Abstract
In the budding yeast Saccharomyces cerevisiae, the Cdc42 effector Ste20 plays a crucial role in the regulation of filamentous growth, a response to nutrient limitation. Using the split-ubiquitin technique, we found that Ste20 forms a complex with Vma13, an important regulatory subunit of vacuolar H+-ATPase (V-ATPase). This protein-protein interaction was confirmed by a pulldown assay and coimmunoprecipitation. We also demonstrate that Ste20 associates with vacuolar membranes and that Ste20 stimulates V-ATPase activity in isolated vacuolar membranes. This activation requires Ste20 kinase activity and does not depend on increased assembly of the V1 and V0 sectors of the V-ATPase, which is a major regulatory mechanism. Furthermore, loss of V-ATPase activity leads to a strong increase in invasive growth, possibly because these cells fail to store and mobilize nutrients efficiently in the vacuole in the absence of the vacuolar proton gradient. In contrast to the wild type, which grows in rather small, isolated colonies on solid medium during filamentation, hyperinvasive vma mutants form much bigger aggregates in which a large number of cells are tightly clustered together. Genetic data suggest that Ste20 and the protein kinase A catalytic subunit Tpk2 are both activated in the vma13Δ strain. We propose that during filamentous growth, Ste20 stimulates V-ATPase activity. This would sustain nutrient mobilization from vacuolar stores, which is beneficial for filamentous growth.
INTRODUCTION
The Rho-type GTPase Cdc42 regulates cell morphology and signal transduction in eukaryotic cells (11, 17, 40). In the budding yeast Saccharomyces cerevisiae, the p21-activated kinase Ste20 is one of the best-characterized effectors of Cdc42 that triggers these pathways. Ste20 promotes polarized growth, exit from mitosis, and cell death (1, 16, 53). It also regulates vacuolar inheritance, mRNA decay, and the synthesis and uptake of sterols (2, 28, 29, 68). Furthermore, Ste20 activates distinct mitogen-activated protein kinase (MAPK) cascades that control filamentous growth, mating, and osmotic-stress responses (26, 30, 38, 44, 45, 49). Consistent with these functions, Ste20 can be found in the cytoplasm and at the plasma membrane at sites of polarized growth, including the bud cortex and tips of mating projections (25, 43).
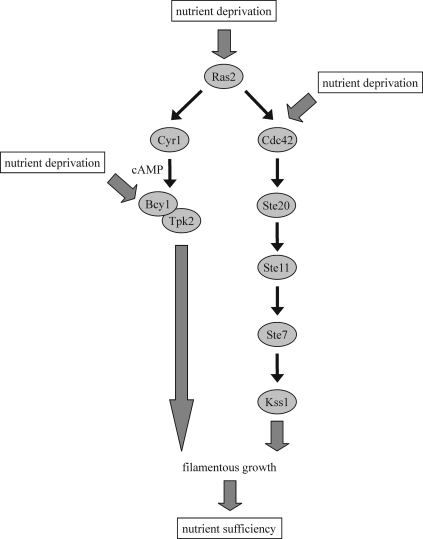
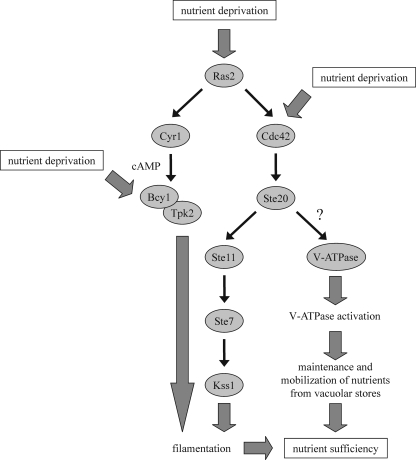
Upon nutrient limitation, budding yeast cells can switch to filamentous growth (13, 49). This is characterized by cell elongation, a switch to unipolar budding, increased cell-cell adhesion, increased cell-substratum adhesion, and the ability to penetrate a solid medium, such as agar. Together, these mechanisms allow the cells to forage for nutrients. By mechanisms that are not fully understood, nutrient sensors trigger at least two signaling pathways: a MAPK module activated by Ste20 and a cyclic AMP (cAMP)-dependent protein kinase A (PKA) pathway. The nutrient sensors may relay their signal to Ras2, which activates a cascade that consists of Cdc42, Ste20, the MAPK kinase kinase Ste11, the MAPK kinase Ste7, and the MAPK Kss1 (Fig. 1) (30, 36, 49). In addition, Ras2 activates the adenylate cyclase Cyr1, which stimulates cAMP production (Fig. 1) (59). Increased cAMP levels result in PKA activation by dissociating the inhibitory cAMP-binding regulatory subunit Bcy1 from the catalytic subunits Tpk1, Tpk2, and Tpk3 (60, 61). Tpk2 is an important activator of filamentation, whereas Tpk1 and Tpk3 function as inhibitors (39, 50). It has also been reported that nutrient sensors may activate the MAPK cascade and the PKA pathway independently of Ras2 (33, 42, 65). Stimulation of the filamentous-growth MAPK cascade by the plasma membrane-bound Sho1-Msb2 complex, for instance, does not seem to involve Ras2 (5). In addition to the MAPK and PKA pathways, the 5′-AMP-dependent kinase Snf1 is involved in filamentation, as well (4, 24).
Fig 1.
The MAPK and PKA pathways in filamentation. See the text for details. The precise molecular signaling mechanisms are not clear. In addition to the activation of the filamentous-growth MAPK and PKA pathways by Ras2, as shown here, it has been reported that these branches can be stimulated in a Ras2-independent manner, as well. Our data suggest that the loss of V-ATPase activity in vma mutants strongly stimulates both Ste20 and Tpk2, probably because these cells are unable to efficiently store and mobilize nutrients in vacuoles.
Vacuolar H+-ATPases (V-ATPases) are ATP-driven proton pumps that acidify organelles, such as the lysosome, the Golgi apparatus, and endosomes, in all eukaryotic cells (12, 21). Acidification of the lysosome-like vacuole in budding yeast is implicated in metabolite storage, autophagy, and ion homeostasis (27). The electrochemical gradient established by the V-ATPase is exploited by numerous secondary transporters, and the acidic vacuolar pH is optimal for proteolysis of proteins arriving by autophagy or endocytosis. As a result, the vacuole is well suited for storage of nutrients when they are plentiful and for mobilization during periods of depletion (67). The importance of V-ATPases in the plant tonoplast for nutrient storage was recently highlighted by the discovery that the vacuolar subunit isoforms of the Arabidopsis V-ATPase are essential for nutrient storage (23).
