. 6B, panels b′ to d′). Without nuclear degradation, the total number of nuclei in a pair should increase from four (one MIC and one pa-MAC in each cell) at the pair formation (PF) stage to a maximum of 16 (one pa-MAC, three meiotic products, and four postzygotic nuclei in each cell) at the postzygotic (PZ) mitosis stage. Indeed, as shown in Fig. 6C, the nuclear numbers of CKO–ATG8-2 pairs reached 16, with some of them even exceeding this number. In contrast, the nuclear number of control pairs never reached 16, mainly due to the degradation of meiotic products (Fig. 6C). These results showed that the meiotic products in CKO–ATG8-2 cells were not degraded as efficiently as those in normal cells. The presence of some CKO–ATG8-2 pairs with more than 16 nuclei may have been due to possible replication of those meiotic products not destined to become the gametic nuclei.
Following the separation of pairs, the delayed nuclear degradation phenotype was still apparent (Fig. 6C). At these stages (PS I and PS II), the nuclear number of normal cells decreased from five (one old MAC, two new MIC, and two new MAC) to three (one new MIC and two new MAC) by degrading the pa-MAC first and then one of the two new MICs (Fig. 2A, PS I and PS II stages). In contrast, most CKO–ATG8-2 cells retained the old MAC and both new MICs. However, after extended periods (Fig. 6A, conjugation at 24 to 72 h), all the nuclei that were destined to be degraded (one old MAC, one of the two new MICs, three meiotic products, and extra replicating meiotic products) were eventually eliminated from CKO–ATG8-2 cells. These results showed that the extra nuclei phenotype of CKO–ATG8-2 cells was caused by severely delayed nuclear degradation, suggesting a critical role for ATG8-2, and thus autophagy, in PND in Tetrahymena during conjugation.
Development of new macronuclei was not affected by the delay in nuclear degradation.
To determine whether ATG8-2 mutation also affected other developmental events, we examined the formation of the new MAC (Fig. 2A) in cells with extra nuclei. We classified those mutant cells into the meiosis, MA I, MA II, and PS I stages by comparing them with normal cells (Fig. 6B). The percentages of control or CKO–ATG8-2 cells at different developmental stages were determined (Fig. 6A). Because of the 2-hour delay in pair formation, the developmental progression of mutant cells was slower than that of normal cells by about 2 h. However, both mutant and normal cells required about 6 h to develop and form prominent new MAC from the zygotic nucleus (Fig. 6A [asterisks for normal cells and rhombuses for ATG8-2 knockout cells]). This result showed that the development of new MAC occurred on time despite the delay in nuclear degradation, indicating the lack of a critical role for autophagy or nuclear degradation in new macronuclear development.
No acidification occurred, but DNA digestion remained in the old MAC of CKO–ATG8-2 cells.
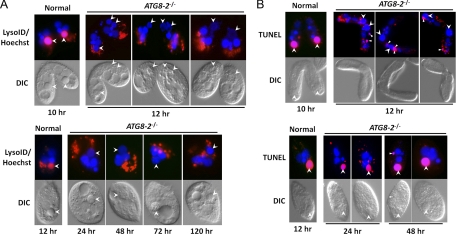
The execution of autophagy is achieved through the fusion of autophagosomes with lysosomes to form acidic organelles, autolysosomes. The acidified event of autophagosomes can be examined by staining with Lyso-ID Red dye. To examine whether acidification occurred in the old MAC of CKO–ATG8-2 cells, we stained mating cells with Lyso-ID Red dye. As shown in Fig. 7A, the old MAC of normal cells showed strong Lyso-ID Red dye staining patterns. On the contrary, the old MAC of mutant cells showed no or background level of Lyso-ID Red dye staining (about 100 cells were examined at each time point), indicating that no pronounced acidification occurred in the old MAC of CKO-ATG8-2 cells. We also measured the size of the old MAC in mutant and normal pairs at the MA II stage and found it to be 6.97 μm ± 0.93 μm (n = 83, from three individual experiments) and 5.41 μm ± 0.56 μm (n = 47, from three individual experiments), respectively. This result showed that the old MAC in the mutant could still undergo some degree of compaction without autophagy, indicating that autophagy is not required for the initiation step of nuclear compaction.
