Abstract
Recent studies indicate that mitochondrial functions impinge on cell wall integrity, drug tolerance, and virulence of human fungal pathogens. However, the mechanistic aspects of these processes are poorly understood. We focused on the mitochondrial outer membrane SAM (Sorting and Assembly Machinery) complex subunit Sam37 in Candida albicans. Inactivation of SAM37 in C. albicans leads to a large reduction in fitness, a phenotype not conserved with the model yeast Saccharomyces cerevisiae. Our data indicate that slow growth of the sam37ΔΔ mutant results from mitochondrial DNA loss, a new function for Sam37 in C. albicans, and from reduced activity of the essential SAM complex subunit Sam35. The sam37ΔΔ mutant was hypersensitive to drugs that target the cell wall and displayed altered cell wall structure, supporting a role for Sam37 in cell wall integrity in C. albicans. The sensitivity of the mutant to membrane-targeting antifungals was not significantly altered. The sam37ΔΔ mutant was avirulent in the mouse model, and bioinformatics showed that the fungal Sam37 proteins are distant from their animal counterparts and could thus represent potential drug targets. Our study provides the first direct evidence for a link between mitochondrial function and cell wall integrity in C. albicans and is further relevant for understanding mitochondrial function in fitness, antifungal drug tolerance, and virulence of this major pathogen. Beyond the relevance to fungal pathogenesis, this work also provides new insight into the mitochondrial and cellular roles of the SAM complex in fungi.
INTRODUCTION
Human fungal pathogens, of which Candida albicans is the most common, can cause life-threatening infections in immunocompromised patients. Fungal infections have become more prevalent in recent decades, and despite improvement in therapies, such as the introduction of the echinocandins into clinical practice in 2002 (56), mortality from systemic fungal infections remains high and is commonly in the range of ≈40% (54, 57). Currently used antifungal drugs inhibit a limited number of cellular functions: membrane integrity (azoles and polyenes), cell wall biogenesis (echinocandins), or DNA synthesis (flucytosine). Each of the classes of antifungals has its disadvantages in terms of development of resistance, toxicity, or availability/route of administration (60). Identification and characterization of cellular pathways other than those targeted by the current drugs is likely to lead to the development of better antifungal therapies. Recent studies indicate that mitochondrial pathways hold promise in this respect, as crippling mitochondrial function leads to hypovirulence of C. albicans (2, 3, 52; reviewed in reference 64). Moreover, mitochondrial mutants in several fungal species are more sensitive to antifungal drugs, in particular those targeting the cell wall, such as the echinocandins (12, 14, 16, 30). However, very few mitochondrial factors have been studied in any detail in C. albicans, or for that matter, any other human fungal pathogen. A deeper understanding of the cellular and virulence functions of mitochondrial factors in human fungal pathogens is warranted.
In this report, we focused on the mitochondrial outer membrane SAM (Sorting and Assembly Machinery) complex in C. albicans. In the model yeast Saccharomyces cerevisiae, the SAM complex is required for biogenesis of the mitochondrial outer membrane, primarily the insertion of β-barrel proteins (11, 22, 70). The core SAM complex is composed of five subunits—the β-barrel proteins Sam50 and Mdm10, with Sam50 as the central channel component of the complex, as well as Sam35, Sam37, and Mim1, which function in the binding and releasing of proteins interacting with the SAM complex; however, their precise biochemical functions are not well understood (reviewed in reference 11). In addition to roles in protein insertion into the mitochondrial outer membrane, the SAM complex functions in phospholipid homeostasis (16, 25). The proposed role is in trafficking of phospholipids between the endoplasmic reticulum (ER) and mitochondrial membrane systems (16). A recent screen using S. cerevisiae performed in our lab found several mitochondrial mutants with increased susceptibility to the echinocandin caspofungin, including mutants deleted for two subunits of the SAM complex, SAM37 and MDM10 (16). In the same study, we deleted SAM37 in C. albicans and found that the mutant was sensitive to caspofungin, similar to what we observed in S. cerevisiae (16). The sensitivity of mitochondrial mutants to drugs that disrupt the cell wall is indicative of links between mitochondrial function and cell wall integrity (CWI) (12, 14, 16, 30). However, in most cases, these links have been made solely based on the sensitivity of mitochondrial mutants to cell wall inhibitors, which could in principle stem from indirect effects of mitochondrial dysfunction on cell growth and metabolism. In other words, whether the mitochondrial mutants have an actual cell wall defect is largely uncharacterized.