The V-ATPase is a membrane-bound multisubunit enzyme complex composed of two distinct domains (12, 21). The V1 sector, which resides on the cytoplasmic side of the membrane, hydrolyzes ATP and contains 8 different subunits, whereas the membrane-embedded V0 sector, which is composed of 6 different subunits, translocates protons across the membrane. Among eukaryotes, V-ATPases are highly conserved in both their overall structure and the sequences of individual subunits. V-ATPase activity is tightly regulated. A major regulatory mechanism is the dissociation of V-ATPase into its V0 and V1 sectors. In budding yeast, glucose depletion induces rapid and reversible V-ATPase disassembly (20). This process is regulated in part by the Ras/cAMP/PKA pathway (3), whereas a heterotrimeric complex termed RAVE is required for reassembly of V-ATPase complexes (52, 55). Other proteins, including glycolytic enzymes, such as aldolase, have also been implicated in regulated dissociation of V-ATPases (32). Recently, Dechant et al. (6) proposed that a decrease in cytosolic pH, which is a consequence of glucose starvation, triggers V-ATPase disassembly. The dissociated V0 and V1 sectors are both inactive (41, 69). This inactivation is critical, in particular, for the V1 sector, because release of an uncoupled ATPase into the cytosol could deplete cellular energy stores. Vma13, the V-ATPase subunit H, plays an important role in silencing ATP hydrolysis by the V1 sector. Cytosolic V1 complexes from cells lacking VMA13 have significant ATPase activity, and this activity can be silenced by addition of bacterially expressed H subunit (41, 7). Vma13 is also the only subunit that is not required for the full assembly of V-ATPase. In the absence of VMA13, V0-V1 complexes assemble and are transported to the vacuole, but these complexes are inactive (15). These results suggest that Vma13 functions as an activator in the fully assembled V-ATPase and an inhibitor of detached V1 sectors.
In this work, we show evidence for Ste20 binding to Vma13 and for localization of Ste20 at vacuolar membranes. We also find that Ste20 positively regulates V-ATPase activity in vivo, and this activation is independent of V-ATPase assembly. Furthermore, deletion of VMA13 or other V-ATPase subunits leads to a strong increase in agar invasion, possibly due to defective nutrient storage and mobilization in these cells. We propose that during filamentous growth, Ste20 not only triggers a MAPK cascade, but, in parallel, activates the V-ATPase, facilitating mobilization of intracellular nutrient reserves.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
All yeast strains used in this study are listed in Table 1. The strains are in the Σ1278b background (PPY966), with the exception of strains used for vacuolar assays. For vacuole isolation, wild-type SF838-5Aα was used. Yeast strains were constructed using PCR-amplified cassettes (18, 31) and were grown in 1% yeast extract, 2% peptone, 2% dextrose (YPD) or synthetic complete (SC) medium. For induction of the GAL1 promoter, yeast cells were grown in YP medium with 3% raffinose instead of glucose. Galactose (final concentration, 2%) was added to induce the GAL1 promoter. To compare the growth rates between strains, cells were grown overnight in liquid YPD medium. Serial dilutions starting from 104 cells were then spotted on YPD plates and incubated at 30°C for 2 days.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source or reference |
|---|---|---|
| MLY65 | PPY966 vma13Δ::His3MX6 | This study |
| MLY125 | PPY966 vma2Δ::His3MX6 | This study |
| MLY126 | PPY966 vma3Δ::His3MX6 | This study |
| MLY150 | PPY966 ste20Δ::hphNT1 vma2Δ::His3MX6 | This study |
| MLY151 | PPY966 ste20Δ::hphNT1 vma3Δ::His3MX6 | This study |
| MLY159 | PPY966 tpk2Δ::His3MX6 | This study |
| MLY161 | PPY966 tpk2Δ::His3MX6 ste20Δ::hphNT1 | This study |
| MLY166 | PPY966 tpk2Δ::klTRP1 vma13Δ::His3MX6 | This study |
| MLY167 | PPY966 tpk2Δ::klTRP1 ste20Δ::hphNT1 vma13Δ::His3MX6 | This study |
| MLY168 | PPY966 vma13Δ::His3MX6 ste7Δ::klTRP1 | This study |
| MLY170 | PPY966 ste20Δ::His3MX6 vma13Δ::hphNT1 | This study |
| MLY177 | PPY966 vma13Δ::His3MX6 ste11Δ::klTRP1 | This study |
| MLY208 | PPY966 STE20-3HA-His3MX6 | This study |
| PPY966 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 | 57 |
| SHY26 | PPY966 ste11Δ::His3MX6 | This study |
| SHY45 | PPY966 ste7Δ::His3MX6 | This study |
| THY697 | PPY966 ste20Δ::hphNT1 | 58 |
All constructs used in this work are listed in Table 2. To obtain 2μm plasmids containing STE20-3HA and STE20K649A-3HA under the control of the STE20 promoter, a C-terminal STE20-3HA fragment was amplified by PCR using pRS316-pGAL1-STE20-3HA and pRS316-pGAL1-STE20K649A-3HA, respectively, as templates and the following primers: 5′-TCCTCCGTCGACAAGTGA and 5′-ATGCGGGCCCAACAGCTATGACCATGAT. PCR products were digested with SalI and ApaI and ligated into SalI/ApaI-cut pRS425-STE20.
Table 2.
Plasmids used in this study
| Name | Genotype | Source or reference |
|---|---|---|
| pAK6 | pADNX carrying pADH1-NUbiquitin-VMA13 | This study |
| pCT44 | pADNX carrying pADH1-NUbiquitin-EXO70 | This study |
| pKA86 | pRS316 carrying pGAL1-STE20-3HA | 57 |
| pKK19 | pRS316 carrying myc-VMA13 | 41 |
| pTH197 | pRS313 carrying pMET25-STE20-CUbiquitin-RURA3 | 57 |
| pTH263 | pRS425 carrying STE20 | This study |
| pTH380 | pRS425 carrying STE20-3HA | This study |
| pTH381 | pRS425 carrying STE20K649A-3HA | This study |
| pTH383 | pADNX carrying pADH1-NUbiquitin-VMA2 | This study |
| pTH385 | pRS313 carrying pMET25-STE14-CUbiquitin-RURA3 | This study |
| pTH386 | pADNX carrying pADH1-NUbiquitin-VMA3 | This study |
| MBP-Vma13 | pMALpAse carrying VMA13 | 7 |
Split-ubiquitin technique.
The split-ubiquitin screen using STE20 as bait is described by Tiedje et al. (57). For the interaction assays, 104 cells carrying the split-ubiquitin plasmids were spotted on SC medium lacking histidine and leucine to select for the plasmids or onto SC medium lacking histidine and leucine and supplemented with 0.5 g/liter 5-fluoroorotic acid (5-FOA) to monitor protein interactions. The 5-FOA plates also lacked methionine and cysteine to induce expression of the STE20 and STE14 fusion genes under the control of the MET25 promoter. The plates were grown for 2 days at 30°C.
Biochemical interaction of Vma13 and Ste20.
Maltose-binding protein (MBP)-tagged Vma13 was expressed and purified from bacteria as described previously (7), but with the following modifications. Transformed cells were grown to an A600 of 1 at 30°C. These cultures were then saved as frozen 1-ml stocks in 50% glycerol at −80°C, and 1 ml of frozen stock culture was added to 1 liter of LB containing 125 μg/ml ampicillin and 2 g glucose to grow the cells for MBP-Vma13 purification. The soluble fraction from the bacterial cell lysate was bound to a suspension of amylose resin (New England BioLabs), and the resin was washed with at least 20 volumes of cold TBSE (20 mM Tris-HCl, pH 7.2, 150 mM NaCl, 0.5 mM EDTA). Purified MBP-Vma13 was eluted in five 1-ml washes of TBSE containing 100 mM maltose.
