Abstract
The Saccharomyces cerevisiae SEN1 gene codes for a nuclear, ATP-dependent helicase which is embedded in a complex network of protein-protein interactions. Pleiotropic phenotypes of mutations in SEN1 suggest that Sen1 functions in many nuclear processes, including transcription termination, DNA repair, and RNA processing. Sen1, along with termination factors Nrd1 and Nab3, is required for the termination of noncoding RNA transcripts, but Sen1 is associated during transcription with coding and noncoding genes. Sen1 and Nrd1 both interact directly with Nab3, as well as with the C-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II. It has been proposed that Sen1, Nab3, and Nrd1 form a complex that associates with Rpb1 through an interaction between Nrd1 and the Ser5-phosphorylated (Ser5-P) CTD. To further study the relationship between the termination factors and Rpb1, we used two-hybrid analysis and immunoprecipitation to characterize sen1-R302W, a mutation that impairs an interaction between Sen1 and the Ser2-phosphorylated CTD. Chromatin immunoprecipitation indicates that the impairment of the interaction between Sen1 and Ser2-P causes the reduced occupancy of mutant Sen1 across the entire length of noncoding genes. For protein-coding genes, mutant Sen1 occupancy is reduced early and late in transcription but is similar to that of the wild type across most of the coding region. The combined data suggest a handoff model in which proteins differentially transfer from the Ser5- to the Ser2-phosphorylated CTD to promote the termination of noncoding transcripts or other cotranscriptional events for protein-coding genes.
INTRODUCTION
In eukaryotes, protein-coding genes and some noncoding genes are transcribed by RNA polymerase II (Pol II). Transcription occurs in three highly ordered stages: initiation, elongation, and termination. Order is maintained in part through the complex interplay between factors that bind to the C-terminal domain (CTD) of Rpb1, the largest subunit of Pol II (21, 64). The CTD is composed of heptad repeats with the consensus sequence Y1S2P3T4S5P6S7 (12). The number of repeats varies in different organisms. In the yeast Saccharomyces cerevisiae, the CTD is composed of 26 heptad repeats, whereas there are 52 repeats in the CTD in humans (62).
All three serine residues in the heptad repeats of the CTD are dynamically phosphorylated and dephosphorylated during the transcription cycle. The pattern of the phosphorylation of the CTD coordinates the events of the transcription cycle by serving as a binding scaffold for proteins needed at each stage of transcription, with the phosphorylation state of the CTD determining which factors bind and in what order. The process has been likened to a code that orchestrates the ordered assembly of factors required for transcription and associated processes (6, 10, 13, 41, 43).
During initiation, a hypophosphorylated Pol II is recruited to the promoter and the preinitiation complex is formed (38). Once 8 to 9 nucleotides have been transcribed (48), the CTD is phosphorylated at Ser5 by the transcription factor IIH (TFIIH) kinase Kin28 (28, 47). The high levels of phosphorylated Ser5 promote the association of CTD binding proteins needed for capping, histone modification, and early termination (23, 28, 40, 47, 61). Kin28 also phosphorylates the CTD at Ser7 when Pol II is close to the 5′ end of genes (2, 26).
As transcription continues and enters productive elongation, the levels of phosphorylated Ser5 decrease and levels of phosphorylated Ser2 increase (24, 28, 36, 54). The CTD kinases Bur1 and CTDK1 phosphorylate the CTD at Ser2 (31, 35), and this change in phosphorylation state allows for the association of other factors involved in histone remodeling, RNA processing, and termination (7, 32, 42). Overall, about 40 factors that interact with the CTD during the transcription cycle differ in their requirements for the status of the phosphorylation of the CTD (10, 14, 43, 44, 62, 65).
Sen1, an ∼252-kDa nuclear superfamily I RNA/DNA helicase, is a CTD-binding protein (25, 55, 56). Mutations in SEN1 result in pleiotropic phenotypes, including defects in transcription, DNA repair, RNA processing, and snRNP assembly (15, 16, 45, 49–53, 55, 56, 63). Furthermore, Sen1 is part of a broad network of protein-protein interactions (39, 55) in which individual protein-protein interactions direct Sen1 to function in different transcriptional and/or cotranscriptional events (16). The N-terminal segment upstream of the ATP-helicase motif contains domains that interact not only with Rpb1 but also with Rad2, a DNA repair enzyme, Rnt1, an enzyme involved in RNA processing, and SmD3, a component of the Sm complex required for intron splicing (1, 8, 9, 17, 19, 20, 30). Mutations that disrupt specific interactions of Sen1 with Rpb1 or Rnt1 cause specific phenotypic defects in transcription termination and RNA processing (16).
Interactions with Glc7, a protein phosphatase subunit of the cleavage/polyadenylation factor, and Nab3, a heterogeneous ribonucleoprotein, require domains in a C-terminal segment of Sen1 (11, 39, 51). Sen1 acts in concert with Nab3 and Nrd1, a serine-arginine (SR)-like CTD-associated factor (51), in the termination of the transcription of nonpolyadenylated, noncoding Pol II transcripts (4, 16, 39, 51, 52, 61). Sen1 binds directly to Nab3, which binds directly to Nrd1, but Sen1 and Nrd1 do not physically interact (11, 39, 51, 55).
The proper termination of most primary transcripts coding for snoRNAs and snRNAs is believed to require a tripartite complex consisting of Nrd1, Nab3, and Sen1 and the CTD of Pol II (16, 27, 51, 60). The formation of mature 3′ ends of the primary transcripts for noncoding RNAs depends on the nuclear exosome for 3′ end trimming, which in turn requires the efficient Nrd1-dependent termination of transcription (3, 59). Nrd1 binding to the Ser5-phosphorylated CTD of Rpb1 coupled with the binding of Nrd1 and Nab3 to an RNA consensus element are necessary to promote termination (27, 51, 60).
To further understand how protein-protein interactions influence the function of Sen1 in transcription and related events that occur during gene expression, we focused on the interactions of Sen1 with different phosphorylated forms of the CTD of Rpb1 as well as the interactions between Sen1, Nrd1, and Nab3 with respect to the CTD. To accomplish this, we made use of a mutation in Sen1, sen1-R302W, that specifically impairs the interaction between Sen1 and Rpb1 (16). The results suggest that Nrd1 and Sen1 interact with different phosphorylated forms of the CTD and that Sen1 associates with Rpb1 by at least two mechanisms involving either direct binding to the Ser2-phosphorylated CTD or through indirect interaction with the Ser5-phosphorylated CTD as part of the Nrd1-Nab3-Sen1 complex.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Yeast strains and plasmids are described in Table S1 in the supplemental material. In strains DUY1397, DUY1531, and 46α, the SEN1 open reading frame (ORF) was modified by inserting DNA coding for a C-terminal TAP or cMyc13 epitope tag (18, 34, 53). The mutant allele sen1-R302W results in a substitution of tryptophan (W) for arginine (R) at amino acid position 302 (see Fig. 2B). To create sen1-R302W in plasmids pKC122 and pKC173, AGA in the coding strand (nucleotides 1904 to 1906) was changed to TGG by site-directed mutagenesis (Agilent) in plasmids pKW7 and pJF9, respectively, which carry wild-type SEN1. The 26-mer CTD containing alanine substitutions at Ser7 (pGAD-S2S5A7) was obtained by custom synthesis (MWG Operon). pKC174 and pKC180 contain the Nrd1 and Nab3 ORFs cloned into pGBDU-C1. pKC175, pKC178, pKC182, and pKC190 were created by inserting DNA coding for an N-terminal GST or HA epitope tag in the NRD1 or NAB3 ORF and placing the tagged ORFs under the control of the ADH1 promoter. Epitope-tagged Sen1 was determined to be functional through its ability to complement the temperature-sensitive phenotype of a strain carrying sen1-1 (63). Epitope-tagged Nrd1 and Nab3 were shown to be stably expressed and to engage in functional interactions, indicating the production of functional proteins. Media and conditions for the growth of yeast were described previously (55). Alleles carrying point mutations were integrated into the genome by two-step gene replacement (5, 46) and confirmed by DNA sequence analysis.
