Abstract
Hemicellulose, the second most abundant plant biomass fraction after cellulose, is widely viewed as a potential substrate for the production of liquid fuels and other value-added materials. Degradation of hemicellulose by filamentous fungi requires production of many different enzymes, which are induced by biopolymers or its derivatives and regulated mainly at the transcriptional level through transcription factors (TFs). Neurospora crassa, a model filamentous fungus, expresses and secretes enzymes required for plant cell wall deconstruction. To better understand genes specifically associated with degradation of hemicellulose, we applied secretome and transcriptome analysis to N. crassa grown on beechwood xylan. We identified 34 secreted proteins and 353 genes with elevated transcription on xylan. The xylanolytic phenotype of strains with deletions in genes identified from the secretome and transcriptome analysis of the wild type was assessed, revealing functions for known and unknown proteins associated with hemicellulose degradation. By evaluating phenotypes of strains containing deletions of predicted TF genes in N. crassa, we identified a TF (XLR-1; xylan degradation regulator 1) essential for hemicellulose degradation that is an ortholog to XlnR/XYR1 in Aspergillus and Trichoderma species, respectively, a major transcriptional regulator of genes encoding both cellulases and hemicellulases. Deletion of xlr-1 in N. crassa abolished growth on xylan and xylose, but growth on cellulose and cellulolytic activity were only slightly affected. To determine the regulatory mechanisms for hemicellulose degradation, we explored the transcriptional regulon of XLR-1 under xylose, xylanolytic, and cellulolytic conditions. XLR-1 regulated only some predicted hemicellulase genes in N. crassa and was required for a full induction of several cellulase genes. Hemicellulase gene expression was induced by a combination of release from carbon catabolite repression (CCR) and induction. This systematic analysis illustrates the similarities and differences in regulation of hemicellulose degradation among filamentous fungi.
INTRODUCTION
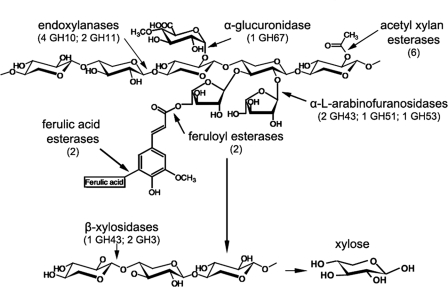
Hemicellulose, the second most abundant component of renewable biomass, is a complex heterogeneous polysaccharide composed mainly of xylose and arabinose. The xylan backbone contains d-xylose as its monomeric unit, its side groups can be replaced with hexose and sugar acids, and it varies from grass to wood (reviewed in reference 4). The heterogeneous nature of xylan means that it requires a variety of enzymes for degradation, including endoxylanases, which cleave the β-1,4 glycosidic linkage in the xylan backbone (glycoside hydrolase [GH] families 10 and 11); arabinofuranosidases, which remove arabinose side chains (GH43, GH51, and GH53); β-xylosidases (GH43 and GH3), which release xylose from xylo-oligosaccharides; acetyl xylan esterases, which remove acetyl groups from the xylan backbone; and feruloyl and ferulic acid esterases, which remove ferulic acid from xylan side chains (Fig. 1; the number of genes for each enzyme class predicted in the Neurospora crassa genome is noted in the figure) (for a review, see reference 17).
Fig 1.
Schematic outline showing the functional coordination of hemicellulose degradation enzymes (mainly xylan) (17). The numbers of related enzymes were retrieved by searching the genome annotation of N. crassa.
A large number of filamentous fungi are able to degrade plant cell wall material by secretion of cellulase and hemicellulase enzymes. The filamentous ascomycete fungus Neurospora crassa has been used as a model laboratory organism for many years (13). In nature, N. crassa is found only on burnt plant material, primarily grasses, including sugarcane, which is closely related to Miscanthus (41, 62). Previously, we showed that N. crassa is able to express and secrete many plant cell wall-degrading enzymes when grown on ground Miscanthus stems and crystalline cellulose (57). There are 23 predicted cellulase genes and 19 predicted hemicellulase genes in the genome of N. crassa (33), plus additional genes with annotated functions associated with degradation and utilization of hemicellulose (Fig. 1). In this study, we investigated what genes/proteins are specifically associated with hemicellulose degradation, by performing transcriptome and secretome analysis of N. crassa grown on xylose and xylan substrates and assaying phenotypic consequences of strains carrying deletions in genes encoding secreted proteins predicted to be involved in xylan degradation. We further assessed the phenotype of 34 strains containing deletions of predicted transcription factor (TF) genes that showed induction when N. crassa is exposed to xylan and identified a predicted ortholog of xlnR/xyr1 (NCU06971; xlr-1). In Hypocrea jecorina (Trichoderma reesei) and Aspergillus spp., the transcriptional regulator XYR1/XlnR, respectively, regulates expression of both hemicellulase and cellulase genes (12, 31, 34, 38, 51, 52, 61). Loss-of-function mutants in xlnR in Aspergillus niger exhibit strongly reduced xylanolytic activities (64). Strains carrying a deletion of xyr1 in H. jecorina or Aspergillus oryzae xlnR mutant strains are affected in both xylanase and cellulase activity (38, 51). However, deletion of xlnR in Fusarium oxysporum affected only xylanase activity (8). In this study, we evaluated the phenotype of an N. crassa strain carrying a deletion of xlr-1 and determined the XLR-1 transcriptional regulon under xylose, xylan, and cellulose conditions. Our results contribute to unraveling the molecular mechanisms used by N. crassa to degrade hemicellulose and have aided in the identification of important genes/proteins associated with hemicellulose deconstruction, which could benefit future industrial engineering strategies.
MATERIALS AND METHODS
N. crassa strains and growth conditions.
The Neurospora crassa wild-type (WT) strain (FGSC 2489), the xlr-1 gene deletion strains (Δxlr-1) (FGSC 11066 and 11067), and other gene deletion strains used in this article were obtained from the Fungal Genetics Stock Center (FGSC) (35). N. crassa was grown on 1× Vogel's salts (65) with 2% (wt/vol) carbon sources (sucrose, beechwood xylan, xylose, or Avicel) at 25°C and 220 rpm with constant light, unless otherwise indicated. Beechwood xylan and Avicel PH 101 were purchased from Sigma-Aldrich (catalog numbers X4252-100G and 11365).
Transcriptional profiling and data analysis.
Ten-day-old conidia of the WT or Δxlr-1 strain were inoculated as 106 conidia/ml into 100 ml 1× Vogel's salts minimal medium (MM) (2% sucrose) and grown for 16 h at 25°C with constant light. Mycelia were centrifuged and washed with 1× Vogel's only medium and then transferred into 100 ml 1× Vogel's salts with 2% carbon source (sucrose, beechwood xylan, xylose, or Avicel) for an additional 4 h. Mycelia were harvested and immediately added to 1 ml of TRIzol reagent (Invitrogen) and zirconia-silica beads (0.2 g, 0.5-mm diameter; Biospec Products). Cells were disrupted using a MiniBead Beater instrument (Biospec Products) at maximum speed for 30 s 3 times in succession. Total RNA was isolated according to the manufacturer's instructions and treated with DNase I (Turbo DNA-free kit; Ambion). RNA was subsequently used for either microarray or quantitative reverse transcription-PCR (qRT-PCR) experiments (see below).
For microarray experiments, the Pronto kit (catalog no. 40076; Corning) was used according to the manufacturer's specifications for cDNA synthesis and labeling. Ten micrograms of total RNA was used per sample. Cy3 and Cy5 dye swaps were used for each sample to avoid bias. Microarray hybridization and data analysis were done as previously described (28, 57, 58). Images were acquired by using a GenePix 4000B scanner (Axon Instruments), and GenePix Pro6 software was used to quantify hybridization signals and collect the raw data. Normalized expression values were analyzed by using the BAGEL (Bayesian analysis of gene expression levels) software program (59, 60). All profiling data are available as Table S1 in the supplemental material and also at the GEO database (GSE34098; http://www.ncbi.nlm.nih.gov/geo/info/linking.html).
Proteomics sample preparation and mass spectrometry (MS).
The N. crassa WT strain (FGSC 2489) was grown on 2% beechwood xylan medium for 4 or 7 days. Final cultures were centrifuged, and the resulting culture supernatants were filtered through a 0.22-μm filter (Corning). Culture supernatants were concentrated 10 times with 10-kDa-molecular-mass-cutoff PES spin concentrators (Millipore). Two to three micrograms of protein was used to generate peptide sample as described in reference 54.