Ste20-hemagglutinin (HA) was isolated from wild-type cells transformed with the plasmid pKA86 carrying HA-tagged STE20 under the control of the GAL1/10 promoter after 3 h of galactose induction. The cells were lysed, and cytosolic fractions were prepared as described previously (56). Each sample was incubated with 100 μl of protein A-Sepharose beads (a 40% suspension in phosphate-buffered saline-bovine serum albumin [PBS-BSA]) and 100 μg of anti-HA antibody (monoclonal antibody 16B12 from Covance Research Products) at 4°C for 1 h. Transformed cells that were not induced with galactose were treated in parallel as a negative control. The beads were washed with lysis buffer (50 mM Tris-HCl, 30 mM KCl, 30 mM NaCl, 0.3 mM EDTA) three times. Purified MBP-Vma13 was then incubated with Ste20-HA-bound beads (or beads from the uninduced control) at 4°C with gentle shaking for 3 h. The beads were washed three times with TBSE, and then the bound protein was eluted, separated by SDS-PAGE, and transferred to nitrocellulose as described previously (7). The level of MBP-Vma13 bound to the beads was detected on immunoblots with anti-MBP antibody (New England BioLabs).
For the coimmunoprecipitation experiment, 200 ml STE20-3HA cells carrying either pRS316-myc-VMA13 (pKK19) or the empty pRS316 were grown to an A600 of 1. Cells were harvested, washed twice with PBS, and resuspended in 10 ml PBS. DSP (dithiobis [succinimidylpropionate]) was added from a freshly prepared 20 mM stock solution in dimethyl sulfoxide (DMSO) to a final concentration of 1 mM. The reaction mixture was incubated for 30 min at room temperature. The reaction was stopped by adding Tris, pH 7.5, to a final concentration of 20 mM and incubation for 15 min at room temperature. After washing with lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10 mM EDTA, 1 mM EGTA, 5% glycerol, 1% Triton X-100), the cells were disrupted with glass beads in lysis buffer and clarified by centrifugation at 13,000 rpm for 5 min. The protein concentration was determined using Bradford protein assay solution. myc-Vma13 was immunoprecipitated by adding mouse monoclonal anti-myc antibody (9E10; Santa Cruz Biotechnology) and protein G-Sepharose (GE Healthcare). The resin was washed three times with lysis buffer, resuspended in 2× SDS sample buffer, and analyzed by immunoblotting. Mouse monoclonal anti-HA (12CA5) was obtained from Roche Diagnostics, and secondary antibodies were from Jackson ImmunoResearch Laboratories.
Filamentation assays.
For agar invasion assays, 104 cells of an overnight YPD culture were spotted on a YPD plate and grown for 3 days at 30°C. The plates were photographed before and after being rinsed under a stream of deionized water.
For the single-cell invasive-growth assay, cells were grown to stationary phase in SC medium, washed twice with water, and spread onto synthetic complete medium lacking glucose at a concentration of 104 cells/plate. The plates were incubated at 30°C for 18 h and analyzed microscopically with a Zeiss Axiovert 200 M fluorescence microscope. Images were captured using a Zeiss AxioCam MRm charge-coupled-device (CCD) camera.
The formation of cell clusters was analyzed by spotting 103 cells of an overnight YPD culture on a YPD plate and incubating the plate for 3 days at 30°C. The strains were examined microscopically after the plates were washed with water.
Vacuole isolation and biochemical analysis.
Wild-type (SF838-5Aα) cells or the congenic ste20Δ::kanMX strain were transformed with a multicopy plasmid carrying STE20, STE20-3HA, or STE20K649A-3HA (STE20KD-3HA). The cells were grown to log phase in SC buffered to pH 5 with 50 mM morpholineethanesulfonic acid (MES), converted to spheroplasts, and lysed, and then vacuolar vesicles were isolated by Ficoll density gradient centrifugation as described previously (48). ATPase activity was determined by coupled enzyme assay, and activity sensitive to 100 nM concanamycin A was taken as V-ATPase activity (48). For immunoblot analysis, vacuolar vesicles were solubilized, separated by SDS-PAGE, and transferred to nitrocellulose as described previously (56). V-ATPase subunits were detected with mouse monoclonal antibodies 10D7 (anti-V0 a subunit), 8B1 (anti-V1 A subunit), 13D11 (anti-V1 B subunit), and 7 A2 (anti-V1 C subunit) (22). STE20-HA and STE20KD-3HA were detected with monoclonal antibody 16B12 (Covance). Activities were compared between strains using the two-sample t test assuming equal variance in Microsoft Excel.
RESULTS
Ste20 interacts with the V-ATPase subunit Vma13.
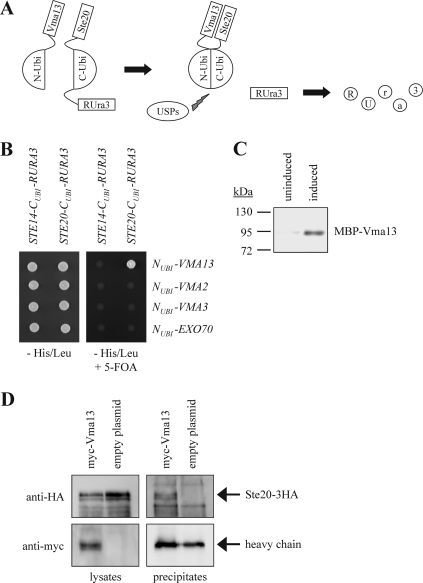
In an effort to identify regulators and targets of Ste20, we screened for proteins that bind to Ste20 using the split-ubiquitin system (57). This technique is based on the ability of the N- and C-terminal halves of ubiquitin to form a native-like ubiquitin (Fig. 2A) (19). Ubiquitin-specific proteases present in the cytosol and nucleus recognize the reconstituted ubiquitin, but not its halves, and cleave off a reporter protein, which had been linked to the C-terminal half of ubiquitin. The assay described here employs RUra3, a modified Ura3 with an arginine as the first amino acid, as the reporter (63). The freed RUra3 is rapidly degraded because arginine is a destabilizing residue in the N-end rule pathway. Therefore, interaction between two proteins fused to the N- and C-terminal halves of ubiquitin, respectively, results in nongrowth on medium lacking uracil. Conversely, growth on 5-FOA medium indicates a protein-protein interaction because 5-FOA is converted by Ura3 into a toxic compound. Using this technique, we identified the V-ATPase subunit Vma13 as an interactor of Ste20 (Fig. 2B). Ste20 does not bind to either Vma2, a component of the V1 domain, or the V0 subunit Vma3 in this assay, indicating that the observed Ste20-Vma13 interaction is specific (Fig. 2B).
Fig 2.