Fig 7.
Degenerating nuclei of ATG8-2 knockout cells showed DNA damage but no acidification. (A) Lyso-ID Red dye staining of ATG8-2 knockout mating cells. Living mating cells at 10 and 12 h (wild type) or at 12, 24, 48, 72, and 120 h (knockout) of conjugation were stained with Lyso-ID Red dye for acidified structures and Hoechst for nuclei. (B) TUNEL assay results of ATG8-2 knockout mating cells. Mating cells after conjugation for 10 and 12 h (wild type) or at 12, 24, and 48 h (knockout) after conjugation were fixed in 2% paraformaldehyde and then incubated with a TdT (terminal deoxynucleotidyl transferase) mixture solution (Roche Applied Science, Indianapolis, IN) after permeabilization. Images were taken by using a conventional fluorescence microscope. Arrowheads, old macronuclei; triangles, degrading meiotic products.
Because the extra nuclei of CKO–ATG8-2 cells eventually disappeared, we wondered whether DNA degradation still occurred in the old MAC of CKO–ATG8-2 cells even without acidification. We employed a TUNEL assay to detect DNA degradation. As shown in Fig. 7B, the old MAC of both mutant and normal mating cells showed strong TUNEL-positive staining, suggesting that DNA degradation occurred in the old MAC of CKO–ATG8-2 cells. However, 90% of the TUNEL signal was detected after pair separation (at the PS II stage but not at the MA II stage, as in normal cells), indicating a significant delay of DNA degradation in knockout mutants. These data also implied that ATG8-directed autophagy is not the only machinery responsible for degradation of the old MAC in Tetrahymena.
DISCUSSION
Programmed nuclear death of the parental (old) macronucleus is a special feature of the binucleated species Tetrahymena thermophila. How these nuclei are degraded is an interesting question that remains mostly unanswered. During PND, DNA degradation is perhaps the most prominent molecular event that has received some attention. The autophagy machinery has also been suggested to play a role, possibly for delivering mitochondrial endonuclease to the old MAC (1, 2, 33), but its exact nature has not been clearly established. In fact, the role of autophagy in destroying an entire nucleus has not been known in any other organism. Thus, the study of autophagy genes with regard to the PND process of Tetrahymena should expand our general understanding of autophagy. Through the study of two ATG8 genes, we have demonstrated a prominent role for autophagy in Tetrahymena PND and revealed several interesting features of this process.
Autophagy-related genes in Tetrahymena.
Autophagy has been known to be the process that recycles cytoplasmic compartments by using the double membrane structure, the autophagosome, to fuse with the vacuole/lysosome for degradation (64). This process has been extensively studied in the unicellular eukaryote Saccharomyces cerevisiae. Autophagy-related process also have been observed in other unicellular organisms, e.g., trichomonads (5), trypanosomatid protozoa, and amoebae. The trypanosomatid protozoan Leishmania major has one Atg3, Atg5, Atg7, Atg12, and Atg16 proteins, two Atg4 proteins, and remarkably, 26 Atg8-related proteins (7, 58, 59). In two other trypanosome species, Trypanosoma cruzi and Trypanosoma brucei, one Atg3, Atg7, and Atg16 proteins and two Atg4 and Atg8 proteins have been found (4, 23). In the soil amoeba Dictyostelium discoideum, 10 autophagy genes, ATG1 to ATG9 and ATG12, have been found in its genome (44, 45). Most of the above species contain the entire Atg8 ubiquitin-like conjugation system but lack parts of the Atg12 conjugation machinery (Atg5, Atg10, Atg12, and Atg16). In this study, we also identified autophagy genes in the Tetrahymena genome by an in silico analysis based on their similarity to yeast genes (see Table S1 in the supplemental material). The result indicates the loss of Atg12 and Atg16 of the Atg12 conjugation machinery, suggesting that Tetrahymena may harbor only a concise form of autophagy. However, it should be noted that some Atg genes might have escaped detection by this method due to the evolutionary distance between yeast and Tetrahymena.