Mass spectrometry was performed (QB3/Chemistry Mass Spectrometry Facility at UC Berkeley) as previously described (54). Trypsin-digested proteins were analyzed using a tandem mass spectrometer (MS/MS) connected in-line with an ultra-high-performance liquid chromatograph (UPLC). Protein-Lynx Global Server software (Waters) was used for analyzing the resulting data from liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of trypsin-digested proteins. The processed data were searched against the N. crassa database (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
Phylogenetic analyses.
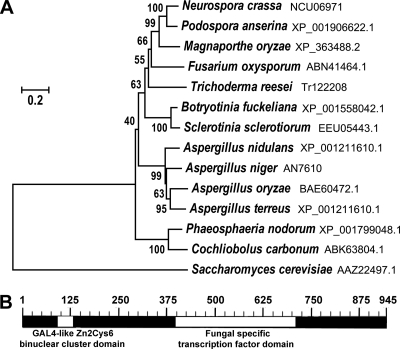
The predicted orthologs of xlr-1 in N. crassa (NCU06971) were retrieved from the NCBI and JGI (for H. jecorina) databases based on amino acid similarity. XLR-1 members from Podospora anserina (XP_001906622.1), Phaeosphaeria nodorum (XP_001799048.1), Cochliobolus carbonum (ABK63804.1), Magnaporthe oryzae (XP_363488.2), Fusarium oxysporum (ABN41464.1), Trichoderma reesei (Tr122208), Botryotinia fuckeliana (XP_001558042.1), Sclerotinia sclerotiorum (AAZ75672.1), Saccharomyces cerevisiae (EEU05443.1 and AAZ22497.1), Aspergillus oryzae (BAE60472.1), Aspergillus terreus (XP_001211610.1), and Aspergillus niger (AN7610) were chosen, and the neighbor-joining tree was made with MEGA4.1. Alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Plasmid construction, transformation, and complementation.
Genomic DNA from a wild-type strain was isolated as described in the work of Lee et al. (S. B. Lee, M. G. Milgroom, and J. W. Taylor, available at http://www.fgsc.net/fgn35/lee35.pdf). To complement the Δxlr-1 strain, a plasmid harboring the open reading frame of NCU06971 under constitutive promoter ccg-1 (36) was constructed according to the method described in references 54 and 55. Primers containing the ligation-independent cloning (LIC) adapter were NCU06971AF (5′-tacttccaatccaatgcaATGTTGTCTAATCCGCTTCACCGGTTC-3′) and NCU06971R (5′-ctcccactaccaatgccGAGGGCCAAGCCAGTTCCATCGCCGG-3′) (lowercase indicates sequence of LIC adapters). The final plasmid was sequenced by the UC Berkeley DNA Sequencing Facility.
One microgram of plasmid DNA was transformed into the his-3;Δxlr-1 a strain (obtained by a cross between FGSC 6103 and FGSC 11066), and constructs were targeted to the his-3 locus by homologous recombination. Correct integration at the his-3 locus was confirmed by green fluorescent protein (GFP) fluorescence and PCR. To recover homokaryotic strains, His+ GFP+ transformants were crossed with the xlr-1 gene deletion strain (FGSC 11067). Progeny were selected for histidine prototrophy and GFP fluorescence and screened for complementation of Δxlr-1 by evaluating growth on beechwood xylan and assessing xylanase activity.
Enzyme activity measurements.
Total secreted protein was determined by using a Bio-Rad DC protein assay kit (Bio-Rad). Endoxylanase activity in culture supernatants of N. crassa strains growing on beechwood xylan medium for 4 days was measured with an azoxylan kit (Megazyme; S-AXBL). A 3,5-dinitrosalicylic acid (DNS) assay measuring the reducing sugar, which indicates the total xylanase activity in culture supernatants, was determined by adapting the method from the work of Bailey et al. (3). A xylose standard (in H2O) curve was used to calibrate xylose concentration. A 900-μl substrate solution (beechwood xylan, 10 mg/ml, in 50 mM sodium acetate solution, pH 5.0, autoclaved for 20 min) was incubated at 50°C for 10 min. One hundred microliters of culture supernatant as well as standards was added to the substrate solution, mixed well, and incubated at 50°C for another 5 min. Then, the resulting solution was centrifuged for 10 min at 3,400 rpm. A 75-μl DNS solution was added to a PCR plate. Five microliters of solution from the above reaction mixture was added to the PCR plate containing DNS solution and mixed well. The PCR plate was heated to 99°C in a PCR machine for 5 min. Samples were transferred to a clear flat-bottomed plate, and absorbance at 540 nm was measured by a plate reader (Biospec). Xylose concentration was calculated according to the standard curve.
Quantitative reverse transcription-PCR.
Total RNA was isolated as described above. Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed according to the manufacturer's instructions (Invitrogen). Primers used to detect expression of hemicellulase genes are shown in Table S2 in the supplemental material. Three replicates were performed per experiment. Experimental setup and data analyses were done as previously described (14). Expression of the actin (NCU04173) gene was used as an endogenous control for all experiments.
RESULTS
Xylan induces hemicellulase but not cellulase gene expression in N. crassa.
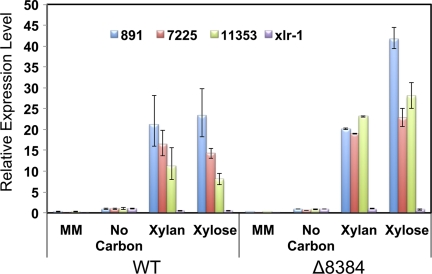
To complement transcriptional profiling data from N. crassa grown on Miscanthus and crystalline cellulose, we assessed transcriptional profiles of N. crassa grown on a hemicellulose substrate, beechwood xylan, and its major hydrolytic product, xylose (57). To directly assess inductive mechanisms associated with utilization of hemicellulose, WT N. crassa (FGSC 2489) was pregrown in sucrose MM for 16 h, washed, and then transferred into medium containing 2% beechwood xylan, 2% sucrose, or 2% xylose for 4 h. RNA was then extracted from mycelia, and expression profiles were analyzed with full-genome oligonucleotide arrays developed for N. crassa (see Fig. S1 in the supplemental material) (28, 58). A total of 353 genes were significantly induced by beechwood xylan compared to sucrose MM conditions (Fig. 2; also see Table S1). Functional category analysis (47) showed that the most enriched gene group was C-compound and carbohydrate metabolism (92 genes; see Table S1), including 8 of the 19 predicted hemicellulase genes in the genome (NCU01900, gh43-2, arabinofuranosidase; NCU02343, gh51-1, arabinofuranosidase; NCU05924, gh10-1, endoxylanase; NCU05965, arabinase; NCU07225, gh11-2, endoxylanase; NCU08189, gh10-2, endoxylanase; NCU09652, gh43-5, β-xylosidase; and NCU11198, gh53-1, arabinogalactan endo-β-galactosidase) (Fig. 1). Three genes, gh51-1 (arabinofuranosidase), gh10-2 (endoxylanase), and gh43-5 (β-xylosidase) (Fig. 1), showed expression levels that were increased over 200-fold. Since the main product of xylan degradation is d-xylose or xylose derivatives, the xylose metabolism pathway genes (10, 26) were also enriched among the 353 genes, including xylose reductase (NCU008384, xyr-1), xylitol dehydrogenase (NCU00891, xdh-1), and d-xylulose kinase (NCU11353, xyk-1), and 7 genes encoding the pentose-phosphate pathway (PPP) enzymes transketolase (NCU01328), 2-deoxy-d-gluconate 3-dehydrogenase (NCU01904), transaldolase (NCU02136), fructose-bisphosphate aldolase (NCU04401), xylulose-5-phosphate phosphoketolase (NCU06123), oxidoreductase (NCU09821), and ribose-5-phosphate isomerase (NCU10107). Other genes that showed increased expression levels with annotation associated with plant cell wall degradation included NCU00130 (gh1-1; β-glucosidase), three β-galactosidase genes (NCU00642, NCU00810, and NCU04623), NCU09041 (l-xylulose reductase), NCU00643 (l-arabinitol 4-dehydrogenase), two d-arabinitol 2-dehydrogenase genes (NCU02097 and NCU20791), NCU03188 (d-arabino-1,4-lactone oxidase), and four esterase genes (NCU05159, NCU00173, NCU05751, and NCU08752).
Fig 2.
Venn diagram of comparison of transcriptomes and secretomes of the wild-type strain on different carbon sources. (A) Overlap among genes that exhibit a statistically significant 2-fold increase in expression levels when a wild-type strain (FGSC 2489) was transferred to xylan or xylose from a 16-h sucrose culture (MM). (B) Overlap among genes that exhibit a statistically significant 2-fold increase in expression levels in FGSC 2489 when transferred to xylan or Avicel from 16-h sucrose culture (MM). (C) Overlap of secretomes identified from FGSC 2489 growing on beechwood xylan for 4 days versus that growing on Avicel for 7 days (Avicel secretome data are from reference 57).