Ste20 binds to Vma13. (A) The split-ubiquitin system. The N-terminal and C-terminal halves of ubiquitin (N-Ubi and C-Ubi) alone do not assemble. If Vma13, fused to N-Ubi, binds to Ste20, linked to C-Ubi, the two halves of ubiquitin are brought into close proximity. This reconstituted quasinative ubiquitin is recognized by ubiquitin-specific proteases (USPs), which cleave off the reporter RUra3, which is fused to Ste20. The released RUra3, a modified version of the enzyme Ura3 with an arginine at the extreme N terminus, is targeted for degradation by the enzymes of the N-end rule. Since Ura3 converts 5-FOA to the toxic 5-fluorouracil, 5-FOA resistance indicates an interaction between Ste20 and Vma13. (B) Ste20 interacts with Vma13 using the split-ubiquitin system. Cells (104) of the indicated plasmid combinations were spotted either onto medium lacking histidine and leucine to select for the plasmids or onto medium lacking histidine and leucine and supplemented with 5-FOA to monitor protein interactions. The unrelated genes STE14 and EXO70 served as negative controls. (C) Ste20 interacts with Vma13 in a pulldown assay. Wild-type cells were transformed with a plasmid carrying 3HA-tagged STE20 (STE20-3HA) under the control of the GAL1/10 promoter. Transformed cells were grown in raffinose medium to relieve glucose repression and either left in raffinose (uninduced) or transferred to galactose medium (induced) for 3 h. Ste20-HA was immunoprecipitated from yeast cell lysate with anti-HA antibody, followed by protein A-Sepharose. MBP-tagged Vma13 purified from E. coli was added directly to the immunoprecipitates on protein A-Sepharose. After incubation and washing of the beads to remove unbound protein, binding was assessed by SDS-PAGE and immunoblotting with anti-MBP antibody. (D) Ste20 binds to Vma13 in vivo. STE20-3HA cells carrying either a pRS316 plasmid with myc-VMA13 or the empty pRS316 were lysed, and equal amounts of protein extract were precipitated with anti-myc antibodies. The immunoprecipitates were analyzed by immunoblotting using antibodies against the HA and myc epitopes, respectively. myc-Vma13 runs at the same height as the heavy chain. However, a stronger signal indicates that the protein was precipitated.
To confirm the protein-protein interaction between Ste20 and Vma13 by an independent approach, Ste20-3HA was expressed from the inducible GAL1/10 promoter and immunoprecipitated from a yeast cell lysate. The immunoprecipitates bound to protein A-Sepharose beads were incubated with MBP-Vma13 purified from Escherichia coli. In this assay, MBP-Vma13 was specifically pulled down by Ste20-3HA in an induced sample but was not bound to the beads in an uninduced sample (Fig. 2C).
Finally, binding of HA-tagged Ste20 to myc-tagged Vma13 was confirmed by immunoprecipitation from yeast cells using anti-myc antibodies. Ste20-3HA coprecipitated with myc-Vma13 but was not detected in immunoprecipitated protein from cells that do not express myc-Vma13 (Fig. 2D). Thus, Ste20 forms a complex with Vma13 in vivo.
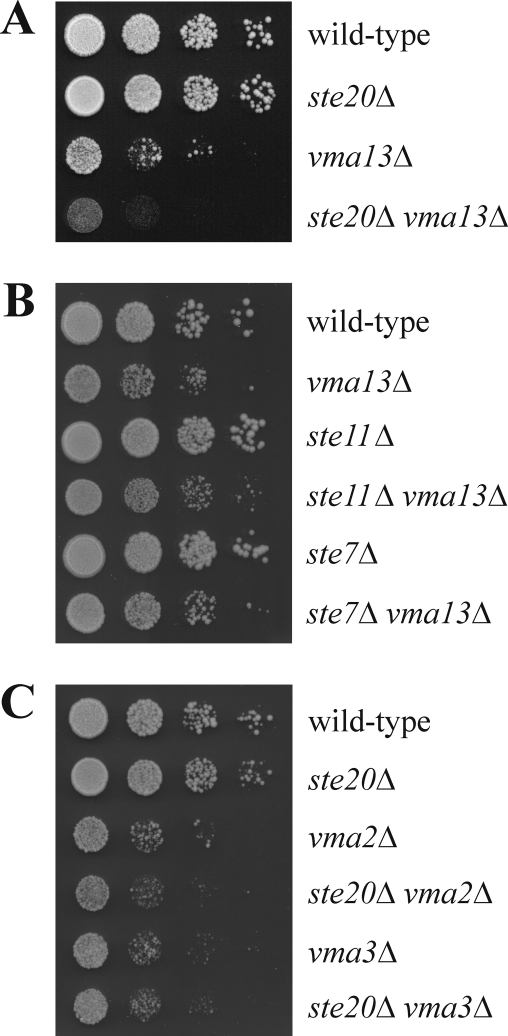
We next analyzed genetic interactions between STE20 and VMA13. As shown previously, the growth rate of vma13Δ cells was reduced in comparison to the wild type (Fig. 3A) (15). Notably, additional deletion of STE20 in the strain exacerbated this growth defect (Fig. 3A), suggesting that STE20 and VMA13 act in parallel to support normal cell growth. In contrast, there is no synthetic growth defect between vma13Δ and ste11Δ and ste7Δ, two mutants in the filamentation and pheromone response MAPK pathways activated by Ste20 (Fig. 1 and 3B). We also examined genetic interactions between STE20 and the V-ATPase subunit genes VMA2 and VMA3. Deletion of either VMA2 or VMA3 resulted in a growth defect comparable to that of the vma13Δ strain. However, the loss of STE20 in these strains had no effect on the growth rate (Fig. 3C). Thus, the genetic interaction between VMA13 and STE20 is remarkably specific.
Fig 3.
Genetic interaction between STE20 and VMA13. (A) STE20 deletion exacerbates the growth defect of vma13Δ cells. Serial dilutions (1:10) of the indicated strains were spotted on YPD and incubated for 2 days at 30°C. (B) Deletion of STE11 and STE7, respectively, does not affect the growth rate of the vma13Δ strain. Growth was assessed by serial dilutions as in panel A. (C) STE20 does not interact genetically with VMA2 or VMA3. Growth was assessed by serial dilutions as in panel A.
Activation of V-ATPase by Ste20.