Another notable result is the finding of 60 ATG1/ULK1/UNC-51 candidate genes in Tetrahymena. Recent studies have suggested that multiple Atg1 proteins could have distinctive cellular functions through differences in their specificities in interacting with various binding proteins (12, 13, 19, 21, 24, 27). Since Tetrahymena carries such a large number of ATG1 genes, it would be interesting to find out how they may participate in the regulation of various aspects of autophagy in this one-celled organism.
Autophagosome formation in Tetrahymena.
Long-lived proteins or entire organelles can be degraded by autophagy through the specialized double membrane structure the autophagosome. The formation of the autophagosome starts with an isolated membrane structure, the phagophore, at the phagophore assembly site (PAS) (60). A core autophagy protein, Atg8, plays a crucial role in this process (25, 47). The carboxyl-terminal glycine residue of newly synthesized Atg8 is exposed by an autophagic cysteine protease, Atg4 (22). The glycine-exposed Atg8 is then conjugated to a lipid moiety, PE, by the activating enzyme Atg7 and the conjugating enzyme Atg3 (52, 53). The Atg12 conjugation machinery, the Atg12-Atg5-Atg16 complex, facilitates the conjugation of Atg8 to the membrane of the phagophore (20). The phagophore elongates its membrane through fusion with other phagophores mediated by the potential hemifusion function of Atg8 (60). Therefore, Atg8 localizes to the preautophagosomal structure and the mature autophagosome during autophagy (28, 31). Atg8 on the leaflet of the outer-most double lipid bilayer of the autophagosome can be deconjugated by the same processing enzyme, Atg4 (31). Some Atg8 trapped on the inner-most leaflet is eventually degraded after the autophagosome fuses with lysosomes (31, 51).
Previous studies have been based largely on yeast or mammalian systems. In this study, we investigated the role of Atg8 in the free-living protozoan Tetrahymena. Using the GFP-tagged Atg8, we found that the perinuclear distribution pattern of this protein resembled the process of autophagosome formation (Fig. 2C, 3B, and 4). These results suggest that the perinuclear GFP-positive structure is either the preautophagosomal structure or autophagosome. Previously, a lysosome wrapping model was proposed to explain how the macronucleus are transformed into an acidified organelle and degraded (34). It suggested that lysosomes, instead of phagophores, were located at the periphery of the old MAC, and a lysosomal enzyme, acid phosphatase, was detected to support this idea. However, the detection of this lysosomal enzyme occurred relatively late in conjugation, at the MA II stages, when the old MAC has already been moved to the posterior end of the mating cell. In this study, we found the wrapping of nucleus by Atg8 in the earlier MA I stages, in which the old MAC still positioned in the anterior part of the cells (Fig. 4). We observed neither the surrounding perinuclear lysosomes nor the acidification of the old MAC in this stage. Therefore, lysosome wrapping occurs after autophagosome formation and appears to be a later step in nuclear degradation in Tetrahymena.
Function of Atg8 proteins in Tetrahymena.
There is only one Atg8 protein in yeast, but there are eight ATG8 orthologous proteins in mammalian systems that belong to the LC3, GABARAP, or GATE-16 families. Functional diversification among proteins from these three families has been proposed (54). A recent study suggested that LC3s could work in the step of phagophore elongation and that the GABARAP/GATE-16 subfamily of proteins could function in the later autophagosome completion step (56). From our ATG8 knockout experiments, we also found clear differences between ATG8-2 and ATG8-65 of Tetrahymena. ATG8-65 alone appears to be required in starvation, but both ATG8-2 and ATG8-65 are important for nuclear degradation. During nuclear degradation, it is possible that Atg8-65p may function in the initiation and expansion of phagophore to form early autophagosomes and that Atg8-2p may specifically be involved in the fusion between autophagosomes at several peripheral regions of the old MAC to complete the huge autophagic structure. In this case, genetic loss of either ATG8-2 or ATG8-65 would lead to a significant delay of old MAC degeneration. Whether this is the case requires additional molecular analysis.
Elimination of the macronucleus in Tetrahymena.