Continuous growth of N. crassa on cellulose induces both cellulase and hemicellulase gene expression (57). To assess induction by cellulose directly, WT N. crassa (FGSC 2489) was pregrown in sucrose MM for 16 h, washed, and then transferred into medium containing 2% Avicel for 4 h. Transcriptome analysis showed that gene expression levels of 343 genes were significantly increased (Fig. 2; see also Table S1 in the supplemental material), including 12 cellulase genes (NCU00762, gh5-1; NCU00836, gh61-7; NCU01050, gh61-4; NCU02240, gh61-1; NCU02916, gh61-3; NCU03328, gh61-6; NCU05057, gh7-1; NCU07190, gh6-3; NCU07340, cbh-1; NCU07898, gh61-13; NCU08760, gh61-5; and NCU09680, cbh-2). Expression of nine hemicellulase genes was induced (NCU02855, gh11-1; NCU05955, gh74-1; NCU07326; NCU01900, gh43-2; NCU05924, gh10-1; NCU07255, gh11-2; NCU08189, gh10-2; NCU09652, gh43-5; and NCU11198, gh53-1). Of these nine hemicellulase genes, three (NCU02855, gh11-1; NCU05955, gh74-1; and NCU07326, gh43-4) were significantly induced only by growth in Avicel, while two hemicellulase genes (NCU02343, gh51-1, alpha-l-arabinofuranosidase A, and NCU05965 gh74-1, putative arabinase) were specifically induced only by xylan. Importantly, no predicted cellulase genes were significantly induced by exposure of N. crassa to xylan.
In Aspergillus, xylose serves as an inducer for some cellulase and hemicellulase genes (23, 34). In N. crassa, transcriptional profiling analysis revealed that only 30 genes showed significantly increased expression levels when a 16-h-grown MM culture was transferred to xylose medium for 4 h (Fig. 2; see also Table S1 in the supplemental material). Fifteen of these 30 genes were also induced by xylan, including a single endoxylanase (NCU08189, gh10-2), two xylose metabolism genes (NCU00891, xdh-1, and NCU08384, xyr-1), and two sugar transporter genes (NCU04963 and NCU05897). In an expression profile comparison of three Aspergillus species grown under xylose conditions, the differential expression of 23 genes was conserved (2). Of the genes that were significantly induced by xylose in N. crassa and Aspergillus, the differential expression of only two xylose metabolism-related genes (xdh-1, NCU00891, and xyr-1, NCU08384) was conserved.
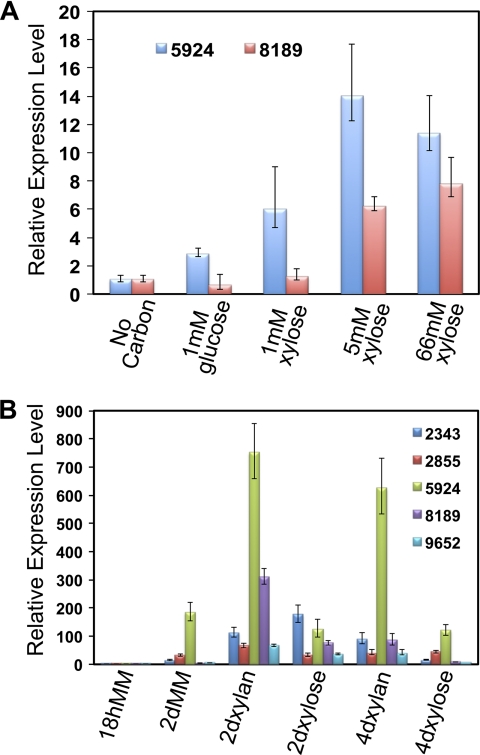
In H. jecorina, it has been reported that xylose can function as both a repressor and an inducer of xylanase expression, depending on concentration (30). In A. niger, xylose concentrations above 1 mM repress expression of xylanase genes (16). We therefore assessed whether increasing concentrations of xylose repress xylanase gene expression in N. crassa. However, using concentrations of xylose identical to those in the work of Mach-Aigner et al. (30) (0 mM to 66 mM), quantitative RT-PCR data showed that the expression levels of xylanase genes (NCU08189, gh10-2, and NCU05924, gh10-1) increased with the increasing xylose concentration (Fig. 3A), suggesting that xylose does not function to repress xylanase gene expression in N. crassa.
Fig 3.
Hemicellulase gene expression levels of the wild-type strain when exposed to xylose or xylan. (A) The wild-type strain (FGSC 2489) was pregrown in MM for 18 h, washed, and then transferred into MM without any carbon source or MM with 1 mM glucose, 1 mM xylose, 5 mM xylose, or 66 mM xylose as the sole carbon source. Gene expression levels of NCU05924 (endoxylanase, gh10-1) and NCU08189 (endoxylanase, gh10-2) were determined by quantitative RT-PCR (see Materials and Methods). (B) FGSC 2489 was grown in MM-2% sucrose for 18 h or 2 days or grown in MM-2% xylan or MM-2% xylose medium for 2 or 4 days. RNA was extracted from samples, and qRT-PCR was performed as indicated in Materials and Methods. Expression levels of NCU05924 (gh10-1), NCU08189 (gh10-2), NCU02343 (arabinofuranosidase, gh510-1), NCU02855 (endoxylanase, gh11-1), and NCU09652 (β-xylosidase, gh43-5) were determined. Expression of the actin (NCU04173) gene was used as an endogenous control for all experiments. Error bars indicate errors of 3 replicates.
Our transcriptional profiling experiments assessed inductive mechanisms associated with xylan utilization. To evaluate expression levels of hemicellulase genes during continuous growth on xylan and xylose, we performed quantitative RT-PCR to monitor the expression changes in five major hemicellulase genes over time (18 h, 2 days, and 4 days). Expression of actin (NCU04173) was used as an intracellular control, and all expression levels were normalized to 18-h gene expression levels in MM. Expression levels for NCU02343 (gh51-1), NCU02855 (gh11-1), and NCU05924 (gh10-1) were slightly induced under starvation conditions (e.g., 2 days in MM, sucrose is utilized after ∼24 h [28]). Similarly, a slight induction of all five hemicellulase genes occurred in 2-day xylose cultures (Fig. 3B). However, four of the five hemicellulase genes (NCU02855, gh11-1; NCU05924, gh10-1; NCU08189, gh10-2; and NCU09652, gh43-5) showed high expression levels during continuous growth in xylan (Fig. 3B). Expression levels of gh10-1 (endoxylanase) were very high on xylan medium (751-fold in 2-day xylan and 625-fold in 4-day xylan). These data indicate that induction of hemicellulase genes in N. crassa responds to a combination of relief from carbon catabolite repression (CCR) and the presence of an inducer (xylan).
In summary, exposure of N. crassa to xylan induced the expression of 8 hemicellulase genes, but no predicted cellulase genes, while exposure to Avicel induced both cellulase and hemicellulase gene expression, with some predicted hemicellulase genes responding significantly only to a cellulose signal.
Secretome analysis of N. crassa grown on beechwood xylan and characterization of strains with deletions in genes involved in hemicellulose degradation.
To identify extracellular proteins associated with xylan degradation, we analyzed the secretome of N. crassa growing on beechwood xylan using a shotgun proteomics approach. Multiple supernatants from both 4- and 6-day-old beechwood xylan cultures were analyzed by liquid chromatography nano-electrospray ionization tandem mass spectrometry (LC-MS; see Materials and Methods). A total of 34 proteins were identified with confidence by LC-MS (Fig. 2C; see also Table S3 in the supplemental material). Eighteen proteins were predicted to be secreted (SignalP 3.0) (6, 37), including 4 hemicellulases (NCU02343, GH51-1; NCU07225, GH11-2; NCU08189, GH10-2; and NCU09170, GH43-4) (Fig. 1), 3 hypothetical proteins (NCU05137, NCU08171, and NCU07143), 7 additional proteins assigned to GH families, and 4 proteins with annotation for other metabolic functions. The remaining 16 proteins contained 3 glycoside hydrolases (NCU04395, NCU05974, and NCU07067). Other proteins were either cell wall-associated protein (NCU08936) (32) or predicted to be involved in intracellular hemicellulose utilization (e.g., NCU08384 and NCU09491). A comparison of the N. crassa xylan secretome with that identified on Avicel (57) showed that 13 proteins were shared by both secretomes, including 2 hemicellulases (NCU07225, GH11-2, and NCU08189, GH10-2), 3 hypothetical proteins (NCU05137, NCU00798, and NCU07143), 3 cell wall proteins (NCU08171, NCU05974, and NCU08936), 4 other proteins related to hydrolases (NCU09024, NCU09175, NCU04395, and NCU09491), and a glucanosyltransferase (NCU08909) (Fig. 2C; see also Table S3).