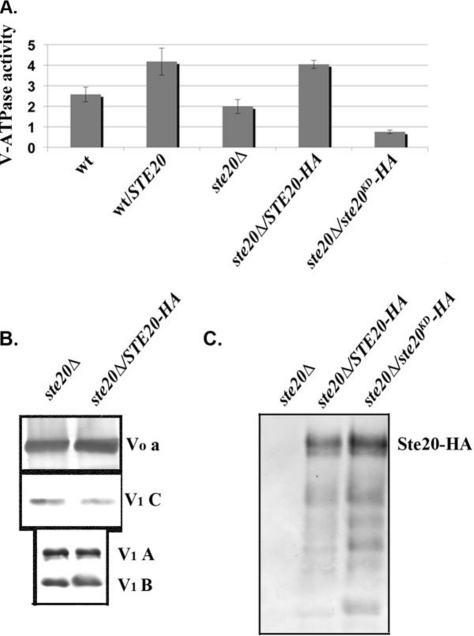
Since Ste20 binds to Vma13, it is tempting to speculate that Ste20 regulates V-ATPase activity. To test this, we first isolated vacuolar vesicles from wild-type and ste20Δ cells and cells overexpressing STE20 and assessed concanamycin A-sensitive ATPase activity. STE20 expressed from its own promoter or the GAL1/10 promoter is active, allowing assessment of its activity in the absence of triggers, such as nutrient deprivation (43, 64). As shown in Fig. 4A, the specific ATPase activity in vacuolar vesicles isolated from a ste20Δ strain was not significantly different from the activity in wild-type vacuolar vesicles (P > 0.05). Overexpression of STE20 in the wild-type strain appeared to give higher ATPase activity, but the difference from wild-type activity was not statistically significant (P = 0.14). However, we also transformed ste20Δ cells with STE20-3HA and STE20K649A-3HA, a kinase-dead allele of Ste20 (64), and then isolated vacuolar vesicles. As shown in Fig. 4A, overexpressed STE20-3HA activates V-ATPase activity to an extent comparable to the untagged STE20; this activity was also significantly different from those of both the ste20Δ and wild-type strains (P ≤ 0.01 for each strain). In contrast, the inactive STE20KD-3HA does not activate the V-ATPase. We next asked whether differences in V-ATPase activity in the ste20Δ mutant and STE20-overexpressing strains could be accounted for by differences in V-ATPase subunit levels at the vacuole. Immunoblots of vacuolar vesicles from ste20Δ and STE20-3HA-overexpressing strains were probed for V0 subunit a and V1 subunits A, B, and C, as shown in Fig. 4B. There were no consistent differences in the levels of these subunits in the wild-type, ste20Δ, or STE20-overexpressing cells over several independent vacuole preparations. The ratio of V1 to V0 subunit levels at the vacuole provides an established measure of V-ATPase assembly (3, 8, 20), so these results also imply that Ste20 does not change V-ATPase activity by altering the level of V1-V0 assembly. The levels of alkaline phosphatase, a vacuolar membrane protein that is transported independently of the V-ATPase, also showed no significant differences (data not shown). In order to determine whether Ste20 is localized to the vacuole, we also probed immunoblots of vacuolar vesicles for Ste20-3HA (Fig. 4C). Both the wild-type Ste20-HA and the kinase-dead mutant ste20K649A-3HA (ste20KD-HA) are localized to vacuolar membranes, even though the kinase-dead mutant does not support activation of the V-ATPase. These results indicate that Ste20 kinase activity is required for V-ATPase activation and that the kinase could exert its activity at the vacuolar membrane.
Fig 4.
Ste20 localizes to the vacuole and activates V-ATPase activity. (A and B) Vacuolar vesicles were isolated from wild-type cells, wild-type cells transformed with a multicopy plasmid expressing STE20 from its native promoter (wt/STE20), a ste20Δ mutant, and the ste20Δ mutant transformed with a multicopy plasmid bearing either 3HA-tagged STE20 (STE20-HA) or a 3HA-tagged kinase-dead mutant of STE20 (ste20KD-HA). (A) Concanamycin A-sensitive ATPase activity in the isolated vacuolar vesicles is expressed as μmol ATP hydrolyzed/min/mg vacuolar protein. For each sample, the mean ± standard error of the mean (SEM) for four to seven independent vacuolar preparations is shown. (B) Vacuolar vesicles from the ste20Δ and STE20-3HA-overexpressing cells were solubilized, and equivalent quantities of vacuolar protein were separated by SDS-PAGE and transferred to nitrocellulose, followed by immunoblotting with antibodies to subunit a of the V-ATPase V0 sector and subunits A, B, and C of the V-ATPase V1 sector. (C) Localization of wild-type and kinase-dead HA-tagged Ste20 to vacuolar membranes. Equal concentrations of vacuolar protein were separated by SDS-PAGE as in panel B and then probed for the presence of Ste20-HA on immunoblots. The predicted molecular mass of Ste20-3HA is indicated.
Finally, we tested whether direct phosphorylation of Vma13 could account for the activation of V-ATPase activity by Ste20. However, although bacterially expressed Vma13 could bind to Ste20 in pulldowns, there was no phosphorylation of Vma13 under these conditions. As a result, we cannot yet directly attribute V-ATPase activation to direct phosphorylation by Ste20.
V-ATPase activity and filamentous growth.
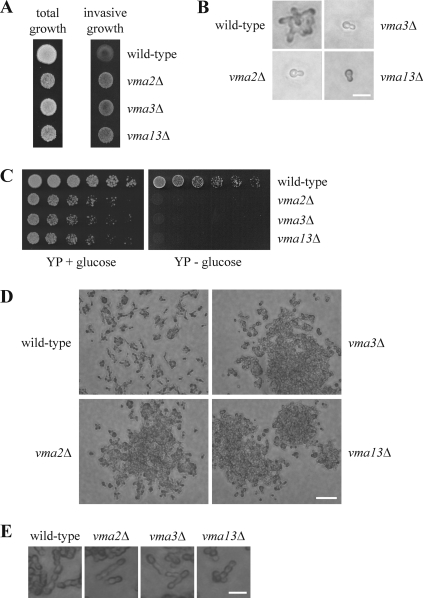
Since STE20 and VMA13 interact genetically and at the protein level, we examined whether the V-ATPase contributes to processes that are regulated by Ste20. vma13Δ cells have normal morphology and form a mating projection in response to pheromone (data not shown). We also examined invasive growth. For this assay, cells are spotted at very high density on the rich YPD medium. Under these conditions, nutrients are locally used up quickly. As a consequence, filamentation is induced and cells penetrate the agar. This agar invasion can easily be assessed by washing off cells that grow only superficially. In the absence of VMA13, cells exhibited a marked increase in agar invasion (Fig. 5A). Presumably, vma13Δ cells are unable to store and/or mobilize nutrients in the vacuole efficiently due to the lack of V-ATPase activity. Therefore, these cells might be more sensitive to nutrient deprivation. We next asked whether this phenotype was specific to vma13Δ cells or characteristic of a general loss of V-ATPase activity. Like the vma13Δ strain, vma2Δ and vma3Δ cells displayed a growth defect and a hyperinvasive phenotype (Fig. 5A). Therefore, increased agar invasion seems to be a common feature of cells lacking V-ATPase activity and is not specific to vma13Δ.
Fig 5.
Cells lacking V-ATPase activity are hyperinvasive. (A) Deletion of V-ATPase subunits leads to stronger agar invasion. Cells (104) of the indicated strains were spotted on a YPD plate and grown for 3 days at 30°C. Photographs were taken before (total growth) and after (invasive growth) rinsing with water. Note that agar invasion by the vma mutants is markedly stronger than that by the wild type even though these strains have a greatly reduced growth rate. (B) vma mutants fail to form colonies in the absence of glucose. Cells (104) of the indicated strains were spread on SC medium lacking glucose and were incubated for 18 h at 30°C. Bar, 10 μm. (C) Cells lacking V-ATPase activity are unable to grow on YP medium lacking glucose. Serial dilutions (1:3) of the indicated strains were spotted on YP plates with or without glucose and incubated for 2 days at 30°C. (D) vma mutants form cell clusters in response to nutrient deprivation. Cells (103) of the indicated strains were spotted on a YPD plate and grown for 3 days at 30°C. Cells that did not invade the agar were washed off with water. Bar, 50 μm. (E) Morphology of cells grown on agar. The indicated strains were treated as described for panel D but incubated for 3 days at 30°C. Bar, 10 μm.