Fig 2.
sen1-R302W selectively impairs the interaction of Sen1 with Rpb1. (A) Map of the SEN1 gene. The Figure shows the locations of the N-terminal binding regions for Rad2, Rnt1, Rpb1, and SmD3, the ATP helicase region and RNA binding domain, and the C-terminal binding regions for Glc7 and Nab3. (B) Amino acid sequence surrounding the replacement of arginine (R) with tryptophan (W) at position 302. (C) Two-hybrid interactions of GBD-Sen1 and GBD-Sen1-R302W with GAD-Rpb1, GAD-Rad2, and GAD-Rnt1 (see Materials and Methods). Growth on medium lacking histidine or adenine was assayed by drop tests using serial dilutions of mid-log-phase cultures. Growth on medium lacking histidine, leucine, and uracil or leucine, uracil, and adenine indicates the activation of GAL1-HIS3 or GAL2-ADE2 reporter, respectively. (D) Coimmunoprecipitation of Sen1 with Rpb1 (Materials and Methods). Sen1-R302W impairs the binding of Sen1 to Rpb1 in the presence of RNase (+ RNase). Lanes 5 and 6 were previously published (16) but are included for comparison. TAP, tandem affinity purification tag. HA, influenza virus hemagglutinin tag.
Standard two-hybrid interactions.
Plasmids pGBDU-C1 and pGAD-C1 were used to construct fusions for use in the standard two-hybrid system (22, 55). Sets of GAD (Gal4 activation domain) and GBD (Gal4 binding domain) fusions and plasmids representing empty vectors were transformed into strain PJ69-4A, which carries two reporter genes, HIS3 and ADE2, that are under the control of Gal4-inducible GAL1 and GAL2 promoters, respectively (22). Plasmids carrying the GAD fusions express LEU2, and those carrying the GBD fusions express URA3. The GBD-Sen1 and GBD-Sen1-R302W fusions contain GBD fused to the first 565 amino acids of Sen1. GBD-Sen1-R302W contains a mutation causing a substitution of arginine (R) for tryptophan (W) at position 302. The GAD-Rad2 fusion contains DNA coding for amino acids 327 to 672 of Rad2. The GAD-Rpb1 fusion contains DNA coding for amino acids 1497 to 1733 (the C-terminal domain [CTD]) of Rpb1. The GAD-Rnt1 fusion contains DNA coding for amino acids 1 to 471 of Rnt1. Growth rates were assayed by drop tests using serial dilutions of mid-log-phase cultures. Five-microliter drops of undiluted and 5-, 25-, and 125-fold diluted cultures were placed on solid medium and incubated for 3 days at 30°C. The expression of the GAL1-HIS3 reporter was assayed on synthetic dextrose (SD) medium lacking histidine, leucine, and uracil. Growth is expected for all strains on medium lacking uracil and leucine, which selects for the presence of the two-hybrid vectors.
Two-hybrid GAD-CTD-P fusions.
We previously reported the development of a novel two-hybrid system designed to detect interactions between proteins that bind to the CTD of Rpb1 in a manner that depends on the phosphorylation of serine residues in Tyr1Ser2Pro3Thr4Ser5Pro6Ser7, which is repeated 26 times in the CTD of S. cerevisiae (12, 57). Two-hybrid plasmids expressing GAD-CTD-HA (hemagglutinin epitope) contain substitutions of alanine for serine at Ser2, Ser5, or Ser7 within each repeat (pGAD-A2S5S7, pGAD-S2A5S7, and pGAD-S2S5A7). The fusions include 18, 15, and 26 heptapeptide repeats, respectively. The two-hybrid fusions were constructed as described previously (57).
Protein extraction.
Cells were grown in the appropriate minimal medium to an optical density at 600 nm (OD600) of ∼1.0. Cells then were harvested at 4°C at 5,000 rpm for 10 min and washed once with water. The pellet was resuspended in cold lysis buffer (30 mM HEPES, pH 7.5, 200 mM KOAC, 1 mM MgOAC, 1 mM EGTA, pH 8.0, 10% glycerol, 0.05% Tween 20) with phenylmethylsulfonyl fluoride (PMSF), benzamidine hydrochloride, protease inhibitor cocktail (Sigma), and HALT phosphatase inhibitor cocktail (Thermo Fisher Scientific) (33). The cell slurry was added directly to a mortar filled with liquid nitrogen. The liquid nitrogen was allowed to evaporate and the frozen yeast was ground with a pestle until the yeast had the appearance of fine powder. The frozen yeast powder was thawed on ice and then cleared by centrifugation at 18,000 rpm for 2 h at 4°C.
IP.
Yeast lysates were quantified using the Bradford assay (Bio-Rad). A 50-μl aliquot representing the whole-cell extract or input was taken and frozen at −80°C. Equal amounts of protein were used for each set of immunoprecipitations (IP). For all experiments where RNase was used, 10 μg/ml RNase A (Sigma) was added to pre-IP lysates. Lysates were allowed to bind the appropriate antibody overnight, with rotating at 4°C. One hundred-μl protein G Sepharose 4 Fast Flow beads (GE Healthcare) then were added to each IP and allowed to bind the antibody-antigen complex for 2 h, with rotating at 4°C. Beads were collected by centrifugation at 1,500 rpm for 1 min and washed three times with 700 μl lysis buffer. The beads were resuspended in 4× Laemmli sample buffer (29) and heated at 50°C for 20 min. This nonstandard treatment was found to prevent protein aggregation. The suspension was centrifuged for 10 min, and the supernatant was transferred to new tubes and heated at 65°C for 10 min.
Western blotting.