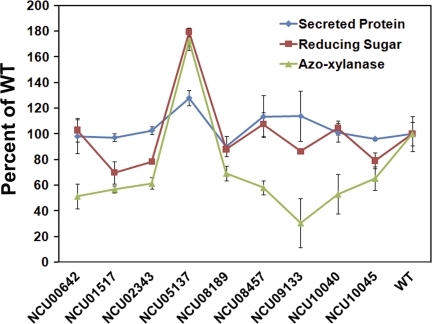
Among the 353 genes that showed a significant increase in expression level when N. crassa was exposed to beechwood xylan, 45 were predicted to be secreted (SignalP 3.0) (6, 37). Combining these 45 proteins with unique proteins identified in the xylan secretome and additional hemicellulases from the 19 predicted in the N. crassa genome, a total of 71 unique proteins could potentially be involved in hemicellulose deconstruction (see Table S4 in the supplemental material). Of the 71 genes encoding these proteins, strains containing individual deletions of 40 genes are available (11); none of them have been characterized with respect to hemicellulose degradation in N. crassa. This data set included strains with mutations in genes encoding secreted proteins of unknown function (13 strains) in addition to a variety of predicted enzymes associated with plant cell wall degradation, as well as lipases and a gene encoding a hydrophobin (see Table S4). The 40 deletion mutants were grown in medium containing sucrose or beechwood xylan as a sole carbon source. All deletion strains grew similarly to wild-type strains on sucrose MM with the exception of the ΔNCU08131 (α-amylase) strain. The culture supernatants from 4-day beechwood xylan cultures of each deletion strain were assayed for total secreted protein, endoxylanase activity, and reducing sugar concentration, which measures the total xylanase activity (Fig. 4; see also Table S5); of the 40 mutants screened, eight showed a significant difference in hemicellulase activity from the WT. For example, a strain carrying a deletion of ncw-1 (NCU05137), encoding a non-cell-wall-anchored protein of unknown function (32), showed increased protein levels (28%) and xylanase activity (73% higher endoxylanase activity and 79% higher total xylanase activity) (Fig. 4). These results are similar to the phenotype of an ΔNCU05137 mutant when grown on Avicel (57). Deletion of one endoxylanase (NCU08189, gh10-2) caused a 31% ± 6% decrease in endoxylanase activity and a 12% ± 2% decrease in total xylanase activity, indicating that GH10-2 is the major endoxylanase in N. crassa. Deletion of another hemicellulase gene (NCU02343, gh51-1; alpha-l-arabinofuranosidase 2) resulted in a slightly decreased level of endoxylanase activity and total xylanase activity (decreases of 39% ± 4% and 22% ± 1%, respectively). Three deletion strains (ΔNCU00642, β-galactosidase; ΔNCU08457, ccg-2, encoding a hydrophobin; and ΔNCU10040, hypothetical protein) showed decreased endoxylanase activity but not total xylanase activity (Fig. 4). A strain with a deletion in a gene encoding glucan 1,4-alpha-glucosidase (NCU01517), a strain carrying a deletion of a potential pectin esterase (NCU10045), and a strain with a deletion of a hypothetical secreted protein (NCU09133) exhibited significantly less endoxylanase and total xylanase activity (Fig. 4; see also Table S5). Notably, the ΔNCU09133 mutant gave very low endoxylanase activity, suggesting that the protein of unknown function encoded by NCU09133 might be an endoxylanase-related protein. Thus, we identified genes encoding proteins with predicted hemicellulase activity that, when deleted, reduced xylanase activity in N. crassa, as well as genes that encode proteins of unknown function (NCU09133, NCU10040, and NCU05137) that, when deleted, reduced or increased xylanase activity. Further characterization of the biochemical function of these proteins and their role in hemicellulose degradation is warranted.
Fig 4.
Enzyme activity of culture supernatants from strains containing deletions of genes related to hemicellulose degradation. Total secreted protein, endoxylanase activity on azoxylan, and reducing sugar concentration in assays with culture supernatants from WT (FGSC 2489) and selected isogenic deletion mutants (missing gene denoted as the NCU gene number in figure) grown on beechwood xylan as a sole carbon source for 4 days. Data from FGSC 2489 were set to 100% (WT levels) (see Table S5 in the supplemental material). Bars represent standard deviations.
An N. crassa xlnR ortholog (xlr-1) is essential for the growth on xylan but not on cellulose.
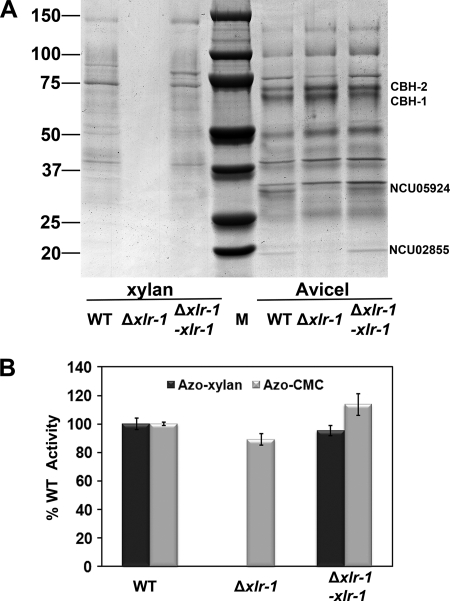
A survey of the transcriptional profiling data (see Table S1 in the supplemental material; also RNA-Seq data, unpublished) revealed that 34 genes encoding predicted transcription factors increased in expression levels (over 2-fold) when N. crassa was exposed to xylan. Culture supernatants from minimal medium and 4-day beechwood xylan cultures of each TF deletion strain were assayed for total secreted protein and xylanase activity. Some mutants (e.g., ΔNCU07788; col-26 [11]) showed a general growth defect under all conditions. Only one transcription factor mutant (ΔNCU06971) showed a severe and specific growth defect on xylan. The N. crassa ΔNCU06971 mutant (here referred to as xylan degradation regulator 1) showed no significant growth defect in MM compared to the wild-type strain but failed to grow on 2% beechwood xylan or xylose as a sole carbon source (see Fig. S2). Neither secreted protein nor endoxylanase activity was detectable when Δxlr-1 mutants were exposed to xylan (Fig. 5A) or xylose (not shown). Although an Δxlr-1 strain was unable to grow on xylan or xylose, it displayed only slightly decreased growth on Avicel and slightly reduced endoglucanase activity on azo-carboxymethyl cellulose (azo-CMC) (89% ± 4% of the level of the WT strain). No obvious difference in the secretome of the Δxlr-1 strain on Avicel compared to that on WT was observed, with the exception of two proteins that were missing in the Δxlr-1 strain (Fig. 5A). The first protein has a molecular mass of ∼36 kDa and is GH10-1 (NCU05924, endoxylanase) (57). The second protein (∼20 kDa) is GH11-1 (NCU02855) (see Fig. S3), the gene for which encodes a predicted endo-1,4-beta-xylanase. The identified cellulases in WT, including highly abundant CBH-1 and CBH-2 (43, 57), were present in the Δxlr-1 strain when grown on Avicel.
Fig 5.
Phenotype and complementation of Δxlr-1 strain. (A) SDS-PAGE gel of wild-type (FGSC 2489), Δxlr-1, and complemented Δxlr-1 (Δxlr-1-xlr-1) strains grown at 25°C for 4 days on beechwood xylan or 7 days on Avicel. Twenty microliters of supernatant was loaded onto a Criterion 4%-15% gradient SDS-PAGE gel. CBH-1 and CBH-2 were present in the secretome of the Δxlr-1 mutant on Avicel. However, there were two bands (NCU05924, gh10-1, and NCU02855, gh11-1) (see Fig. S3 in the supplemental material) (57) missing in the secretome of the Δxlr-1 mutant. Numbers at left are molecular masses in kilodaltons. (B) Endoxylanase (Azo-xylan) and endoglucanase (Azo-CMC) activity of culture filtrates from wild-type (FGSC 2489), Δxlr-1, or Δxlr-1-xlr-1 strains grown at 25°C for 4 days on beechwood xylan (black bars) or for 7 days on Avicel (gray bars).
xlr-1 (NCU06971) encodes a member of a TF family containing a conserved fungal Zn2Cys6 binuclear cluster domain with significant amino acid homology to xlnR/xyr1 (24), sharing 57.6% identity with homologs in H. jecorina, 47.3% identity with A. niger, and 63% identity with Fusarium oxysporum (Fig. 6). In filamentous fungi, such as Aspergillus and H. jecorina, XlnR/XYR1 regulates the expression of some cellulase and hemicellulase genes (7, 8, 31, 34, 38, 52, 63, 64). Constitutive expression of xlnR/xyr1 in A. oryzae and T. reesei causes increased xylanolytic and cellulolytic activity (31, 38), although constitutive expression of xlnR in F. oxysporum did not result in increased xylanase activity (8). An N. crassa strain carrying a constitutively expressed copy of xlr-1-gfp in an xlr-1 deletion background [Δxlr-1 (xlr-1-gfp)] showed similar protein profiles on both xylan and Avicel media (Fig. 5A), and the two missing proteins in the Δxlr-1 strain on Avicel (NCU05924 and NCU02855) were produced in the Δxlr-1 (xlr-1-gfp) strain. Although endoxylanase activity in the Δxlr-1 (xlr-1-gfp) strain was almost identical to that of the WT (95% ± 4% of WT level) (Fig. 5B), the Δxlr-1 (xlr-1-gfp) strain produced 14% ± 8% more endoglucanase activity when grown on Avicel (Fig. 5B). Thus, constitutively expressed xlr-1 complements the phenotype of the Δxlr-1 mutant on both xylan and Avicel media and displays slightly increased cellulolytic activity.