To examine the morphology of vma cells during filamentation, a single-cell invasive-growth assay was performed. For this experiment, cells were grown overnight in liquid SC medium supplemented with glucose. The cells were then washed to remove nutrients and spread onto SC plates lacking glucose. Under these conditions of very severe nutrient deprivation, wild-type cells undergo few divisions and form small colonies of branched and slightly elongated cells, presumably by utilizing stored nutrients (Fig. 5B) (4). In contrast, cells lacking VMA2, VMA3, or VMA13 did not divide (Fig. 5B). Even after more than 48 h of incubation, we observed only single vma cells. This is clearly different from the growth of vma strains on rich medium, where growth is slowed but cells still divide. Therefore, the failure to form colonies cannot be explained by the reduced growth rate of these strains. Instead, this result is consistent with the notion that the vma strains are unable to store and mobilize nutrients efficiently. The complete loss of growth of vma2Δ, vma3Δ, and vma13Δ cells upon glucose removal is not restricted to SC medium. On YPD medium, growth of the vma mutants is reduced, whereas on YP medium lacking glucose, it is completely abolished (Fig. 5C). Even after several days, no growth of the vma mutants was observed.
The increased agar invasion of the vma mutants shown in Fig. 5A could be due to either stronger cell elongation or increased cell-cell adhesion and/or cell-substratum adhesion. Since vma strains did not form microcolonies on SC medium in the absence of glucose (Fig. 5B), we examined the morphology of cells grown on YPD plates. When cells were grown at a very high density, as shown in Fig. 5A, it was extremely difficult to make out individual cells. Therefore, we analyzed cells that were spotted on YPD agar at 1/10 of the normal density. Under these conditions, cells still penetrated the agar. After removing superficially growing cells by rinsing the plate, isolated small colonies were observed for the wild type (Fig. 5D). In contrast, vma mutants formed large cell clusters (Fig. 5D). These contained a much higher number of cells than the wild-type colonies, and the cells seemed to be densely packed together. Notably, these aggregates were not observed when vma strains were grown in liquid culture. Thus, it seems that nutrient deprivation on agar plates induced by high cell density triggers strong cell aggregation in cells lacking V-ATPase activity. This effect could be caused by increased cell-cell and cell-substratum adhesion and would explain the hyperinvasive phenotype of the vma mutants. In line with this model, the morphology of vma2Δ, vma3Δ, and vma13Δ cells was indistinguishable from that of the wild type (Fig. 5E). Thus, it seems that the observed increase in agar invasion is not caused by stronger cell elongation, but rather, by a change in adhesion.
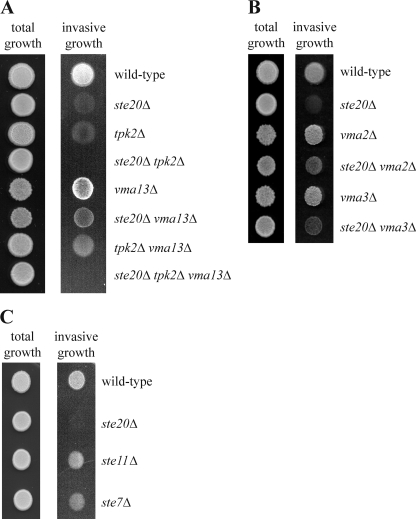
Next, we characterized the link between VMA13 and known components of signaling pathways for agar invasion. Deletion of either STE20 or TPK2, the activating catalytic PKA subunit gene in filamentous growth, resulted in a strong invasive-growth defect (Fig. 6A). In the ste20Δ tpk2Δ strain, agar invasion was completely abolished (Fig. 6A), suggesting that the Ste20 and PKA pathways are the major signaling cascades in filamentation, which is in line with observations of other groups (30, 39, 49, 50) (Fig. 1). Interestingly, VMA13 deletion suppressed the invasive-growth defect of ste20Δ cells (Fig. 6A). The agar invasion defect of ste20Δ cells can also be suppressed by deletion of either VMA2 or VMA3 (Fig. 6B), indicating that loss of V-ATPase activity, not absence of a specific subunit, triggers the underlying signaling pathways. This invasion is possibly mediated by activation of the PKA pathway. VMA13 deletion also rescued the agar invasion defect of the tpk2Δ strain (Fig. 6A), where invasion might be induced by activation of the Ste20-driven MAPK pathway. Notably, the vma13Δ ste20Δ tpk2Δ triple mutant did not exhibit any invasive growth (Fig. 6A). Together, these data suggest that loss of V-ATPase activity can trigger agar invasion by activating both the Ste20 and PKA pathways.
Fig 6.
PKA and MAPK signaling in vma13Δ cells. (A) VMA13 deletion leads to activation of the Ste20 and PKA pathways. Agar invasion by the indicated strains was determined as described in the legend to Fig. 5A. (B) Deletion of either VMA2 or VMA3 partially suppresses the agar invasion defect of ste20Δ cells. The experiment was performed as described in the legend to Fig. 5A. (C) Deletion of STE11 or STE7 results in only a moderate filamentation defect. Agar invasion was determined as described in the legend to Fig. 5A.
Because of the interactions between Ste20 and the V-ATPase described above and the fact that both proteins are involved in invasive growth, we speculated that Ste20 and the V-ATPase may be parts of a novel pathway regulating filamentation. The only known function of Ste20 in filamentous growth is the activation of a MAPK cascade by phosphorylation of the MAPK kinase kinase Ste11 (30, 49, 62, 64). We reasoned that if Ste11 were the only Ste20 target, deletion of STE11 would cause a defect in filamentous growth as severe as that observed in the ste20Δ strain. Importantly, the agar invasion phenotype of ste11Δ cells was much less pronounced than that of cells lacking STE20 (Fig. 6C). Deletion of STE7, which encodes the MAPK kinase that is activated by Ste11 during filamentous growth, similarly led to only a relatively moderate phenotype (Fig. 6C). Ste20 also activates Ste11 and Ste7 during mating. Notably, all three STE genes are essential for this process (data not shown) (14, 26). Thus, the difference in phenotype severity between the ste mutants is specific for filamentous growth and could be explained by additional functional targets of Ste20, such as the V-ATPase. This is in line with the regulation of V-ATPase activity by Ste20 as described above.
DISCUSSION
Regulation of V-ATPase activity by Ste20.
The Cdc42 effector Ste20 is involved in multiple signaling pathways, including a MAPK cascade that triggers filamentation (30, 49). Here, using the split-ubiquitin technique, a pulldown assay, and coimmunoprecipitation, we showed that Ste20 specifically interacts with the V-ATPase subunit Vma13, demonstrating that these proteins bind to each other in vivo and in vitro. We further reported the stimulation of V-ATPase activity by Ste20. It is not clear how Ste20 regulates V-ATPase activity. V0-V1 assembly, a major control mechanism, is not affected by overexpression or deletion of STE20. Vma13, which activates the assembled V-ATPase (15), does not seem to be a phosphorylation target of Ste20. V-ATPase activation by Ste20 requires Ste20 kinase activity, but Vma13 was not phosphorylated in our hands. Therefore, other V-ATPase subunits might be targets of Ste20, with Vma13 serving as a platform for binding.