Proteins were detected by Western blotting as described previously (16). Immunoprecipitated proteins were fractionated on SDS-PAGE gels and transferred to nitrocellulose membranes (GE Healthcare). Membranes were probed with the following antibodies: mouse monoclonal anti-human influenza hemagglutinin (HA) (clone HA-7; Sigma), mouse monoclonal anti-cMyc (clone 9E10; Sigma), rabbit polyclonal anti-glutathione S-transferase (GST) (Sigma), and rabbit polyclonal anti-TAP (Thermo Fisher Scientific). Phosphospecific antibodies against Rpb1p include rat monoclonal anti-RNA polymerase II-CTD-Ser2-P (clone 3E10; Ascenion), anti-RNA polymerase II CTD-Ser5-P (clone 3E8; Ascenion), anti-RNA polymerase II-CTD-Ser7-P (clone 4E12; Ascenion), mouse monoclonal anti-unphosphorylated RNA polymerase II (clone 8WG16; Covance), anti-RNA polymerase II CTD-Ser5-P (clone H14; Covance), and anti-RNA polymerase II CTD-Ser2-P (clone H5; Covance). Secondary antibodies were horseradish peroxidase conjugated (Thermo Fisher Scientific) or fluorescently labeled (LI-COR). Proteins bands were visualized by chemiluminescence (SuperSignal West Pico chemiluminescent substrate; Thermo Fisher Scientific) or by infrared imaging (Odyssey imaging system; LI-COR).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (54), with some modifications. Briefly, anti-Rpb3 (Neoclone) or anti-cMyc (clone 9E10; no. sc-40; Santa Cruz Biotechnology) antibody was added to yeast whole-cell extract (1:167 dilution for anti-Rpb3 and 1:100 dilution for anti-cMyc) and incubated with mixing at 4°C overnight. For TAP tag ChIPs, IgG beads (GE Healthcare) were equilibrated with two 1-ml lysis buffer washes. Forty μl of a 50/50 IgG resin (GE Healthcare) slurry was added to yeast whole-cell extract and incubated with mixing at 4°C overnight. For ligation-mediated PCR, 40 μl of purified ChIP DNA and 5 μl of purified DNA were blunted with T4 DNA polymerase (Fermentas) for 20 min at 12°C. DNA was purified by phenol-chloroform-isoamyl alcohol (25:24:1) using 10 μg of glycogen as the carrier and precipitated with 0.1 volumes of 3 M sodium acetate (NaOAc) and 2.5 volumes of cold ethanol (EtOH). Samples were left at −80°C for 30 min and centrifuged at 15,000 rpm for 15 min at 4°C. The DNA pellet was washed with 10 volumes of 70% cold EtOH and centrifuged at 15,000 rpm for 5 min at 4°C. Liquid was removed, and the dry DNA pellet was resuspended in 25 μl of nano-filtered water (nfH2O; Sigma). Linkers were prepared by mixing 40 μM two oligonucleotides (oJW102, GCGGTGACCCGGGAGATCTGAATTC; oJW103, GAATTCAGATC) in 250 mM Tris HCl, pH 7.9, heating to 95°C for 5 min, and cooling to room temperature. Two μM annealed linkers were ligated onto both ends of the DNA with T4 DNA ligase (Fermentas) overnight at 16°C. ChIP and input DNA with annealed linkers were precipitated from solution with 3 M NaOAc and cold EtOH as described above. The labeling of ChIP and input DNA was carried out by PCR with 0.4 mM dATP, dCTP, dGTP, and 800 μM dTTP. Forty μM Cy3-dUTP and Cy5-dUTP (Enzo Life Sciences) were added to label input and ChIP DNA, respectively. PCR was performed with the following steps: 55°C for 4 min, 72°C for 5 min, 95°C for 2 min, 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 4 min. The last three steps were cycled 31 times for ChIP samples and 33 times for input samples. Samples were purified with the QIAquick PCR purification kit (Qiagen) and eluted from the column twice with 54 μl of elution buffer. Dye incorporation was determined with a Nano Drop 1000 (Thermo Scientific). Microarray design, hybridization, data processing, and data analysis have been previously described (54).
Calculation of Rpb1 occupancy.
The fraction of relative RNA Pol II at protein versus noncoding genes was estimated using RNA Pol II ChIP-chip data (54). Pol II occupancy was averaged for all 6,575 ORFs and 82 Pol II-transcribed small nuclear RNAs and small nucleolar RNAs (Saccharomyces Genome Database). Genes with an average negative Pol II occupancy over the corresponding ORF were discarded (587 ORFs and 2 noncoding genes). The average values of Pol II occupancy for each gene were added together to calculate total occupancy across the genome. A total Pol II ChIP-chip signal of 95.5% was found at protein-coding genes.
RESULTS
Nrd1, Nab3, and Sen1 form a complex.
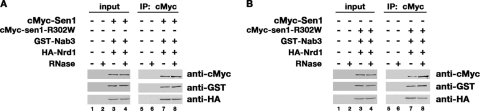
Nrd1-Nab3 and Nab3-Sen1 physically interact in the two-hybrid system (39, 60), suggesting that two binary complexes form or that the two interactions combined lead to the formation of a tripartite complex. If a tripartite complex forms, then the three proteins should copurify. To test this hypothesis, IP experiments were performed using protein extracts from a strain that coexpresses epitope-tagged HA-Nrd1, GST-Nab3, and cMyc-Sen1 from centromeric plasmids and a control strain lacking the plasmids.
Protein extracts were incubated with anti-cMyc antibodies in the absence or presence of RNase. Proteins bound to Sepharose beads were eluted, fractionated by gel electrophoresis, transferred to membranes, and analyzed by Western blotting (Fig. 1A). When membranes were probed with anti-cMyc antibodies, a band migrating at ∼250 kDa, corresponding to cMyc-Sen1, was detected in the strain expressing the epitope-tagged protein (Fig. 1A, lane 7) but not in the control strain (Fig. 1A, lane 5). When the same membranes were probed with anti-GST antibodies, a band migrating at ∼125 kDa, corresponding to GST-Nab3, was detected (Fig. 1A, lane 7), indicating that Nab3 copurifies with Sen1. The same membranes were also probed with anti-HA antibodies. A band migrating at ∼70 kDa, corresponding to HA-Nrd1, was detected (Fig. 1A, lane 7), indicating that Nrd1 also copurifies with Sen1. When the experiment was performed in the presence of RNase, both Nab3 and Nrd1 copurified with Sen1, indicating that the interactions are not dependent on RNA (Fig. 1A, lanes 6 and 8). Taken together, the yeast two-hybrid experiments performed by others (39, 60) and our IP experiments indicate that the three proteins form a complex.
Fig 1.
Sen1, Nrd1, and Nab3 copurify as a complex. (A) Immunoprecipitation experiments performed using protein extracts from a strain that coexpresses epitope-tagged HA-Nrd1, GST-Nab3, and cMyc-Sen1 and a control strain lacking the plasmids. Protein extracts were incubated with anti-cMyc antibodies, and membranes were probed with anti-cMyc, anti-GST, and anti-HA antibodies to detect Sen1, Nab3, and Nrd1, respectively. Experiments were performed in the absence (−) and presence (+) of RNase. The input lanes contain 0.2% of the total lysate. IP lanes contain 20% of the total lysate. (B) Immunoprecipitation experiments were performed as described for panel A, except protein extracts came from a strain that coexpresses epitope-tagged HA-Nrd1, GST-Nab3, and cMyc-Sen1-R302W and a control strain lacking the plasmids.
R302W amino acid substitution disrupts a physical interaction between Sen1 and Rpb1.
Previously reported two-hybrid and IP experiments indicated that two members of the complex, Nrd1 and Sen1, interact physically with the C-terminal domain of Rpb1, the largest subunit of RNA polymerase II (49, 51, 55, 64, 65). The interaction domain in Sen1 was localized to the first 565 amino acids (Fig. 2A) (55). The combined results showing that two of the three members of Nrd1-Nab3-Sen1 interact with Rpb1 suggest a potentially complex interplay with the transcription machinery.
To further examine the Sen1-Rpb1 interaction, conserved, charged residues were identified in the first 565 residues of Sen1 using the phylogenetic comparison of sequences from closely related species of yeast. Mutations in conserved residues that reverse or abolish the charge were constructed by site-directed mutagenesis (Materials and Methods) and screened by two-hybrid analysis for the loss of interaction with three proteins, Rad2, Rnt1, and Rpb1, which are known to interact with Sen1 domains in the first 565 amino acids (Table 1). Most of the mutations either had no effect on any of the interactions or impaired all of the interactions. However, three mutations were identified that specifically disrupt a single interaction. E58K impaired the interaction with Rad2; K128E impaired the interaction with Rnt1; and R302W impaired the interaction with Rpb1 (Fig. 2B). The phenotypic effects of K128E and R302W on the maturation of U5 snRNA and the termination of the transcription of SNR7 (coding for U5 snRNA), respectively, were reported previously (16). In that study it was also reported that sen1-R302W reduces the copurification of Sen1 and Rpb1 by IP in the presence of RNase. In this study, we further characterized the Sen1-Rpb1 interaction and its impairment by sen1-R302W. It was also found that sen1-R302W causes a slight growth impairment. Strain DUY1513, which carries sen1-R302W, has a doubling time in rich undefined media of 2 h, whereas the isogenic SEN1 strain BY4741 has a doubling time of 1.5 h (see Table S1 in the supplemental material).