Fig 6.
Phylogeny and domain structure of XLR-1. (A) XLR-1 is highly conserved across the genomes of most sequenced filamentous ascomycete species. The neighbor-joining tree shows phylogenetic relationships of fungal XlnR/XYR1 proteins and their relationship to N. crassa XLR-1. Bootstrap values are shown, and the scale bar indicates 0.2 substitutions per amino acid residue. (B) The domain structure of N. crassa XLR-1 contains an N-terminal GAL4-like Zn2Cys6 binuclear cluster domain and a C-terminal fungus-specific transcription factor domain.
Identification of the XLR-1 regulon.
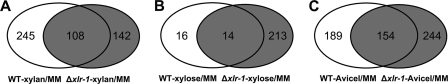
In A. niger, ∼10 genes involved in degradation of xylan and cellulose are regulated by XlnR, including cellobiohydrolase genes (cbhA and cbhB) (23), two endoglucanase genes (eglA and eglB), and xylanase genes (xlnB, xlnC, and xlnD) (63, 64). In addition, a gene involved in xylose metabolism (xyrA; xylose reductase) is also regulated by XlnR (26). In H. jecorina, XYR1 has been shown to regulate some cellulase (cbh1, cbh2, and egl1) and hemicellulase (xyn1, xyn2, bxl1, and abf2 [arabinofuranosidase 2]) genes as well as xyl1 (xylose reductase) (1, 20, 31, 51). Our data indicate that XLR-1 is required for utilization of xylose and hemicellulose in N. crassa and modulates cellulase activity. To identify the XLR-1 regulon in N. crassa, we performed genome-wide transcriptional profiling using the Δxlr-1 strain compared to the WT strain under hemicellulose (xylan and xylose) and cellulose (Avicel) conditions (see Fig. S1 in the supplemental material). When a 16-h sucrose-grown Δxlr-1 culture was transferred to xylan medium for 4 h, 245 genes were dependent on a functional XLR-1 for increased expression levels when exposed to xylan (Fig. 7A; see also Table S1). FunCat analysis of these 245 genes (see Table S1) showed that the C-compound and carbohydrate metabolism group was the most enriched group (P = 7.19e−19), including 11 carbohydrate transporter genes (NCU00988, NCU01132, NCU02188, NCU02238, NCU02582, NCU05853, NCU06138, NCU06305, NCU06358, NCU08114, and NCU09027). In addition, 6 pentose-phosphate pathway (PPP) genes (NCU01328, transketolase; NCU01904, 2-deoxy-d-gluconate 3-dehydrogenase; NCU02136, transaldolase; NCU04401, fructose-bisphosphate aldolase; NCU06123, xylulose-5-phosphate phosphoketolase; and NCU10107, ribose-5-phosphate isomerase), two xylose metabolism genes (xdh-1 and xyr-1), and all 8 of the hemicellulase genes (NCU11198, gh53-1; NCU01900, gh43-2; NCU02343, gh51-1; NCU05924, gh10-1; NCU05965, gh74-1; NCU07225, gh11-2; NCU08189, gh10-2; and NCU09652, gh43-5) showed induction by WT on xylan. Other hemicellulose degradation-related genes (those for mannosidase, galactosidase, arabinitol dehydrogenase, and esterases) were also dependent on a functional xlr-1 for induction (see Table S1). As with WT on xylan medium, no induction of any cellulase genes was observed in the Δxlr-1 mutant on xylan medium. These data indicate that XLR-1 is the major regulator of genes involved in hemicellulose utilization in N. crassa.
Fig 7.
Venn diagrams of transcriptomes from wild-type (FGSC 2489) and Δxlr-1 strains when exposed to different carbon sources. Overlap among genes that exhibit a statistically significant 2-fold increase in expression levels in Δxlr-1 strains compared to FGSC 2489 when transferred to either xylan (A), xylose (B), or Avicel (C) for 4 h from 16-h sucrose cultures (MM).
An additional 108 genes were fully induced in both the Δxlr-1 strain and the WT strain upon exposure to xylan (Fig. 7A), and these included several genes that encode hemicellulose metabolism-related proteins (e.g., NCU04265, β-fructofuranosidase), suggesting that heretofore-unknown transcriptional regulators in N. crassa also play a role in xylan degradation and utilization (see Fig. 10). Of the 142 genes that showed increased expression levels specifically in the Δxlr-1 strain (see Table S1 in the supplemental material), the functional category of protein degradation was slightly enriched (P = 0.0002) (e.g., lysosome-related and vacuolar proteins), suggesting that the Δxlr-1 strain is under stress due to the inability to utilize xylan.
Fig 10.
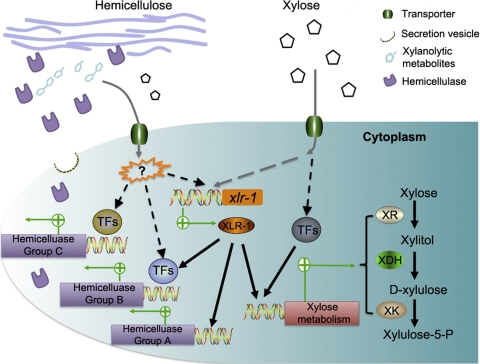
Regulation of genes encoding xylanolytic enzymes in N. crassa. Metabolites released from hemicellulose degradation by basal levels of secreted hemicellulases trigger the transcription and activation of xlr-1 and other unknown transcription factors (TFs). XLR-1 activates the transcription of certain hemicellulase genes (group A) and also functions with other unknown transcription factors to modulate expression of other hemicellulase genes (group B). An unknown TF(s) also induces the expression of separate XLR-1-independent hemicellulase genes (group C). Action of these TFs results in production of hydrolytic enzymes, which are secreted, thereby deconstructing hemicellulose, producing metabolites and additional signaling molecules. Another unknown transcription factor(s) also coordinates with XLR-1 to modulate the regulation of genes associated with xylose metabolism.
Of the 30 genes that showed significantly increased expression levels when the WT was transferred to xylose, 16 genes were xlr-1 dependent (Fig. 7B; see also Table S1 in the supplemental material). Among these 16 genes, xdh-1 and xyr-1 are involved in xylose metabolism. However, only a single endoxylanase (gh10-2, NCU08189) was identified, plus 4 proteins of unknown function. Of note, one of these genes encoded a cell wall-associated protein of unknown biochemical function (ncw-1; NCU05137) (32); deletion of ncw-1 resulted in increased secretion of both hemicellulases and cellulases (Fig. 4) (57). There were 227 genes that showed increased expression levels in the Δxlr-1 strain when transferred from sucrose to xylose medium (Fig. 7B). As with the Δxlr-1 mutant on xylan, genes were enriched in categories such as lipid, fatty acid, and isoprenoid metabolism and energy, indicating that the Δxlr-1 mutant was under starvation due to an inability to utilize xylose.