Ste20 has previously been known to localize to the plasma membrane at sites of polarized growth in the cytoplasm and the nucleus (1, 25, 28, 43). Here, we show that Ste20 is also enriched in vacuolar membranes. This localization of Ste20 is in line with its role in V-ATPase activation and is consistent with observations of other groups. Cdc42 and Bem1, a scaffold protein that brings Cdc42, its activator Cdc24, and Ste20 into close proximity, are also present at vacuolar membranes (9, 10, 37, 46, 66). Thus, it seems that not only Ste20 associates with vacuolar membranes, but all components of the Bem1-Cdc24-Cdc42-Ste20 cascade.
It has been reported that, together with Cla4, another Cdc42 effector and also a p21-activated kinase, Ste20 regulates vacuolar inheritance (2). Thus, Ste20 has at least two distinct vacuolar functions.
Hyperinvasive growth is stimulated in the absence of V-ATPase activity.
Deletion of a number of V-ATPase subunits leads to strongly increased invasive growth relative to wild-type cells. This phenotype is not caused by increased cell elongation. Instead, it correlates with strong cell clustering. vma mutants probably experience acute nutrient deprivation under conditions where wild-type cells remain nutrient replete. The vacuolar pH gradient is required for both uptake and mobilization of nutrients (27, 51). The loss of vacuolar acidification in vma mutants causes defective nutrient storage and mobilization in these strains (54). Since depletion of certain nutrients induces filamentation, it seems most likely that the hyperinvasive phenotype of vma mutants is caused by the failure of these cells to maintain sufficient nutrient stores for times of starvation. As a consequence, cells lacking V-ATPase activity would be much more sensitive to nutrient limitation. Stronger invasive growth might increase the chance of finding extracellular nutrients and would therefore be beneficial for these cells.
Our genetic data suggest that STE20 and TPK2 are both required for the hyperinvasive phenotype of vma13Δ cells, but it is not clear how a nutrient deprivation signal activates these pathways. This is most likely an intracellular signal. Possibly, cells are able to somehow sense the loss of nutrient storage, as suggested for autophagy-deficient yeast (34). In fact, a direct role for the V-ATPase in lysosomal amino acid sensing by mTORC1 in mammals was recently described (70), although this pathway involves Ragulator components not present in yeast. Alternatively, since the V-ATPase plays a crucial role in the homeostasis not only of vacuolar but also of cytosolic pH (35), the altered pH of these compartments may trigger filamentation.
The only function that has been attributed to Ste20 in filamentous growth is the phosphorylation and thereby activation of the MAPK kinase kinase Ste11 (30, 49, 62, 64) (Fig. 7). In this work, we show that ste11Δ and ste7Δ cells exhibit a rather mild invasive-growth defect. In contrast, in cells lacking STE20, agar invasion is almost completely absent. This difference in phenotype severity is filamentation specific. STE20, STE11, and STE7 are all absolutely required for mating (14, 26). A likely explanation for the more pronounced phenotype of ste20Δ cells is that Ste20 has targets beyond Ste11 in filamentous growth. Here, we show that Ste20 can activate V-ATPase activity. Although this observation was obtained in yeast cells grown in nutrient-replete liquid medium, activation of the V-ATPase by Ste20 could be particularly important for filamentous growth on solid surfaces (Fig. 7). When nutrients are limited in cells growing on a solid surface, Ste20 triggers the Ste11-Ste7-Kss1 cascade. Together with other activated pathways, including PKA, this results in cell elongation, substratum invasion, and increased adhesion. Under these conditions, Ste20 could stimulate V-ATPase activity as well, sustaining vacuolar pH gradients during nutrient mobilization by vacuolar antiporters and allowing more efficient mobilization of intracellular reserves required for cell elongation. Thus, filamentous growth and vacuolar mobilization are complementary approaches to deal with starvation. Unfortunately, altered V-ATPase activity during true filamentous growth cannot easily be examined because the cells grow into the agar, precluding biochemical isolation of vacuoles.
Fig 7.
Model of Ste20 and Vma13 functions during filamentous growth. Low nutrient levels in cells growing on a solid surface trigger the PKA and filamentous-growth MAPK pathways. This results in cell elongation, increased adhesion, and growth into the substratum. We propose that, in addition to these signaling events, Ste20 activates V-ATPase, possibly via Vma13. This leads to a lower vacuolar pH, allowing optimal storage and mobilization of nutrients that are required for cell growth. Therefore, filamentous growth and allocation of nutrients complement each other.
Low glucose levels induce filamentation and also trigger disassembly and downregulation of V-ATPase (4, 6, 20). Here, we propose that V-ATPase activity might be increased during filamentous growth. These seemingly contradictory observations can be reconciled. The experiments demonstrating V-ATPase disassembly in response to glucose depletion were carried out in liquid medium (6, 20). Under these conditions, it is likely to be advantageous to reduce growth and to save energy by V-ATPase disassembly. In contrast, for cells growing on a solid medium it may become essential to increase V-ATPase activity and to invade the substratum because there is a chance of finding nutrients. Disassembly of the V-ATPase in response to glucose deprivation is suppressed at high extracellular pH (8), presumably in order to preserve organelle acidification under conditions of alkaline stress. Similarly, nutrient limitation under filamentous-growth conditions may prevent disassembly so that high V-ATPase activity can be maintained.
We found that STE20 deletion specifically exacerbates the growth defect of vma13Δ cells. This observation is somewhat puzzling, since mutations in V-ATPase subunits abolish all V-ATPase activity and generally result in similar growth phenotypes, and it is unlikely that Vma13 functions outside the V-ATPase. Vma13 does play a distinctive role in regulating V-ATPase activity, however, acting as an inhibitor of ATPase activity in cytosolic V1 complexes but as an activator of the intact V-ATPase (41). Because of this regulatory role, vma13Δ mutations show very specific genetic interactions with certain other vma mutations (47), and the genetic interaction with ste20Δ observed here could also be accounted for by an increased need for the silencing or activating properties of Vma13 in this mutant. Further investigation will be necessary to test this possibility.
In summary, we describe a link between V-ATPase and invasive growth and demonstrate the activation of V-ATPase activity by Ste20. Since these components are conserved from yeast to humans, it will be interesting to test whether V-ATPase activity is modulated in a similar way in higher eukaryotes.
ACKNOWLEDGMENTS
We thank Silke Horn, Maureen Tarsio, and Theodore Diakov for excellent technical support.