Table 1.
Effects of sen1 mutations on protein-protein interactionsa
| Amino acid change | Growth status of interacting partner: |
||
|---|---|---|---|
| Rad2 | Rnt1 | Rpb1 | |
| E58K | − | + | + |
| H77D | −/+ | −/+ | −/+ |
| C80Y | − | − | − |
| K128E | + | − | + |
| R139E | − | − | − |
| W156E | + | + | + |
| R197E | − | − | − |
| K203E | + | + | + |
| W250E | − | − | − |
| H281D | + | + | + |
| W300E | − | − | − |
| R302W | + | + | − |
Symbols: +, growth that is indicative of a protein-protein interaction; −, no growth, which is indicative of no observed protein-protein interaction; −/+, diminished growth compared to the wild type, which is indicative of a weaker protein-protein interaction.
The two-hybrid system detects protein-protein interactions when the GAL4 DNA binding domain (GBD) and GAL4 transcriptional activation domain (GAD) in GBD and GAD fusions are brought in proximity due to an interaction, activating reporters that result in growth on appropriate omission media (22). Sen1 was previously reported to exhibit two-hybrid interactions with Rpb1, Rad2, and Rnt1 (55). When GBD-Sen1 fusions coding for the wild-type and R302W amino acid sequences were tested for interaction with GAD-Rpb1, GAD-Rad2, and GAD-Rnt1 fusions, we found that the substitution at position 302 severely reduced growth compared to that of the wild type in strains expressing a GAD-Rpb1 fusion (Fig. 2C, rows 1 and 2) but had no visible effects on growth in strains expressing GAD-Rad2 or GAD-Rnt1 (Fig. 2C, rows 3 and 4 and rows 5 and 6, respectively). For Rpb1 and Rad2, similar results were observed using two different reporters, GAL1-HIS3 and GAL2-ADE2. The Rnt1 interaction was the weakest and was only detected using the GAL1-HIS3 reporter. Overall, the results suggest that R302W affects the specific domain in Sen1 that is involved in the Rpb1 interaction without affecting other nearby interaction domains.
To further characterize the effects of R302W on the Sen1-Rpb1 interaction, IPs were performed in the absence or presence of RNase. Protein extracts from strains coexpressing epitope-tagged HA-Rpb1 (hemagglutinin) with Sen1-TAP or Sen1-R302W-TAP (tandem affinity) were incubated with anti-HA antibodies. Bound proteins were eluted, fractionated by gel electrophoresis, and analyzed by Western blotting using anti-HA and anti-TAP antibodies (Fig. 2D). In the absence of RNase, both Sen1 and Sen1-R302W copurified with Rpb1. However, in the presence of RNase only the wild-type Sen1 protein was detected by Western blotting. The failure of Sen1-R302W to copurify with Rpb1 suggests two possibilities. The physical interaction between Sen1 and Rpb1 might depend on the presence of RNA, presumably nascent transcripts. Alternatively, Sen1 might associate with native transcription complexes, including Rpb1, through two different mechanisms, one due to direct protein-protein interaction with Rpb1 and the other due to indirect RNA-dependent tethering to Rpb1. Copurification with Rpb1 is abolished only when the direct and indirect interactions are abolished.
Sen1 interacts with the Ser2-phosphorylated form of the Rpb1 CTD in a modified two-hybrid system.
Two-hybrid fusions containing CTD repeats were engineered for use in a two-hybrid assay designed to reveal phosphorylation-dependent binding to the CTD. This system was shown previously to faithfully reproduce the known binding requirements of three CTD binding proteins (57). Alanine substitutions were introduced in GBD-CTD two-hybrid fusions separately at Ser2, Ser5, or Ser7 (Fig. 3A). Western blots probed with antibodies that uniquely recognize phosphorylated forms of Ser2, Ser5, and Ser7 (10, 26) indicate that the plasmid pS2S5S7 codes for a GAD-CTD fusion that is phosphorylated at all three positions, whereas alanine substitutions in the fusions expressed from the plasmids pA2S5S7, pS2A5S7, and pS2S5A7 abolish phosphorylation at Ser2, Ser5, and Ser7, respectively (Fig. 3B). Not all two-hybrid fusions exhibited equal levels of phosphorylation. For example, the fusion expressed from plasmid pS2A5S7 showed decreased levels of phosphorylation at Ser2 (Fig. 3B). However, the two-hybrid results described below indicate that phosphorylation at Ser2 is above the threshold required for Sen1 to interact with pS2A5S7 (Fig. 3C). Also, phosphorylation at Ser7 was abolished in the two-hybrid fusions expressed from plasmids pA2S5S7 and pS2A5S7 (Fig. 3B), which is consistent with a previously reported suggestion that phosphorylation at Ser7 depends on phosphorylation at Ser2 and Ser5 (2).
Fig 3.
Interactions with phosphorylated forms of the Rpb1 CTD. (A) Two-hybrid fusions between GAD and sequences coding for CTDs containing alanine substitutions at Ser2, Ser5, and Ser7. (B) Western blotting of total protein from strains expressing the GAD-CTD-HA fusions using antibodies that recognize Ser2-P, Ser5-P, and Ser7-P. (C) Two-hybrid interactions between GBD-Sen1, GBD-sen1-R302W, GBD-Nab3, and GBD-Nrd1 with the GAD-CTD fusions. GAL1-HIS3 served as the reporter. Serial dilutions of overnight cultures were plated, and growth was scored after 3 days of incubation at 30°C.
We tested the ability of Sen1 and Sen1-R302W to interact with the GAD fusions described above (Fig. 3C, rows 1 to 10). Strains coexpressing GBD-Sen1 and the GAD fusions containing phosphorylated Ser2 grew in the absence of histidine, indicating that the GAL1-HIS3 reporter was activated through interactions, (Fig. 3C, rows 1 to 5). However, the alanine substitution at Ser2 prevented growth (Fig. 3C, row 2). This result suggests that Sen1 interacts specifically with the Ser2-phosphorylated form of the CTD in Rpb1. When the same experiment was performed in strains expressing GBD-Sen1-R302W, none of the strains grew in the absence of histidine (Fig. 3C, rows 6 to 10), suggesting that the R302W substitution severely impairs the interaction between Sen1 and the Ser2-phosphorylated CTD.
Since Sen1 can form a tripartite complex with Nab3 and Nrd1 (Fig. 1), we used the system described above to test for interactions with strains coexpressing GBD-Nab3 or GBD-Nrd1 with the GAD-CTD fusions (Fig. 3C, rows 11 to 20). None of the strains expressing GBD-Nab3 grew in the absence of histidine (Fig. 3C, rows 11 to 15), which is consistent with a previous report that Nab3 does not directly interact with the CTD (11). Strains expressing GBD-Nrd1 grew in the absence of histidine (Fig. 3C, rows 16, 17, and 19), except when alanine was substituted for Ser5 (Fig. 3C, row 18). The results are consistent with a previous report that Nrd1 binds to the Ser5-phosphorylated form of the CTD but fails to bind directly to the Ser2-phosphorylated form of the CTD (61). Furthermore, phosphorylation at Ser7 does not appear to be required for the binding of either protein to the CTD. Overall, these results suggest that two components of the tripartite complex, Sen1 and Nrd1, physically interact with different phosphorylated forms of the CTD.