Here, we sought to further understand the roles of mitochondria in cell wall integrity, as well as antifungal drug tolerance and fitness of C. albicans, by studying the sam37ΔΔ mutant. We demonstrate for the first time directly that a C. albicans mitochondrial mutant has altered cell wall structure. Our data suggest that the substantial fitness defect of the C. albicans sam37ΔΔ mutant stems in part from mitochondrial DNA (mtDNA) loss, a cellular function not conserved with S. cerevisiae Sam37, and in part from lower activity of the essential SAM complex subunit Sam35. Consistent with a requirement for C. albicans fitness, we found that SAM37 was required for disease in the mouse model of systemic candidiasis. Finally, we performed extensive bioinformatic analyses to show that the fungal Sam37 proteins significantly diverged from their counterpart in animals and could thus be explored as suitable targets for antifungal drug development.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The C. albicans strains used in this study are derived from BWP17 and are described in reference 16. The wild type (WT) used for all experiments was DAY185, the mutant was sam37Δ::ARG4/sam37Δ::URA3 his1::hisG::pHIS1, and the reconstituted strain was sam37Δ::ARG4/sam37Δ::URA3 his1::hisG::pHIS1-SAM37. The mkc1−/− mutant was from the kinase collection described in the work of Blankenship et al. (5). Strains overexpressing SAM35 were constructed by placing one copy of the gene under the strong constitutive promoter TEF1 using the pCJN498 vector (51). For the calcofluor white staining experiments below (see Fig. 2D), the isogenic wild type and sam37ΔΔ mutant strains with a C-terminal HA tag on the kinase Cek1 were used. All strains are fully prototrophic (URA3+ ARG4+ HIS1+). Standard growth conditions were YPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C. Glycerol was added at 2% instead of glucose for the experiments in Fig. 1D. Standard yeast synthetic complete medium was used. For assessing filamentous growth, strains were grown overnight in YPD at 30°C and then diluted to an optical density at 600 nm (OD600) of 0.1 in filamentation-inducing medium and grown at 37°C (200 rpm) for the times indicated in the figures. The medium was YPD plus 10% serum, Spider (1% nutrient broth, 1% d-mannitol), M199, or N-acetylglucosamine medium (9 g NaCl, 6.7 g yeast nitrogen base, and 0.56 g N-acetylglucosamine per liter). Cells were washed, resuspended in phosphate-buffered saline (PBS), and visualized with a differential interference contrast (DIC) microscope (Olympus IX81). For testing filamentation on plates, cells were streaked on plates containing filamentation medium and incubated at 37°C. The colonies were photographed with a stereo dissecting microscope (Olympus SZX 16).
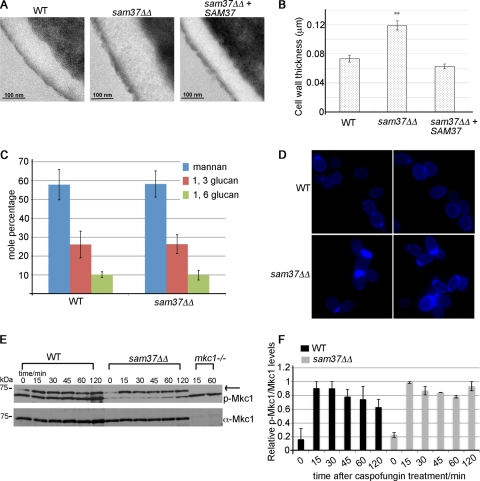
Fig 2.
Sam37 affects cell wall integrity in C. albicans. (A) Transmission electron microscopy showed thicker cell walls in the sam37ΔΔ mutant compared to the wild type and the reconstituted strain. (B) The thickness of the wall was measured for at least 10 individual cells from each of the strains, at three different positions on the cells. Shown are the averages of the measurements and the standard error. (C) The carbohydrate composition of the wall was measured by GC-MS. Shown is the mole percentage average from four or five replicates analyzed over two experiments and the standard deviation. (D) Equal numbers of cells from overnight cultures from the wild type and sam37ΔΔ mutant strains were stained with 250 μg/ml calcofluor white and visualized with fluorescence microscopy using the DAPI filter. Shown are composite pictures from 2 different fields for the mutant and the wild type, respectively. The original micrographs, including bright-field pictures of the same fields, are presented in Fig. S5 in the supplemental material. (E) Cells from the indicated strains were grown to log phase and then treated with 125 ng/ml caspofungin for the indicated times. Whole-cell protein extracts were prepared and proteins separated by SDS-PAGE. Phopsho-Mkc1 was detected using the anti-phospho Erk antibody (p-44/42). Mkc1 was detected by antibodies raised against the protein. (F) Quantification of the phospho-Mkc1 versus total Mkc1 bands was performed using ImageQuant. The highest phospho-Mkc1/total Mkc1 ratio in the wild type or the sam37ΔΔ mutant, respectively, was set to 1, and the other ratios were expressed relative to that. Equivalent results were obtained in several experiments. Shown are the averages of two independent experiments and the standard error.
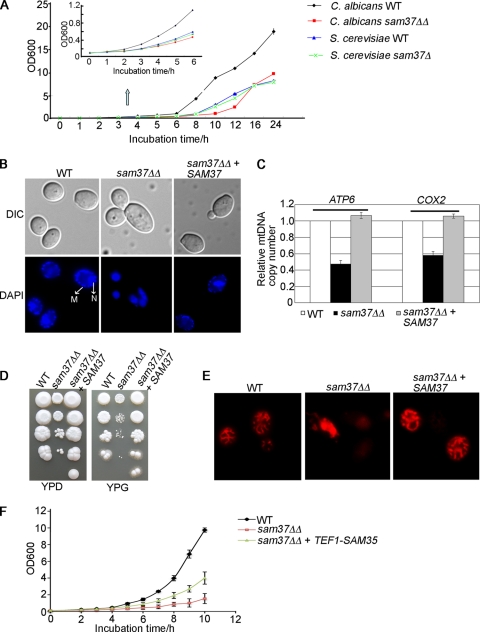
Fig 1.