The degradation steps of autophagy normally depend on fusion with lysosomes, from which the degrading enzymes are derived (62). A lysosomotropic agent, Lyso-ID Red dye, was used to follow this fusion event in Tetrahymena. The Lyso-ID dye-like Lysotracker Red (LTR) is a cationic amphiphilic fluorophore tracer that readily partitions into cells and allows detection of acidic vesicles derived from the lysosomal pathway. Since the staining of Lyso-ID Red dye is not pH dependent, any increase in fluorescence intensity is used to indicate accumulation of the probe within lysosomal structures (15). By analyzing mating cells with complete or incomplete peripheral GFP–Atg8-2p structures, a significant increase of Lyso-ID signals was observed only after the completion of the GFP structure, indicating that the formation of autophagic structure occurred before the old macronucleus became a lysosomal vesicle and trapped this lysosomotropic agent.
Atg8 is important for autophagosome formation and also for its subsequent fusion with lysosomes (26, 30, 46). In our genetic knockout strains, the Lyso-ID staining structures were located far away from the old macronucleus, while they were colocalized with the old MAC in normal cells. In addition, acidification of the old MAC was not detected even at much later time points. Thus, Atg8 in Tetrahymena appears to play a crucial role for the eventual fusion of lysosome with the old MAC, presumably through autophagosome formation.
This lack of lysosome fusion in the knockout mutants would suggest that a putative DNase II-like Tetrahymena lysosomal DNase would encounter difficulties in delivery to the old MAC (26, 49). In addition, without functional autophagy, another putative Tetrahymena mitochondrial endonuclease, which has an activity similar to that of mammalian endonuclease G (EndoG), also cannot be transported into the nucleus through autophagosomes. Remarkably, even without lysosome fusion and acidification, we detected DNA degradation in the old MAC in a TUNEL assay, albeit with some delay. This result suggests that these endonucleases enter the old MAC through a different pathway, or that at least one other nuclease, whose transport is autophagosome-lysosome independent, is responsible for the observed DNA degradation.
Because of the similarity between DNA fragmentation in PND and in mammalian apoptosis (16, 17, 33), a prototypical apoptotic pathway may exist in Tetrahymena and be responsible for nuclear degradation. In the apoptotic process, caspase-activated DNase (CAD) serves a similar function as DNase II or EndoG to cleave DNA (42). It is possible that a putative CAD-like DNase is activated and translocated into the old MAC for DNA degradation during this process. Since a CAD homologue has not been identified in the Tetrahymena genome, this idea cannot be easily tested. Nonetheless, it remains likely that the nuclear degradation process in Tetrahymena requires an apoptosis-like nuclease to complete the DNA degradation process.
In ATG8 knockout cells, the elimination of the old MAC still proceeded during conjugation, but at a very low speed. Even as late as 72 h after the initiation of conjugation, the majority of cells still contained a prominent old macronucleus, while it normally disappears around 16 h. This result provided strong evidence for the importance of ATG8 in nuclear degradation. Nonetheless, the meiotic products and the old MAC were eventually degraded, and viable progeny were produced without ATG8 function. This is not likely due to redundant functions of the two ATG8 genes or other autophagy pathway genes, since lysosome fusion or acidification failed to occur in single mutants. Thus, a digestion mechanism independent of lysosome fusion is apparently in operation for nuclear degradation in the absence of autophagy, although its nature is still unclear. The fact that caspase 1-, 3-, 8-, and 9-like activities have been detected in Tetrahymena (17, 33) suggests the presence of a similar cascade of cysteine proteases, even though to date no candidate genes have been found. Coincidently, caspase 8- and 9-like activities increased at the final stages of degradation of the old MAC (at about 14 h) (33), supporting their possible roles in this process.
In this study, we used Tetrahymena as a model organism to illustrate a novel function of autophagy in this unicellular eukaryote. Through the study of ATG8, the conserved key component of autophagy, we have provided clear evidence that autophagy is involved and probably constitutes the primary machinery for the apoptosis-like nuclear degradation process in Tetrahymena. This finding has helped expand our understanding of autophagy to include the recycling of nuclei, which are among the largest organelles in the cell. To degrade this large structure, ciliates may have evolved special features of autophagy, possibly by combining elements of other cellular process, such as apoptosis, which has been suggested to occur in some protozoa. The dynamic nuclear behavior of Tetrahymena provides an unusually rich source for understanding nuclear degradation, which should advance our understanding of autophagy and its interplay with nuclear differentiation.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from the National Science Council (NSC99-2628-B-001-010-MY3) of Taiwan, Republic of China, and from the Institute of Molecular Biology of Academia Sinica to M.C.Y.