Copurification of Sen1-R302W with Ser2-phosphorylated Rpb1 is severely reduced compared to wild-type Sen1.
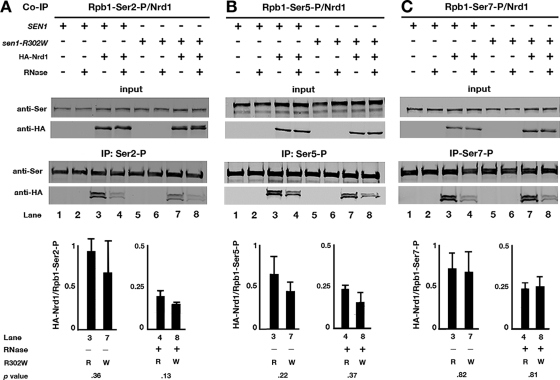
CoIP studies were performed to examine the extent to which Sen1 and Sen1-R302W copurify with each of the three phosphorylated forms of Rpb1. Protein extracts from cells expressing epitope-tagged cMyc-Sen1 or cMyc-Sen1-R302W were incubated with antibodies that specifically recognize the Ser2-, Ser5-, and Ser7-phosphorylated forms of the Rpb1 CTD. Bound proteins were eluted, fractionated by gel electrophoresis, and analyzed by Western blotting using anti-cMyc and anti-Ser-P antibodies (Fig. 4).
Fig 4.
Association of Sen1 with phosphorylated forms of Rpb1. Cells expressing epitope-tagged cMyc-Sen1 or cMyc-sen1-R302W were incubated with phosphospecific antibodies against Rpb1-Ser2-P (A), Rpb1-Ser5-P (B), or Rpb1-Ser7-P (C) in the absence (−) or presence (+) of RNase and analyzed by Western blotting. Membranes were probed with antibodies against the cMyc epitope tag. The amount of copurified Sen1 was measured by calculating the ratio of cMyc-Sen1 or cMyc-Sen1-R302W to the Ser2-, Ser5- and Ser7-phoshorylated forms of the Rpb1 CTD. In cells expressing Sen1-R302W, the addition of RNase results in a drastic decrease in the amount of Sen1 that copurifies with Rpb1-Ser2-P (A). The sen1-R302W mutation had no effect on the amount of Sen1 copurifying with Rpb1-Ser5-P or Rpb1-Ser7-P regardless of RNase being present (B and C). Error bars and P values are based on three experiments.
In the presence of RNase, the relative amount of Sen1-R302W that copurified with the Ser2-phosphorylated form of Rpb1 was significantly reduced (P = 0.01) but not completely eliminated (Fig. 4A). Residual Sen1-R302W is probably due to the copurification of Sen1-R302W with Ser2-Ser5 doubly phosphorylated repeats of the CTD, which are also recognized by the anti-Ser2 antibody used. When similar experiments were conducted using anti-Ser5 and anti-Ser7 for IP, there was no significant reduction in copurification caused by the Sen1-R302W amino acid substitution (Fig. 4B and C).
These results are consistent with the two-hybrid observations (Fig. 3), suggesting that Sen1 binds to the Ser2-phosphorylated form of Rpb1, and that binding is severely reduced by the R302W amino acid substitution. In the absence of RNase, statistically similar amounts of Sen1 and Sen1-R302W copurified with all three of the Rpb1 phosphorylated forms, suggesting that Sen1 is tethered to the transcription machinery by additional mechanisms unrelated to the binding of Sen1 to the Ser2-phosphorylated form of Rpb1.
Nrd1-Nab3-Sen1 complex is not disrupted by Sen1-R302W.
To test whether Sen1-R302W still could associate with Nrd1 and Nab3, we performed IP experiments as described for Fig. 1A, except that protein extracts were from a strain that coexpresses epitope-tagged HA-Nrd1, GST-Nab3, and cMyc-Sen1-R302W from centromeric plasmids and a control strain lacking the plasmids (Fig. 1B). The membranes were probed with anti-cMyc, anti-GST, or anti-HA. Similarly to the experiment described above using cMyc-Sen1 (Fig. 1A), HA-Nrd1 and GST-Nab3 both coimmunoprecipitated with cMyc-Sen1-R302W (Fig. 1B, lanes 7 and 8). This indicates that while R302W impairs the binding of Sen1 to the Ser2-phosphorylated CTD, the amino acid substitution does not appear to affect the assembly or stability of the Nrd1-Nab3-Sen1 complex.
Association of Nrd1 and Nab3 with Rpb1 is not altered by Sen1-R302W.
Since Nrd1 binds to Ser5-phosphorylated Rpb1 (61) and Sen1 binds to the Ser2-phosphorylated form, the tripartite Nrd1-Nab3-Sen1 complex could associate with two different phosphorylated forms of Rpb1. If the complex is bound to phosphorylated Ser2, then the loss of the binding of Sen1-R302W to phosphorylated Ser2 should impair the association of the entire complex with phosphorylated Ser2. If Sen1 is bound to phosphorylated Ser2 by itself without being associated with Nrd1 and Nab3, then R302W should have no effect on copurification of Nrd1 and Nab3 with Rpb1.
To distinguish between these possibilities, IP experiments were performed in strains carrying either SEN1 or sen1-R302W using antibodies that recognize each of the three phosphorylated serine residues in the CTD. The strains expressed either HA-Nrd1 or HA-Nab3 from centromeric plasmids. The IP lysates were analyzed by Western blotting using anti-HA antibodies. In the presence of RNase, sen1-R302W had no significant effect (P = 0.05) on the relative amounts of Nrd1 that associate with Ser2-, Ser5-, or Ser7-phosporylated Rpb1 (Fig. 5A, B, and C, respectively). More Nrd1 associated with Ser2- and Ser7-phosphorylated Rpb1 when RNase was omitted, but no significant change was observed when SEN1 and sen1-R302W were compared (Fig. 5A and C). The results suggest that Sen1 is bound by itself to Ser2-phosphorylated Rpb1 without forming a complex with Nrd1. Using the same strategy, it was also found that sen1-R302W had no effect on the association of Nab3 with any of the phosphorylated forms of Rpb1 in either the presence or absence of RNase (Fig. 6).
Fig 5.
Copurification of phosphorylated forms of Rpb1 with Nrd1. SEN1 or sen1-R302W strains expressing HA-Nrd1 were incubated with phospho-specific antibodies against Rpb1-Ser2-P (A), -Ser5-P (B), or -Ser7-P (C) in the absence (−) or presence (+) of RNase and were analyzed by Western blotting. Membranes were probed with antibodies against HA epitope tag. The amount of copurified HA-Nrd1 was measured by calculating the ratio of HA-Nrd1 to the Ser2-, Ser5-, and Ser7-phoshorylated forms of the Rpb1 CTD. sen1-R302W had no effect on the amount of Nrd1 that copurified with Rpb1-Ser2-P, Rpb1-Ser5-P, or Rpb1-Ser7-P (A, B, and C). Error bars and P values are based on three experiments.
Fig 6.