The project was supported by Deutsche Forschungsgemeinschaft grant HO 2098/3 to T.H. and NIH grant R01 GM50322 to P.M.K.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Ahn SH, et al. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25–36 [DOI] [PubMed] [Google Scholar]
- 2. Bartholomew CR, Hardy CF.2009. p21-activated kinases Cla4 and Ste20 regulate vacuole inheritance in Saccharomyces cerevisiae. Eukaryot. Cell 8:560–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bond S, Forgac M.2008. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J. Biol. Chem. 283:36513–36521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cullen PJ, Sprague GF.2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. U. S. A. 97:13619–13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cullen PJ, et al. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dechant R, et al. 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29:2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diab H, Ohira M, Liu M, Cobb E, Kane PM.2009. Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J. Biol. Chem. 284:13316–13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diakov TT, Kane PM.2010. Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J. Biol. Chem. 285:23771–23778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eitzen G, Wang L, Thorngren N, Wickner W.2002. Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 158:669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eitzen G, Thorngren N, Wickner W.2001. Rho1p and Cdc42p act after Ypt7p to regulate vacuole docking. EMBO J. 20:5650–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Etienne-Manneville S.2004. Cdc42—the centre of polarity. J. Cell Sci. 117:1291–1300 [DOI] [PubMed] [Google Scholar]
- 12. Forgac M.2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929 [DOI] [PubMed] [Google Scholar]
- 13. Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR.1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 14. Hartwell LH.1980. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 85:811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho MN, et al. 1993. VMA13 encodes a 54-kDa vacuolar H+-ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae. J. Biol. Chem. 268:18286–18292 [PubMed] [Google Scholar]
- 16. Höfken T, Schiebel E.2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaffe AB, Hall A.2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 [DOI] [PubMed] [Google Scholar]
- 18. Janke C, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947–962 [DOI] [PubMed] [Google Scholar]
- 19. Johnsson N, Varshavsky A.1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kane PM.1995. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270:17025–17032 [PubMed] [Google Scholar]
- 21. Kane PM.2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kane PM, Kuehn MC, Howald-Stevenson I, Stevens TH.1992. Assembly and tareting of peripheral and integral membrane subunits of the yeast vacuolar H+-ATPase. J. Biol. Chem. 267:447–454 [PubMed] [Google Scholar]
- 23. Krebs M, et al. 2010. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. U. S. A. 107:3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuchin S, Vyas VK, Carlson M.2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leberer E, et al. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M.1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 11:4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li SC, Kane PM.2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793:650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin M, et al. 2009. The Cdc42 effectors Ste20, Cla4 and Skm1 down-regulate the expression of genes involved in sterol uptake by a MAPK-independent pathway. Mol. Biol. Cell 20:4826–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin M, Grillitsch K, Daum G, Just U, Höfken T.2009. Modulation of sterol homeostasis by the Cdc42p effectors Cla4p and Ste20p in the yeast Saccharomyces cerevisiae. FEBS J. 276:7253–7264 [DOI] [PubMed] [Google Scholar]
- 30. Liu H, Styles CA, Fink GR.1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741–1744 [DOI] [PubMed] [Google Scholar]
- 31. Longtine MS, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 32. Lu M, Sautin YY, Holliday LS, Gluck SL.2004. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 279:8732–8739 [DOI] [PubMed] [Google Scholar]
- 33. Lu A, Hirsch JP.2005. Cyclic AMP-independent regulation of protein kinase A substrate phosphorylation by Kelch repeat proteins. Eukaryot. Cell 4:1794–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma J, et al. 2007. An interrelationship between autophagy and filamentous growth in budding yeast. Genetics 177:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martínez-Muñoz GA, Kane P.2008. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283:20309–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mösch HU, Roberts RL, Fink GR.1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5352–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Müller O, Johnson DI, Mayer A.2001. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 20:5657–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Rourke SM, Herskowitz I.1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pan X, Heitman J.1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park HO, Bi E.2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parra KJ, Keenan KL, Kane PM.2000. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275:21761–21767 [DOI] [PubMed] [Google Scholar]
- 42. Peeters T, et al. 2006. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. U. S. A. 103:13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peter M, Neiman AM, Park HO, van Lohuizen M, Herskowitz I.1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046–7059 [PMC free article] [PubMed] [Google Scholar]
- 44. Raitt DC, Posas F, Saito H.2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramer SW, Davis RW.1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 90:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richman TJ, Sawyer MM, Johnson DI.2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1:458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rizzo JM, Tarsio M, Martinez-Munoz GA, Kane PM.2007. Diploids heterozygous for a vma13Δ mutation in Saccharomyces cerevisiae highlight the importance of V-ATPase subunit balance in supporting vacuolar acidification and silencing cytosolic V1-ATPase activity. J. Biol. Chem. 282:8521–8532 [DOI] [PubMed] [Google Scholar]
- 48. Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH.1991. Methods for studying the yeast vacuole. Methods Enzymol. 194:644–661 [DOI] [PubMed] [Google Scholar]
- 49. Roberts RL, Fink GR.1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974–2985 [DOI] [PubMed] [Google Scholar]
- 50. Robertson LS, Fink GR.1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. U. S. A. 95:13783–13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russnak R, Konczal D, McIntire SL.2001. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 276:23849–23857 [DOI] [PubMed] [Google Scholar]
- 52. Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ.2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384–391 [DOI] [PubMed] [Google Scholar]
- 53. Sheu YJ, Barral Y, Snyder M.2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5235–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shimazu M, Sekito T, Akiyama K, Ohsumi Y, Kakinuma Y.2005. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J. Biol. Chem. 280:4850–4857 [DOI] [PubMed] [Google Scholar]
- 55. Smardon AM, Tarsio M, Kane PM.2002. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277:13831–13839 [DOI] [PubMed] [Google Scholar]
- 56. Smardon AM, Kane PM.2007. RAVE is essential for the efficient assembly of the C subunit with the vacuolar H+-ATPase. J. Biol. Chem. 282:26185–26194 [DOI] [PubMed] [Google Scholar]
- 57. Tiedje C, Holland DG, Just U, Höfken T.2007. Proteins involved in sterol synthesis interact with Ste20 and regulate cell polarity. J. Cell Sci. 120:3613–3624 [DOI] [PubMed] [Google Scholar]
- 58. Tiedje C, Sakwa I, Just U, Höfken T.2008. The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol. Biol. Cell 19:2885–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Toda T, et al. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27–36 [DOI] [PubMed] [Google Scholar]
- 60. Toda T, et al. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Toda T, Cameron S, Sass P, Zoller M, Wigler M.1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50:277–287 [DOI] [PubMed] [Google Scholar]
- 62. van Drogen F, et al. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630–639 [DOI] [PubMed] [Google Scholar]
- 63. Wittke S, Lewke N, Müller S, Johnsson N.1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10:2519–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu C, Whiteway M, Thomas DY, Leberer E.1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984–15992 [DOI] [PubMed] [Google Scholar]
- 65. Xue Y, Batlle M, Hirsch JP.1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 17:1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu H, Wickner W.2006. Bem1p is a positive regulator of the homotypic fusion of yeast vacuoles. J. Biol. Chem. 281:27158–27166 [DOI] [PubMed] [Google Scholar]
- 67. Yang Z, Huang J, Geng J, Nair U, Klionsky DJ.2006. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17:5094–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yoon JH, Choi EJ, Parker R.2010. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189:813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang J, Myers M, Forgac M.1992. Characterization of the V0 domain of the coated vesicle (H+)-ATPase. J. Biol. Chem. 267:9773–9778 [PubMed] [Google Scholar]
- 70. Zoncu R, et al. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334:678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]