We appreciate discussions with and suggestions from Chung Wang and all members of the Yao lab, and we thank Aaron Turkewitz, Douglas Chalker, Mochizuki Kazufumi, and Peter Bruns for experimental materials. We thank Sue-Ping Lee and Sue-Ping Tsai (Institute of Molecular Biology, Academia Sinica) for excellent technical assistance in DeltaVision imaging and Heiko Kuhn (English editor, Institute of Molecular Biology, Academia Sinica) for help with editing and improving the manuscript.
Footnotes
Published ahead of print 24 February 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Akematsu T, Endoh H. 2010. Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila. BMC Cell Biol. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akematsu T, Pearlman RE, Endoh H. 2010. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila. Autophagy 6:901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allewell NM, Oles J, Wolfe J. 1976. A physicochemical analysis of conjugation in Tetrahymena pyriformis. Exp. Cell Res. 97:394–405 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez VE, et al. 2008. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283:3454–3464 [DOI] [PubMed] [Google Scholar]
- 5. Benchimol M. 1999. Hydrogenosome autophagy: an ultrastructural and cytochemical study. Biol. Cell 91:165–174 [DOI] [PubMed] [Google Scholar]
- 6. Besteiro S, Williams RA, Coombs GH, Mottram JC. 2007. Protein turnover and differentiation in Leishmania. Int. J. Parasitol. 37:1063–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. 2006. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281:11384–11396 [DOI] [PubMed] [Google Scholar]
- 8. Bruns PJ, Cassidy-Hanley D. 2000. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62:501–512 [DOI] [PubMed] [Google Scholar]
- 9. Calvo-Garrido J, et al. 2010. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 6:686–701 [DOI] [PubMed] [Google Scholar]
- 10. Cassidy-Hanley D, et al. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chalker DL. 2008. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim. Biophys. Acta 1783:2130–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan EY, Kir S, Tooze SA. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282:25464–25474 [DOI] [PubMed] [Google Scholar]
- 13. Chan EY, Longatti A, McKnight NC, Tooze SA. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29:157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng CY, Vogt A, Mochizuki K, Yao MC. 2010. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol. Biol. Cell 21:1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coleman J, et al. 2010. A live-cell fluorescence microplate assay suitable for monitoring vacuolation arising from drug or toxic agent treatment. J. Biomol. Screen. 15:398–405 [DOI] [PubMed] [Google Scholar]
- 16. Davis MC, Ward JG, Herrick G, Allis CD. 1992. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev. Biol. 154:419–432 [DOI] [PubMed] [Google Scholar]
- 17. Ejercito M, Wolfe J. 2003. Caspase-like activity is required for programmed nuclear elimination during conjugation in Tetrahymena. J. Eukaryot. Microbiol. 50:427–429 [DOI] [PubMed] [Google Scholar]
- 18. Endoh H, Kobayashi T. 2006. Death harmony played by nucleus and mitochondria: nuclear apoptosis during conjugation of Tetrahymena. Autophagy 2:129–131 [DOI] [PubMed] [Google Scholar]
- 19. Ganley IG, et al. 2009. ULK1/ATG13/FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284:12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanada T, et al. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282:37298–37302 [DOI] [PubMed] [Google Scholar]
- 21. Hara T, et al. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. 2003. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 278:51841–51850 [DOI] [PubMed] [Google Scholar]
- 23. Herman M, Gillies S, Michels PA, Rigden DJ. 2006. Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy 2:107–118 [DOI] [PubMed] [Google Scholar]
- 24. Hosokawa N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20:1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ichimura Y, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408:488–492 [DOI] [PubMed] [Google Scholar]
- 26. Johansson M, et al. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176:459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung CH, et al. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20:1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabeya Y, et al. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812 [DOI] [PubMed] [Google Scholar]
- 29. Khalfan WA, Klionsky DJ. 2002. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr. Opin. Cell Biol. 14:468–475 [DOI] [PubMed] [Google Scholar]