Copurification of phosphorylated forms of Rpb1 with Nab3. SEN1 or sen1-R302W strains expressing HA-Nab3 were incubated with phosphospecific antibodies against Rpb1-Ser2-P (A), Rpb1-Ser5-P (B), or Rpb1-Ser7-P (C) in the absence (−) or presence (+) of RNase and were analyzed by Western blotting. Membranes were probed with antibodies against HA epitope tag. The amount of copurified HA-Nab3 was measured by calculating the ratio of HA-Nab3 to the Ser2-, Ser5-, and Ser7-phoshorylated forms of the Rpb1 CTD. sen1-R302W did not have an effect on the amount of Nab3 that copurified with Rpb1-Ser2-P, Rpb1-Ser5-P, or Rpb1-Ser7-P (A, B, and C). Error bars and P values are based on three experiments.
sen1-R302W perturbs transcriptional occupancy of Sen1.
Our IP experiments suggest that Sen1 binds to the Ser2-phosphorylated CTD for reasons unrelated to transcription termination and without being associated with Nrd1 and Nab3. However, at odds with this result is the previously reported finding that sen1-R302W causes the transcriptional readthrough of the noncoding gene SNR7 (16). To further explore the basis of the discrepancy, genome-wide occupancy profiles of Sen1 and Sen1-R302W were examined via chromatin immunoprecipitation and tiling microarray analysis that covers the entire yeast genome (ChIP-chip). Specifically, ChIP-chip experiments were performed to assess whether sen1 and sen1-R302W are differentially distributed between different classes of genes (Fig. 7).
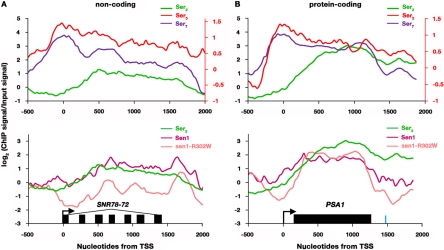
Fig 7.
Strain carrying sen1-R302W causes altered transcriptional occupancy of Sen1. ChIP-chip traces of Ser2-P, Ser5-P, and Ser7-P (top panels) and Sen1-cMyc, Sen1-R302W-TAP, and Ser2-P CTD (bottom panels) over the polycistronic snoRNA cluster SNR78-72 (A) and the protein-coding gene PSA1 (B). Black arrows denote the transcriptional start sites (TSS). Black bars within the transcribed region show the mature snoRNA transcripts in SNR78-72 and the protein-coding region of PSA1. In PSA1, the blue bar denotes the cleavage and polyadenylation site. The scale used for the right y axis corresponds to the Ser5-P profile.
We examined genome-wide Pol II-Ser2-P, Pol II-Ser5-P, Pol II-Ser7-P, Sen1, and Sen1-R302W occupancy and focused on genes with uniformly high levels of Pol II over the transcription unit, as done previously (54) (Fig. 7). SNR78-72 and PSA1 are shown as representative noncoding (Fig. 7A) and protein-coding genes (Fig. 7B), respectively. For the SNR78-72 polycistronic snoRNA gene cluster, Sen1 and Ser2-P profiles coincide across the gene, which is consistent with Sen1 associating with the Ser2-phosphorylated form of Rpb1 (Fig. 7A, bottom). The Sen1-R302W profile also coincides with the Ser2-P profile at the promoter, but it quickly deviates near the transcriptional start site (TSS) (Fig. 7A, bottom). While Ser2-P and Sen1 enrichment is observed at the 5′ end of the gene, Sen1-R302W levels are much lower. Although an increase in Sen1-R302W occupancy is observed over the middle of the transcription unit, Sen1-R302W levels still remain lower than both Sen1 and Ser2-P levels. All three signals decrease toward the 3′ end of the gene.
On PSA1, a protein-coding gene, a different Sen1 profile was observed (Fig. 7B, bottom). Ser2-P, Sen1, and Sen1-R302W profiles coincide at the promoter. Early in transcription, Sen1-R302W shows lower occupancy than Sen1, but it rapidly recovers as transcription proceeds into the PSA1 ORF. Near the end of the ORF, Sen1-R302W occupancy begins to decline more precipitously than that of Sen1, even though Ser2-P levels are still relatively high. All of the signals become diminished at the 3′ end of the gene near the cleavage and polyadenylation site (Fig. 7B, bottom panel).
To assess the global nature of these patterns, the average occupancy profiles of Sen1 and Sen1-R302W were compared across noncoding and protein-coding genes (Fig. 8). The entire transcription unit was divided into five bins across noncoding genes (RNA Pol II-transcribed small nuclear and small nucleolar RNA genes) at least 160 bp in length (Fig. 8A, top and middle) or into 10 evenly scaled bins across 60 protein-coding genes isolated from neighboring elements by at least 400 bp (Fig. 8B, top and middle) (54). For noncoding genes, Sen1-R302W occupancy was significantly lower across the transcription unit (Fig. 8A, middle), which is consistent with the profiles seen for SNR78-72 (Fig. 7). For protein-coding genes (Fig. 8B, middle), Sen1-R302W levels were lower than Sen1 levels in the early regions of the transcription unit, but the levels of mutant Sen1 recover and coincide with the wild type across the transcribed unit up to the point where Ser2-P is maximal (Fig. 8A, top and middle), as was observed for PSA1 (Fig. 7). Beyond that point, the occupancy of the mutant protein declines precipitously compared to that of the wild type (Fig. 8B, middle). The quantification of the data revealed that total Sen1-R302W levels were significantly lower than those of Sen1 on noncoding genes (P = 1.09 × 10−31, α = 0.01) (Fig. 8A, bottom), but there was no significant difference between the wild type and the mutant for protein-coding genes (P = 0.039, α = 0.01) (Fig. 8B, bottom).
Fig 8.
Genome-wide comparison of the distributions of Sen1 and Sen1-R302W. (A) Profiles show the average distribution of Ser2-P and Ser5-P occupancy (top panel) and Sen1 and Sen1-R302W occupancy (middle panel) across noncoding genes (nc genes). The scale used for the right y axis corresponds to the Ser5-P profile. Black arrows denote the transcriptional start site. Bins within the transcribed region are equivalent to 20% of the transcribed region length. The bottom panel shows histograms of average Sen1 and Sen1-R302W occupancy at noncoding genes. Curves are the scaled probability density plots for the observed data. P values (n = 82, α = 0.01) were derived by pairwise t tests. (B) Profiles show the average distribution of Ser2-P and Ser5-P occupancy (top panel) and Sen1 and Sen1-R302W occupancy (middle panel) across protein-coding genes (pc genes). The scale used for the right y axis corresponds to the Ser5-P profile. Black arrows denote the transcriptional start site. Bins within the transcribed region are equivalent to 10% of the transcribed region length. The blue bar represents the cleavage and polyadenylation site. The bottom panel shows histograms of average Sen1 and Sen1-R302W occupancy at protein-coding genes. Curves are the scaled probability density plots for the observed data. P values (n = 60, α = 0.01) were derived by pairwise t tests.
DISCUSSION
Sen1-Rpb1 interactions.
An interaction of Sen1 with the CTD of Rpb1 based on two-hybrid and IP experiments was reported previously (55). However, it was not known at that time whether the phosphorylation of the CTD was important for the Sen1-Rpb1 interaction. In this report, we analyzed the interactions of Sen1 and Sen1-R302W with protein partners using standard two-hybrid assays, a modified two-hybrid assay developed to detect binding to specific phosphorylated forms of the CTD (57), co-IP experiments, and genome-wide analysis using ChIP-chip.