The Δxlr-1 strain was slightly affected in growth on Avicel and has lower endoglucanase activity (Fig. 5B), suggesting that XLR-1 might regulate some cellulase genes under Avicel conditions. We assessed transcriptional profiles of the Δxlr-1 strain compared to those of the WT strain on Avicel. Of the 343 genes induced at least 2-fold in the WT strain under Avicel conditions (Fig. 7C; see also Table S1 in the supplemental material), 154 were induced in both the WT and the Δxlr-1 mutant. These included 5 predicted cellulase genes (NCU01050, gh61-4; NCU02240, gh61-1; NCU07190, gh6-3; NCU07340, cbh-1; and NCU09680, cbh-2) and 2 hemicellulase genes (NCU11198, gh53-1, and NCU09652, gh43-5). The remaining 189 genes increased in expression level only in the WT, suggesting a requirement for functional XLR-1 for fully induced expression levels. Among these 189 genes were 7 cellulase genes (NCU00762, gh5-1; NCU00836, gh61-7; NCU02916, gh61-3; NCU03328, gh61-6; NCU05057, gh7-1; NCU07898, gh61-13; and NCU08760, gh61-5) and 7 hemicellulase genes (NCU01900, gh43-2; NCU02855, gh11-1; NCU05924, gh10-1; NCU05955, gh74-1; NCU07225 gh11-2; NCU07326; and NCU08189, gh10-2). These results suggest that the reduced expression levels of these cellulase genes in the Δxlr-1 mutant may be responsible for the cellulolytic phenotype of the Δxlr-1 mutant (Fig. 5). FunCat analysis of the remaining genes that showed increased expression levels in the Δxlr-1 mutant grown on Avicel showed that genes encoding proteins associated with protein fate, including lysosomal and vacuolar proteins, were enriched. In summary, XLR-1 in N. crassa is essential for induced expression of a subset of predicted hemicellulase genes and modulation of full induction of a subset of cellulase genes under cellulolytic conditions.
By microarray analysis in A. oryzae, AoXlnR was shown to regulate 75 genes during growth on beechwood xylan (38). In our study, we identified 245 genes that showed increased expression levels (at least 2-fold) in the wild type versus the Δxlr-1 strain when exposed to beechwood xylan (Fig. 7A; see also Table S1 in the supplemental material). A total of 19 genes were conserved between the A. oryzae XlnR and N. crassa XLR-1 regulons (Table 1). Thirteen of these conserved genes encode metabolic enzymes, including two xylose metabolism genes (xdh-1 and xyr-1), three hemicellulase genes (NCU01900, arabinofuranosidase, gh43-2; NCU05924, endoxylanase, gh10-1; and NCU09652, β-xylosidase, gh43-5), and two transporter genes (NCU06138 and NCU08114). Other genes encode proteins with a potential role in hemicellulose degradation, and the remaining 6 genes encode hypothetical proteins.
Table 1.
Conserved genes in the Aspergillus oryzae XlnR and Neurospora crassa XLR-1 regulons
| N. crassa gene | A. oryzae genea | Annotation |
|---|---|---|
| NCU00642 | AO090012000445 | Probable beta-galactosidase |
| NCU00891 | AO090038000631 | Probable xylitol dehydrogenase |
| NCU01900 | AO090005000698 | Alpha-N-arabinofuranosidase/alpha-l-arabinofuranosidase |
| NCU03322 | AO090701000345 | Conserved hypothetical protein |
| NCU03639 | AO090001000207 | Probable triacylglycerol lipase precursor |
| NCU05924 | AO090001000208 | Probable endo-beta-1,4-d-xylanase |
| NCU06138 | AO090001000069 | Related to quinate transport protein |
| NCU06143 | AO090001000267 | Conserved hypothetical protein |
| NCU06961 | AO090026000784 | Probable exopolygalacturonase |
| NCU07143 | AO090005000189 | Conserved hypothetical protein |
| NCU07705 | AO090011000944 | Conserved hypothetical protein |
| NCU08114 | AO090003001277 | Related to hexose transporter protein |
| NCU08384 | AO090003000859 | Probable d-xylose reductase |
| NCU08943 | AO090038000426 | Related to 3-oxoacyl-[acyl carrier protein] reductase |
| NCU09652 | AO090701000886 | Probable xylan 1,4-β-xylosidase |
| NCU09705 | AO090020000042 | Conserved hypothetical protein |
| NCU09923 | AO090005000986 | Related to xylan 1,4-β-xylosidase |
| NCU09924 | AO090011000141 | Conserved hypothetical protein |
| NCU10110 | AO090010000515 | Related to 3-hydroxyisobutyrate dehydrogenase |
Data are from reference 38.
In previous studies, the functional XlnR binding sites, identified by motif searching or biochemical experiments, contain either 5′-GGCTAA-3′, 5′-GGCTGA-3′, or 5′-GGCTAG-3′ (9, 15, 34, 38, 63, 64). Andersen et al. also proposed an additional XlnR binding motif, 5′-GGNTAAA-3′, in 3 Aspergillus species when grown on xylose medium (2). These motifs are common in predicted 1-kbp upstream regions of the N. crassa genome (64.8% of predicted promoter regions in the whole genome contain a GGCTRR motif), and none of the profiling data sets showed a statistically significant enrichment for any of these binding motifs. Therefore, we performed motif searching in the 1-kbp upstream sequences of the 19 conserved XlnR/XLR-1-regulated genes. As summarized in Table S6 in the supplemental material, 18 of these genes contain at least one of these motifs and some contain multiple motifs. Further characterization of these promoters and full-genome studies of XLR-1 DNA binding in vivo (by chromatin immunoprecipitation-sequencing [ChIP-seq]) will help to identify bona fide XLR-1 binding sites across the N. crassa genome in response to both xylan and Avicel.
The utilization of xylan and xylose requires expression of xylose metabolism pathway genes in N. crassa, such as NCU08384 (xyr-1, xylose reductase), NCU00891 (xdh-1, xylitol dehydrogenase), and NCU11353 (xyk-1, d-xylulose kinase). In T. reesei, xylose utilization and xylose reductase are required for full induction of xylanase gene expression (29, 30), and a T. reesei strain containing a deletion of xylitol dehydrogenase (xdh1) resulted in an ∼50% decrease in growth rate on xylose (49). We therefore evaluated whether blocking xylose metabolism in N. crassa would result in feedback to xlr-1 expression. A strain carrying a deletion of xylose reductase (xyr-1) failed to grow at all on either d-xylose or xylan medium (data not shown), indicating that other pathways are unable to compensate for loss of this enzyme. Via transfer experiments from MM to xylose or xylan, the Δxyr-1 strain showed increased expression levels of xdh-1 and xyk-1 and NCU07225 (gh11-2, endoxylanase), especially under xylose conditions, but did not show a significant difference in xlr-1 expression levels under either condition (Fig. 8).
Fig 8.
Gene expression levels of the wild-type strain (FGSC 2489) and a xyr-1 (NCU08384; xylose reductase) deletion strain under different growth conditions. Strains were pregrown in MM-sucrose for 18 h, washed, and transferred into minimal medium without any carbon source or with 2% sucrose (MM), 2% xylose, or 2% xylan as a sole carbon source for an additional 4 h. NCU00891 encodes xylitol dehydrogenase, NCU11353 encodes xylulose kinase, and NCU07225 encodes an endoxylanase. RNA was extracted from these samples, and qRT-PCR for these genes and xlr-1 was performed as indicated in Materials and Methods. Error bars indicate errors for 3 replicates.
Xylanolytic substrates and carbon catabolite repression regulate xlr-1 expression.
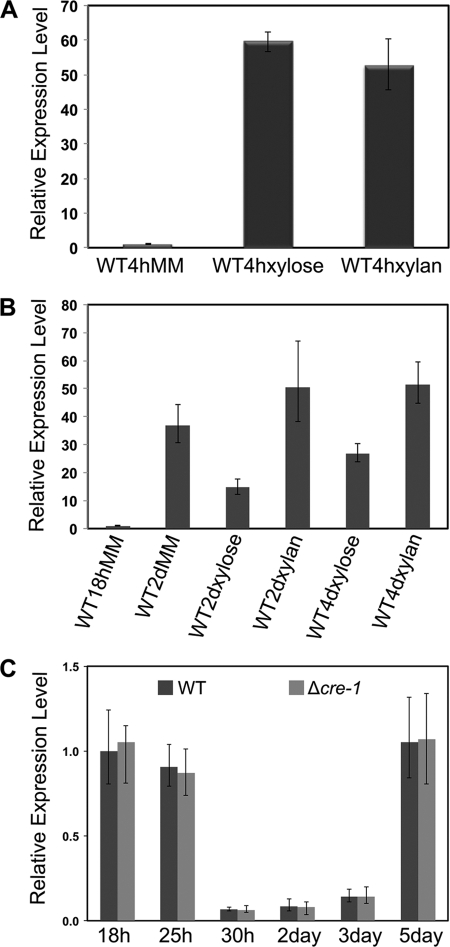
In T. reesei, CRE1 has been proposed to regulate the transcription of xlnR and xylanases through a “double-lock” mechanism (31, 52). In addition, Portnoy et al. found that the full induction of xyr1 requires the positive action of CRE1 in T. reesei (44). In N. crassa, under Avicel conditions, a strain carrying a deletion of cre-1 shows increased expression levels of several hemicellulase genes (54). These data suggest that induction in response to xylan and CCR mediated by CRE-1 plays a role in the regulation of plant cell wall-degrading enzymes. In support of this hypothesis, the expression level of xlr-1 increased under starvation and inducing conditions (Fig. 9A and B). To determine whether this increase in expression of xlr-1 is dependent upon CRE-1, we assessed the expression level of xlr-1 in WT and Δcre-1 strains grown on Avicel (which induces both cellulase and hemicellulase gene expression) over a 5-day growth period (Fig. 9C). No difference in expression levels of xlr-1 between the Δcre-1 strain and the WT was detected, indicating that the response of xlr-1 to CCR in N. crassa is apparently not mediated via transcriptional repression by CRE-1.