- 30. Kimura S, Noda T, Yoshimori T. 2008. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33:109–122 [DOI] [PubMed] [Google Scholar]
- 31. Kirisako T, et al. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151:263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klionsky DJ. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- 33. Kobayashi T, Endoh H. 2003. Caspase-like activity in programmed nuclear death during conjugation of Tetrahymena thermophila. Cell Death Differ. 10:634–640 [DOI] [PubMed] [Google Scholar]
- 34. Lu E, Wolfe J. 2001. Lysosomal enzymes in the macronucleus of Tetrahymena during its apoptosis-like degradation. Cell Death Differ. 8:289–297 [DOI] [PubMed] [Google Scholar]
- 35. Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. 2006. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem. Biophys. Res. Commun. 339:485–489 [DOI] [PubMed] [Google Scholar]
- 36. Michels PA, Bringaud F, Herman M, Hannaert V. 2006. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763:1463–1477 [DOI] [PubMed] [Google Scholar]
- 37. Mijaljica D, Prescott M, Devenish RJ. 2006. Endoplasmic reticulum and Golgi complex: contributions to, and turnover by, autophagy. Traffic 7:1590–1595 [DOI] [PubMed] [Google Scholar]
- 38. Mijaljica D, Prescott M, Devenish RJ. 2007. Nibbling within the nucleus: turnover of nuclear contents. Cell. Mol. Life Sci. 64:581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizushima N, Levine B. 2010. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12:823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mochizuki K. 2008. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 425:79–83 [DOI] [PubMed] [Google Scholar]
- 41. Mpoke S, Wolfe J. 1996. DNA digestion and chromatin condensation during nuclear death in Tetrahymena. Exp. Cell Res. 225:357–365 [DOI] [PubMed] [Google Scholar]
- 42. Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. 2003. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 10:108–116 [DOI] [PubMed] [Google Scholar]
- 43. Nilsson JR. 1984. On starvation-induced autophagy in Tetrahymena. Carlsberg Res. Commun. 49:323–340 [Google Scholar]
- 44. Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278:17636–17645 [DOI] [PubMed] [Google Scholar]
- 45. Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. 2004. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 279:15621–15629 [DOI] [PubMed] [Google Scholar]
- 46. Pankiv S, et al. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paz Y, Elazar Z, Fass D. 2000. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275:25445–25450 [DOI] [PubMed] [Google Scholar]
- 48. Punta M, et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samejima K, Earnshaw WC. 2005. Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 6:677–688 [DOI] [PubMed] [Google Scholar]
- 50. Shang Y, et al. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 99:3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84–91 [DOI] [PubMed] [Google Scholar]
- 52. Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. 2002. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 277:13739–13744 [DOI] [PubMed] [Google Scholar]
- 53. Tanida I, Tanida-Miyake E, Ueno T, Kominami E. 2001. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 276:1701–1706 [DOI] [PubMed] [Google Scholar]
- 54. Tanida I, Ueno T, Kominami E. 2004. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36:2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terman A, Gustafsson B, Brunk UT. 2007. Autophagy, organelles and ageing. J. Pathol. 211:134–143 [DOI] [PubMed] [Google Scholar]
- 56. Weidberg H, et al. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29:1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weiske-Benner A, Eckert WA. 1987. Differentiation of nuclear structure during the sexual cycle in Tetrahymena thermophila. Differentiation 34:1–12 [Google Scholar]
- 58. Williams RA, Tetley L, Mottram JC, Coombs GH. 2006. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol. Microbiol. 61:655–674 [DOI] [PubMed] [Google Scholar]
- 59. Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH. 2009. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 5:159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xie Z, Nair U, Klionsky DJ. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19:3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yakisich JS, Kapler GM. 2004. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ. 11:1146–1149 [DOI] [PubMed] [Google Scholar]
- 62. Yang Z, Klionsky DJ. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yao MC, Chao JL. 2005. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 39:537–559 [DOI] [PubMed] [Google Scholar]
- 64. Yorimitsu T, Klionsky DJ. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2):1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.