Standard two-hybrid tests indicate that sen1-R302W reduces the interaction with Rpb1 based on the growth-dependent activation of the reporter GAL1-HIS3 or GAL2-ADE2. However, interactions with two other partners, Rad2 and Rnt1, were not affected by sen1-R302W when the interactions were monitored using the same reporters. These results are consistent with a previous report based on co-IP experiments showing that sen1-R302W had no effect on the Sen1-Rnt1 interaction (16). The binding defect caused by sen1-R302W therefore appears to be specific for Rpb1.
The nature of the interaction between Sen1 and Rpb1 was further illuminated using a series of two-hybrid fusions designed to discriminate which phosphorylated form of the CTD is required for Sen1 binding (57). The GAD-CTD fusions have two important features: all three serine residues are phosphorylated, and alanine substitutions in each repeat at Ser2, Ser5, or Ser7 abolish phosphorylation at the appropriate serine residue. Using these fusions, we found that Sen1 interacts with Rpb1 only when Ser2 is phosphorylated. Furthermore, the R302W substitution abolished the interaction even when Ser2 is phosphorylated. Overall, the results suggest that Sen1 preferentially binds to the Ser2-phosphorylated form of the CTD of Rpb1 and that sen1-R302W impairs the interaction.
Immunoprecipitation experiments designed to examine the extent to which Sen1 or Sen1-R302W copurifies with Rpb1 revealed another level of complexity to the Sen1-Rpb1 interaction that two-hybrid tests were unable to address. When HA-tagged Rpb1 was immunoprecipitated, the copurification of Sen1 was insensitive to RNase, indicating that the interaction is most likely due to a protein-protein interaction. However, RNase was required to impair the Rpb1-Sen1-R302W interaction (Fig. 2). Thus, Sen1 is tethered to Rpb1 by at least two modes: through the Ser2-P CTD mark and through RNA. The results were corroborated by additional experiments using anti-Ser-P-specific antibodies to immunoprecipitate Rpb1 (Fig. 4). In these experiments, the R302W substitution significantly reduced the level of copurification only when RNase was present.
Overall, the combined results suggest that Sen1 interacts with Rpb1 through at least two independent mechanisms. One depends on a direct protein-protein interaction between Sen1 and Rpb1. We propose that the other depends on RNA tethering to actively transcribing RNA polymerase II. Although we cannot be sure, the RNAs responsible for tethering are presumably nascent transcripts. Since Sen1 is a helicase with a nucleic acid binding domain, Sen1 might bind to nascent transcripts while simultaneously engaging in direct binding to the Ser2-phosphorylated CTD of Rpb1.
Sen1-Nab3-Nrd1 interactions with Rpb1.
Previous reports indicate that Nrd1 binds to the Ser5-phophorylated CTD (61). Consistently with this report, our phosphorylated GAD-CTD fusions indicate that substitution of serine at position 5 with alanine abolishes the Nrd1-CTD interaction. Additionally, TAP purification and mass spectrometry have confirmed that Sen1 associates with Nrd1 and Nab3 along with several other proteins involved in transcription termination (39, 60). Using IP, we provide additional confirmation that all three proteins copurify in the presence of RNase as a native complex. Taken together, the results suggest an interesting scenario where two components of the Sen1-Nab3-Nrd1 complex interact with two different phosphorylated forms of the CTD, suggesting the possibility of two different ways Sen1 and/or the tripartite complex could be bound to the CTD.
Because sen1-R302W impairs the interaction of Sen1 with the Ser2-phosphorylated CTD, we performed experiments to ask whether impaired binding affects the stability of the tripartite complex. Immunoprecipitation experiments indicate that this is not the case, since Nab3 and Nrd1 copurify equally well in the presence of wild-type or mutant Sen1. If the tripartite complex can associate with the CTD through the binding of Sen1 to the Ser2-phosphorylated CTD, then sen1-R302W should reduce the extent of copurification of all three proteins when anti-Ser2 antibodies are used to IP Rpb1. The results show that sen1-R302W significantly reduces the copurification of mutant Sen1 in an anti-Ser2 IP in the presence of RNase, but Nab3 and Nrd1 were not significantly reduced. None of the three proteins were reduced in IPs using anti-Ser5 or anti-Ser7 antibodies. These results suggest that Sen1 is bound alone when associated with the Ser2-phosphorylated CTD. This would not preclude that it might also be bound as part of the tripartite complex associated with the CTD through Nrd1 binding to the Ser5-phosphorylated CTD. Two modes of Sen1 association with Rpb1 could reflect other functions for Sen1 in DNA repair (37) or RNA processing (16, 55, 58). However, it was previously reported that sen1-R302W causes a defect in the termination of SNR7 coding for U5 snRNA, leading to the accumulation of a readthrough transcript (16). Thus, the binding of Sen1 must be important for the termination of transcription of noncoding genes like SNR7.
Coding versus noncoding genes.
To clarify the complexities, we performed ChIP-chip experiments to examine the distribution of Sen1 across all genes. For noncoding genes, the analyses show that the average level of wild-type Sen1 occupancy parallels the level of Ser2-phosphorylated Rpb1. The sen1-R302W mutation causes a global decrease in occupancy across the entire length of noncoding genes. Given that sen1-R302W has been shown to affect transcription termination of the noncoding SNR7 gene (16), the reduced occupancy is most likely related to the role of Sen1 in termination, as described below.
For protein-coding genes, the average occupancy of Sen1 across most of the length of the coding regions is relatively unaffected by sen1-R302W. However, the close examination of wild-type and mutant profiles reveals reduced occupancy of sen1-R302W within the first 500 bp upstream of the transcription start site as well as reduced occupancy near the ends of ORFs. The normal levels of occupancy across most of the coding region could reflect the presence of Sen1 as part of a Sen1-Nab3-Nrd1 tripartite complex bound through Nrd1 to Ser5-P, while the precipitous decline in sen1-R302W levels observed late in transcription, where phosphorylated Ser5 is in slow decline but phosphorylated Ser2 is still near its peak, is most likely due to the impaired binding of mutant Sen1 to Ser2-P. Reduced occupancy early in transcription suggests a transient recruitment defect, but the underlying connection between this effect and the impaired binding of Sen1-R302W to Ser2-P is not yet clear, since the levels of Ser2 phosphorylation are low and Sen1-R302W has no obvious effect on the assembly or stability of the tripartite termination complex.
Multiple modes of binding and multiple functions for Sen1.
Our results, combined with the results of others, support the view that Sen1 associates with the CTD of Rpb1 in at least two ways, as a subunit of the Nrd1-Nab3-Sen1 complex bound to the Ser5-phosphorylated CTD and through the direct interaction of Sen1 with the Ser2-phosphorylated CTD. When differences between coding and noncoding genes are considered, two models of Sen1 association with Rpb1 can be predicted (Fig. 9A). For both protein-coding and noncoding genes, the Nrd1-Nab3-Sen1 complex could associate with Pol II early in transcription through the binding of Nrd1 with the Ser5-phosphorylated CTD (61). Our data support the view described in Fig. 9 that Sen1 may bind to the Ser2-phosphorylated CTD by itself during the transcription of protein-coding genes or in complex with the other partners during the transcription of noncoding genes. This model is consistent with previous reports that the tripartite termination complex is not generally involved in the termination of protein-coding genes and might be released when Ser5 is dephosphorylated (61). An inherent feature of the model is that a hand-off occurs during transcription where all three proteins (noncoding genes) or Sen1 alone (coding genes) transfers from a Ser5- to a Ser2-phosphorylated CTD (Fig. 9B).
Fig 9.