Fig 9.
xlr-1 expression levels under xylose, xylan, and Avicel growth conditions. (A) xlr-1 expression was monitored by qRT-PCR in the WT strain (FGSC 2489) (see Materials and Methods). The WT strain was grown in MM-sucrose for 18 h, washed, and then transferred into medium with 2% sucrose (MM), 2% xylose, or 2% xylan as a sole carbon source for an additional 4 h prior to RNA extraction. (B) Expression levels of xlr-1 were monitored by qRT-PCR in the WT when grown continuously in 2% sucrose-MM (18 h and 2 days), 2% xylose medium (2 and 4 days), and 2% xylan medium (2 and 4 days) prior to RNA extraction. (C) xlr-1 expression was monitored by qRT-PCR in WT and Δcre-1 strains when grown in medium containing 2% Avicel as a sole carbon source for 18, 25, and 30 h and 2, 3, and 5 days. RNA was extracted from samples, and qRT-PCR was performed as indicated in Materials and Methods. Error bars indicate errors for 3 replicates.
DISCUSSION
Current models for plant cell wall structure include extensive cross-linking of cellulose microfibrils by hemicellulose (50). Thus, a filamentous fungus will come into contact with a complex mixture of hemicellulose, cellulose, pectin, proteins, and other components of plant cell walls and under natural conditions may need to exquisitely regulate enzyme production both temporally and spatially for optimum plant cell wall deconstruction. It is likely that filamentous fungi, such as N. crassa, respond to a variety of inducer molecules that affect expression of plant cell wall-degrading enzymes. In this study, we show that N. crassa responds to the presence of cellulose (Avicel) by inducing both cellulase and hemicellulase gene expression but that exposure to xylan induces only hemicellulase gene expression. In addition, exposure to Avicel induces some hemicellulase genes to a much higher expression level than does exposure to xylan. In Trichoderma and Aspergillus species, hemicellulase and cellulase gene expressions are coregulated (23, 26, 40, 46). In A. niger, the xylanolytic and cellulolytic systems are coregulated via the inducer d-xylose (23, 26, 40), while in H. jecorina, at least four inducers are known, although none of them triggers the expression of all major cellulase and hemicellulase genes (reviewed in reference 52). For example, xylan induces endoglucanase (egl1) and several xylanase genes (xyn1 and xyn2), while sophorose induces cellulase-encoding genes cbh1, cbh2, and egl1 as well as bgl1 and bgl2 (19, 27, 48). These observations and our data suggest cross talk between inducer molecules and regulatory pathways that are involved in deconstruction of plant cell walls in filamentous fungi.
In this work, we identified 353 genes in N. crassa that were significantly induced by xylan but only 30 genes that were induced by exposure to xylose. Although none of the xylanolytic genes was essential for growth of N. crassa on xylan, mutations in a number of them affected xylanase activity. These observations indicate some redundancy among enzymes associated with hemicellulose degradation, similar to those identified with cellulose degradation (57). Within the 353-gene set, 19 permease/transporter genes were induced on xylan (see Table S1 in the supplemental material), and their full induction required a functional xlr-1. Of these 19 transporters, five have been functionally tested for transport of d-glucose and d-xylose when expressed in a mutant of Saccharomyces cerevisiae devoid of 20 sugar transporter genes (18). The expression of one transporter (NCU04963) allowed S. cerevisiae to transport d-glucose and d-xylose, while heterologous expression of another transporter (NCU00821) allowed xylose transport only. An additional transporter (NCU08114), whose expression is induced by Avicel, was shown to transport cellodextrins in both S. cerevisiae and N. crassa (21, 57). Further research on characterizing the specificity of transporters identified in this study could potentially have beneficial impacts in engineering organisms for utilization of degradation products of hemicellulose, as was previously shown for cellodextrin transporters (21).
In this study, we identified a transcription factor, xlr-1, which is required for utilization of xylose and xylan. In N. crassa, a strain carrying a deletion of the xyr1/xlnR homolog, xlr-1, abolished growth in both xylan and xylose but only slightly affected growth in Avicel and cellulase activity. Consistent with this phenotype, transcriptional profiling showed that xlr-1 is required for induction of hemicellulase and xylose metabolism genes and modulated the expression levels of some cellulase genes. Significantly, no cellulase genes showed an absolute requirement for XLR-1 for induction. Similarly in Fusarium oxysporum and Fusarium graminearum, strains containing a deletion of an xlr-1 homolog were impaired in both xylose and xylan utilization (7, 8) but were unaffected in cellulase activity when grown on plant cell walls. These findings imply that an unknown and uncharacterized transcription factor(s) in Neurospora and Fusarium is important for the induced expression of genes encoding cellulases in response to the presence of cellulose. Many genes within the XlnR regulon in A. oryzae (38) and the XLR-1 regulon in N. crassa encode proteins with unknown biochemical functions. Comparative analyses to identify genes encoding conserved proteins will allow the identification of new biochemical activities associated with hemicellulose degradation, as has been recently reported for cellulose degradation (5, 42, 45).
Regulation of hemicellulase gene expression has focused on induction by different inducers and transcriptional regulation mediated by XYR1/XlnR (25, 31, 34, 38, 51, 63, 64; also reviewed in references 52 and 61). In Aspergillus and H. jecorina, xlnR/xyr1 is also regulated by carbon catabolite repression (CCR) (16, 44, 53, 56). In N. crassa, transcription of most hemicellulase genes is via induction by xylanolytic molecules and is regulated via xlr-1 and/or other transcription factors. However, the hemicellulolytic system is also responsive to CCR (54). CRE-1-mediated CCR regulates the expression level of some, but not all, hemicellulase genes in N. crassa under Avicel conditions (54). Our data indicate that xlr-1 is regulated by a combination of induction and derepression (Fig. 8 and Fig. 9A and B) and that xlr-1 is also subject to non-CRE-1-mediated CCR (Fig. 9C). These observations imply that other mechanisms in addition to CRE-1 regulate CCR in filamentous fungi, similar to what has been described for S. cerevisiae (22). Further studies into the interplay between CCR and induction of plant cell wall-degrading enzymes will undoubtedly reveal additional regulatory mechanisms associated with plant cell wall deconstruction in filamentous fungi.
N. crassa can use a wide variety of carbon sources. This requires an optimal adaption to the environment by synthesizing transporters and secreting enzymes, which are induced by specific substrates or metabolites. For N. crassa to grow on plant cell wall material, a low constitutive level of extracellular or cell wall-associated enzymes releases metabolites from plant biomass (57) (Fig. 10). These metabolites are transported into the cell and serve as inducers directly or are converted to signal molecules required for triggering transcription of genes encoding plant cell wall-degrading enzymes (regulated by XLR-1 and other transcription factors) (Fig. 10). Our results show that XLR-1 is required for expression of many hemicellulase genes and works both independently (group A) and synergistically with other unknown transcription factors (group B) to activate and modulate gene expression for plant cell wall-degrading enzymes. Other transcription factors independently regulate genes required for plant cell wall deconstruction (Fig. 10; group C). Additional mechanisms to activate and enhance the expression or activity of either transcription factors or enzymes, for example, by phosphorylation, as has been recently been shown for XlnR (39), or via feedback inhibition, also play a regulatory role in plant cell wall deconstruction. Although xlr-1 in N. crassa is essential for growth on hemicellulose, it is not essential for utilization of cellulose and only modulates the expression of some cellulolytic genes. These data imply that uncharacterized transcription factors play a role in cellulose degradation in N. crassa.
In summary, our study reveals similarities but also differences in the induction and transcriptional regulation of genes involved in hemicellulose utilization in N. crassa compared to T. reesei and Aspergillus. The most obvious commonality is that N. crassa is able to express a broad range of enzymes to degrade hemicellulose, and both xylan and cellulose induce the expression of hemicellulase genes. However, N. crassa compartmentalizes induction of plant cell wall-degrading enzymes, such that XLR-1 regulates the expression of genes encoding proteins required for utilization of hemicellulose, as well as modulating the expression of some cellulolytic genes. Further identification and characterization of novel transcription factors in N. crassa associated with breakdown of cellulose and additional cell wall components will allow the development of a comprehensive model for regulatory aspects associated with plant cell wall deconstruction. The identification of regulons controlled by such transcription factors will identify proteins important for signaling as well as for enzymatic degradation and utilization of plant biomass that will be important for implementing engineering strategies for optimal plant cell wall deconstruction using tailored enzyme cocktails.
Supplementary Material
ACKNOWLEDGMENTS
The work in this paper was funded by a grant to N.L.G. from the Energy Biosciences Institute (EBI). C.T. was partially supported by National Basic Research Program of China grant 2011CB707403.
We thank Sam Coradetti for data on screening transcription factor deletion strains. We thank members of the Glass laboratory, especially James Craig, Trevor Lee Starr, and Sam Coradetti, for critical comments.
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Akel E, Metz B, Seiboth B, Kubicek CP. 2009. Molecular regulation of arabinan and L-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryot. Cell 8:1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersen MR, et al. 2008. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. U. S. A. 105:4387–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey MJ, Biely P, Poutanen K. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257–270 [Google Scholar]
- 4. Bastawde KB. 1992. Xylan structure, microbial xylanases, and their mode of action. World J. Microbiol. Biotechnol. 8:353–368 [DOI] [PubMed] [Google Scholar]
- 5. Beeson WT, Phillips CM, Cate JH, Marletta MA. 2012. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134:890–892 [DOI] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 7. Brunner K, Lichtenauer AM, Kratochwill K, Delic M, Mach RL. 2007. Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr. Genet. 52:213–220 [DOI] [PubMed] [Google Scholar]
- 8. Calero-Nieto F, Di Pietro A, Roncero MI, Hera C. 2007. Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 20:977–985 [DOI] [PubMed] [Google Scholar]
- 9. Calero-Nieto F, Hera C, Di Pietro A, Orejas M, Roncero MI. 2008. Regulatory elements mediating expression of xylanase genes in Fusarium oxysporum. Fungal Genet. Biol. 45:28–34 [DOI] [PubMed] [Google Scholar]
- 10. Chiang C, Knight SG. 1960. Metabolism of d-xylose by moulds. Nature 188:79–81 [DOI] [PubMed] [Google Scholar]
- 11. Colot HV, et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coutinho PM, et al. 2009. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet. Biol. 46(Suppl. 1):S161–S169 [DOI] [PubMed] [Google Scholar]
- 13. Davis RH, Perkins DD. 2002. Neurospora: a model of model microbes. Nat. Rev. Genet. 3:397–403 [DOI] [PubMed] [Google Scholar]
- 14. Dementhon K, Iyer G, Glass NL. 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot. Cell 5:2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Vries RP, van de Vondervoort PJ, Hendriks L, van de Belt M, Visser J. 2002. Regulation of the alpha-glucuronidase-encoding gene (aguA) from Aspergillus niger. Mol. Genet. Genomics 268:96–102 [DOI] [PubMed] [Google Scholar]
- 16. de Vries RP, Visser J, de Graaff LH. 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150:281–285 [DOI] [PubMed] [Google Scholar]
- 17. Dodd D, Cann IKO. 2009. Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy 1:2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du J, Li S, Zhao H. 2010. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol. Biosyst. 6:2150–2156 [DOI] [PubMed] [Google Scholar]
- 19. Fowler T, Brown RD., Jr 1992. The bgl1 gene encoding extracellular beta-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 6:3225–3235 [DOI] [PubMed] [Google Scholar]
- 20. Furukawa T, et al. 2009. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 46:564–574 [DOI] [PubMed] [Google Scholar]
- 21. Galazka JM, et al. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86 [DOI] [PubMed] [Google Scholar]
- 22. Gancedo JM. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gielkens MM, Dekkers E, Visser J, de Graaff LH. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require D-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goncalves RD, Cupertino FB, Freitas FZ, Luchessi AD, Bertolini MC. 2011. A genome-wide screen for Neurospora crassa transcription factors regulating glycogen metabolism. Mol. Cell. Proteomics 10:M111.007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hasper AA, Trindade LM, van der Veen D, van Ooyen AJ, de Graaff LH. 2004. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 150:1367–1375 [DOI] [PubMed] [Google Scholar]
- 26. Hasper AA, Visser J, de Graaff LH. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression. Mol. Microbiol. 36:193–200 [DOI] [PubMed] [Google Scholar]
- 27. Ilmen M, Saloheimo A, Onnela ML, Penttila ME. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kasuga T, et al. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mach-Aigner AR, Gudynaite-Savitch L, Mach RL. 2011. L-arabitol is the actual inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 77:5988–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mach-Aigner AR, Pucher ME, Mach RL. 2010. D-xylose as a repressor or inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 76:1770–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mach-Aigner AR, et al. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74:6554–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maddi A, Bowman SM, Free SJ. 2009. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol. 46:768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martinez D, et al. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553–560 [DOI] [PubMed] [Google Scholar]
- 34. Marui J, et al. 2002. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 35:157–169 [DOI] [PubMed] [Google Scholar]
- 35. McCluskey K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52:245–262 [DOI] [PubMed] [Google Scholar]
- 36. McNally MT, Free SJ. 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14:545–551 [DOI] [PubMed] [Google Scholar]
- 37. Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 38. Noguchi Y, et al. 2009. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 85:141–154 [DOI] [PubMed] [Google Scholar]
- 39. Noguchi Y, Tanaka H, Kanamaru K, Kato M, Kobayashi T. 2011. Xylose triggers reversible phosphorylation of XlnR, the fungal transcriptional activator of xylanolytic and cellulolytic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 75:953–959 [DOI] [PubMed] [Google Scholar]
- 40. Omony J, de Graaff LH, van Straten G, van Boxtel AJB. 2011. Modeling and analysis of the dynamic behavior of the XlnR regulon in Aspergillus niger. BM Syst. Biol. 5:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perkins DD, Turner BC, Barry EG. 1976. Strains of Neurospora collected from nature. Evolution 30:281–313 [DOI] [PubMed] [Google Scholar]
- 42. Phillips CM, Beeson WT, Cate JH, Marletta MA. 2011. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6:1399–1406 [DOI] [PubMed] [Google Scholar]
- 43. Phillips CM, Iavarone AT, Marletta MA. 2011. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J. Proteome Res. 10:4177–4185 [DOI] [PubMed] [Google Scholar]
- 44. Portnoy T, et al. 2011. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics 12:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quinlan RJ, et al. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108:15079–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Royer JC, Nakas JP. 1990. Interrelationship of xylanase induction and cellulase induction of Trichoderma longibrachiatum. Appl. Environ. Microbiol. 56:2535–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruepp A, et al. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saloheimo M, Kuja-Panula J, Ylosmaki E, Ward M, Penttila M. 2002. Enzymatic properties and intracellular localization of the novel Trichoderma reesei beta-glucosidase BGLII (cel1A). Appl. Environ. Microbiol. 68:4546–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seiboth B, Hartl L, Pail M, Kubicek CP. 2003. D-xylose metabolism in Hypocrea jecorina: loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded L-arabinitol-4-dehydrogenase. Eukaryot. Cell 2:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Somerville C, et al. 2004. Toward a systems approach to understanding plant cell walls. Science 306:2206–2211 [DOI] [PubMed] [Google Scholar]
- 51. Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stricker AR, Mach RL, de Graaff LH. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78:211–220 [DOI] [PubMed] [Google Scholar]
- 53. Stricker AR, Steiger MG, Mach RL. 2007. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 581:3915–3920 [DOI] [PubMed] [Google Scholar]
- 54. Sun J, Glass NL. 2011. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6:e25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun J, et al. 2011. Expression and characterization of the Neurospora crassa endoglucanase GH5-1. Protein Expr. Purif. 75:147–154 [DOI] [PubMed] [Google Scholar]
- 56. Tamayo EN, et al. 2008. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 45:984–993 [DOI] [PubMed] [Google Scholar]
- 57. Tian C, et al. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 106:22157–22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tian C, Kasuga T, Sachs MS, Glass NL. 2007. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 6:1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Townsend JP. 2004. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics 5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Townsend JP, Hartl DL. 2002. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 3:RESEARCH0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsukagoshi N, Kobayashi T, Kato M. 2001. Regulation of the amylolytic and (hemi-)cellulolytic genes in aspergilli. J. Gen. Appl. Microbiol. 47:1–19 [DOI] [PubMed] [Google Scholar]
- 62. Turner BC, Perkins DD, Fairfield A. 2001. Neurospora from natural populations: a global study. Fungal Genet. Biol. 32:67–92 [DOI] [PubMed] [Google Scholar]
- 63. van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Peij NN, Visser J, de Graaff LH. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131–142 [DOI] [PubMed] [Google Scholar]
- 65. Vogel HJ. 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13:42–46 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.