Alternative models for the association of Sen1 with Rpb1. The Figure shows alternative ways Sen1 might associate with the Ser2-phosphorylated C-terminal domain (CTD) when protein-coding and noncoding genes are being transcribed. For both sets of genes, it is assumed that Sen1 associates with the Ser5-phosphorylated CTD through its interaction with Nab3-Nrd1 near gene promoters. Later in transcription, the complex transfers to increasing levels of Ser2-P through its interaction with Sen1 (A, left, and B). Once Ser2-P reaches the higher levels found in coding genes, Nrd1-Nab3 dissociates and Sen1 can remain bound (A, right, and B). The black arrow in panel B denotes the transcriptional start site, and the blue bar denotes the cleavage and polyadenylation site.
The underlying basis of the different transfer events proposed to occur for coding versus noncoding genes is not yet clear. One idea is suggested by the interaction between Sen1 and the protein phosphatase Glc7 (39). If Sen1 itself undergoes a phosphorylation-dephosphorylation cycle that is linked in some way to levels of Ser2-P, then different modes of transfer from Ser5-P to Ser2-P might be coupled to high levels of Ser2-P for protein-coding genes versus low levels for noncoding genes.
The binding of Sen1 by itself to Ser2-P during the transcription of protein-coding genes might serve a purpose other than termination. For example, it could play a role in DNA repair given that Sen1 binds to Rad2, a DNA excision enzyme involved in repair, and that sen1 mutants cause synthetic growth defects in response to DNA-damaging agents when combined with deletions of genes required for repair (55). Also, Sen1 was recently implicated in protection from DNA damage caused by R-loops (37). Further studies will be required to test this model.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the University of Wisconsin College of Agricultural and Life Sciences, School of Medicine and Public Health, National Science Foundation grant MCB 0744017 (M.R.C.), National Science Foundation Career Award 0747197 (A.Z.A.), and NIH Genome Sciences Training Grant T32 HG002760 (J.R.).
Footnotes
Published ahead of print 27 January 2012
This article is Laboratory of Genetics paper number 3646.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Abou Elela S, Ares M., Jr 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akhtar MS, et al. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allmang C, et al. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arigo JT, Eyler DE, Carroll KL, Corden JL. 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23:841–851 [DOI] [PubMed] [Google Scholar]
- 5. Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346 [DOI] [PubMed] [Google Scholar]
- 6. Buratowski S. 2003. The CTD code. Nat. Struct. Biol. 10:679–680 [DOI] [PubMed] [Google Scholar]
- 7. Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chanfreau G, Elela SA, Ares M, Jr, Guthrie C. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chanfreau G, Legrain P, Jacquier A. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975–988 [DOI] [PubMed] [Google Scholar]
- 10. Chapman RD, et al. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780–1782 [DOI] [PubMed] [Google Scholar]
- 11. Conrad NK, et al. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corden JL. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383–387 [DOI] [PubMed] [Google Scholar]
- 13. Corden JL. 2007. Transcription. Seven ups the code. Science 318:1735–1736 [DOI] [PubMed] [Google Scholar]
- 14. Dahmus ME. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009–19012 [DOI] [PubMed] [Google Scholar]
- 15. DeMarini DJ, Winey M, Ursic D, Webb F, Culbertson MR. 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2154–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finkel JS, Chinchilla K, Ursic D, Culbertson MR. 2010. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics 184:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fromont-Racine M, Rain JC, Legrain P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277–282 [DOI] [PubMed] [Google Scholar]
- 18. Ghaemmaghami S, et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 19. Habraken Y, Sung P, Prakash L, Prakash S. 1993. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature 366:365–368 [DOI] [PubMed] [Google Scholar]
- 20. Higgins DR, Prakash L, Reynolds P, Prakash S. 1984. Isolation and characterization of the RAD2 gene of Saccharomyces cerevisiae. Gene 30:121–128 [DOI] [PubMed] [Google Scholar]
- 21. Ingles CJ, Himmelfarb HJ, Shales M, Greenleaf AL, Friesen JD. 1984. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc. Natl. Acad. Sci. U. S. A. 81:2157–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanin EI, et al. 2007. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim H, et al. 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17:1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim HD, Choe J, Seo YS. 1999. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38:14697–14710 [DOI] [PubMed] [Google Scholar]
- 26. Kim M, Suh H, Cho EJ, Buratowski S. 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284:26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim M, et al. 2006. Distinct pathways for snoRNA and mRNA termination. Mol. Cell 24:723–734 [DOI] [PubMed] [Google Scholar]
- 28. Komarnitsky P, Cho EJ, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lamontagne B, Tremblay A, Abou Elela S. 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20:1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee JM, Greenleaf AL. 1997. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 272:10990–10993 [DOI] [PubMed] [Google Scholar]
- 32. Licatalosi DD, et al. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101–1111 [DOI] [PubMed] [Google Scholar]
- 33. Lindstrom DL, Hartzog GA. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Longtine MS, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 35. Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176–27183 [DOI] [PubMed] [Google Scholar]
- 36. Mayer A, et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272–1278 [DOI] [PubMed] [Google Scholar]
- 37. Mischo HE, et al. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 41:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nechaev S, Adelman K. 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nedea E, et al. 2008. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29:577–587 [DOI] [PubMed] [Google Scholar]
- 40. Ng HH, Robert F, Young RA, Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709–719 [DOI] [PubMed] [Google Scholar]
- 41. Patturajan M, Wei X, Berezney R, Corden JL. 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 18:2406–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perales R, Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36:178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Phatnani HP, Greenleaf AL. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922–2936 [DOI] [PubMed] [Google Scholar]
- 44. Phatnani HP, Jones JC, Greenleaf AL. 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43:15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rasmussen TP, Culbertson MR. 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6885–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scherer S, Davis RW. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. U. S. A. 76:4951–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schroeder SC, Schwer B, Shuman S, Bentley D. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sidorenkov I, Komissarova N, Kashlev M. 1998. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell 2:55–64 [DOI] [PubMed] [Google Scholar]
- 49. Steinmetz EJ, Brow DA. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steinmetz EJ, Brow DA. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. U. S. A. 95:6699–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Steinmetz EJ, Conrad NK, Brow DA, Corden JL. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327–331 [DOI] [PubMed] [Google Scholar]
- 52. Steinmetz EJ, Ng SB, Cloute JP, Brow DA. 2006. Cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 26:2688–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Steinmetz EJ, et al. 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24:735–746 [DOI] [PubMed] [Google Scholar]
- 54. Tietjen JR, et al. 2010. Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17:1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ursic D, Chinchilla K, Finkel JS, Culbertson MR. 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 32:2441–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ursic D, DeMarini DJ, Culbertson MR. 1995. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol. Gen. Genet. 249:571–584 [DOI] [PubMed] [Google Scholar]
- 57. Ursic D, Finkel JS, Culbertson MR. 2008. Detecting phosphorylation-dependent interactions with the C-terminal domain of RNA polymerase II subunit Rpb1p using a yeast two-hybrid assay. RNA Biol. 5:1–4 [DOI] [PubMed] [Google Scholar]
- 58. Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 25:4778–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Hoof A, Lennertz P, Parker R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vasiljeva L, Buratowski S. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21:239–248 [DOI] [PubMed] [Google Scholar]
- 61. Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. 2008. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 15:795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. West ML, Corden JL. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Winey M, Culbertson MR. 1988. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics 118:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Young RA, Davis RW. 1983. Yeast RNA polymerase II genes: isolation with antibody probes. Science 222:778–782 [DOI] [PubMed] [Google Scholar]
- 65. Yuryev A, et al. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. U. S. A. 93:6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.