Sam37 is required for mtDNA stability, wild-type mitochondrial morphology, and fitness of C. albicans. (A) Growth curves of C. albicans and S. cerevisiae wild types (WT) and isogenic sam37 deletion mutants. Overnight cultures were diluted to an OD600 of 0.1, and growth was monitored by measuring optical density of the cultures over time. (B) Nuclear and mitochondrial DNA was stained with DAPI. Nuclear DNA stained brightly, while the mitochondrial nucleoids displayed punctuate cytoplasmic staining. N, nucleus; M, mitochondrial DNA. (C) Mitochondrial DNA loss in the sam37ΔΔ mutant was confirmed by quantitative PCR, using primers for amplification of the nuclear ACT1 gene or the mtDNA genes COX2 and ATP6. Levels of the mtDNA genes were normalized to the levels of ACT1. Shown are averages from three independent cultures and the standard deviation. (D) Growth of the indicated strains on YPD (2% glucose) or YPG (2% glycerol) plates was assessed by plating 10-fold serial dilutions starting at an OD600 of 0.5 and incubating the plates at 30°C for 3 to 5 days. (E) Mitochondrial morphology was evaluated after staining with Mitotracker Red. The wild type presents with a tubular network of mitochondria, while the sam37ΔΔ mutant contains aggregated organelles. (F) Growth curve in YPD for the sam37ΔΔ mutant and sam37ΔΔ + TEF1-SAM35 overexpression strain. The experiments were performed as in panel A. Shown are averages from two or three independent experiments and the standard error.
Antifungal susceptibility tests.
MICs were determined using the broth microdilution method according to CLSI guidelines M27-A3. Drug concentrations ranged from 0.0005 to 8 μg/ml for caspofungin; 0.032 to 16 μg/ml for amphotericin B; 0.125 to 64 μg/ml for fluconazole, nystatin, and Congo red; and 0.025 to 128 μg/ml for calcofluor white. One-hundred-microliter portions of 2-fold serial dilutions of the drugs prepared in RPMI-1640 were added into wells of 96-well plates. Exponentially grown cultures were diluted in RPMI-1640 to a density of ∼1 × 103 to 5 × 103 CFU/ml, and 100 μl was added to each well. All plates were incubated for 48 h at 37°C. Although CLSI M27-A3 suggests 24 h of incubation to obtain caspofungin MICs, 48 h of incubation compensates for the slow growth of sam37ΔΔ mutant strain. Fungal growth was examined visually with the aid of a mirror reader. The MIC was defined as the concentration resulting in complete growth inhibition for amphotericin B and an inhibition of at least 50% of fungal growth for the other drugs, corresponding to a score of two in the CLSI M27-A3 protocol. The minimal fungicidal concentration (MFC) assays were performed for the optically clear wells after MIC determination. CFU counting was done by plating an aliquot onto YPD plates followed by an incubation at 37°C for 48 h. The MFC was defined as the lowest concentration killing 99% of the initial inoculum. In addition to standard assays, modified MICs and MFCs of caspofungin were also tested, with YPD or synthetic complete medium replacing RPMI.
Cell wall analysis and Western blots.
Preparation of cell walls and analysis of the carbohydrate composition were performed as described previously (16). The mole percentage of each polysaccharide was estimated from 1,4-linked glucosamine for chitin, 1,3-linked glucopyranose for 1,3-β-glucan, 1,6-linked glucopyranose for 1,6-β-glucan, and addition of all mannopyranosyl derivatives for mannan. For mannan and glucan, at least four replicates were analyzed in two independent experiments. For chitin, two biological replicates for the wild type and three clones of the mutant, respectively, were analyzed in a single experiment. For assaying cell wall integrity pathway activation, yeast strains were grown to log phase and then treated with 125 ng/ml caspofungin for the times indicated in Fig. 2. Whole-protein extracts were prepared by trichloroacetic acid (TCA) precipitation, and Western blotting was performed. Phospho-Mkc1 was detected using the mouse monoclonal anti-phospho-p44/42 mitogen-activated protein (MAP) kinase antibody (Cell Signaling). Mkc1 was detected by an anti-Mkc1 antibody generously provided by Jesus Plà and Elvira Román (61). The bands corresponding to phospho-Mkc1 or total Mkc1 were quantified with ImageQuant.
Microscopy.
For bright-field and fluorescence microscopy, an Olympus IX81 microscope was used and photographs were taken with the Olympus cell^M software. For visualizing nuclear and mitochondrial DNA, cells were grown to mid-log phase, fixed with 70% ethanol for 10 min, and then stained with 5 μg/ml DAPI (4′,6-diamidino-2-phenylindole) for 10 min. Mitotracker straining of mitochondria was done as described previously (16). Calcofluor white (250 μg/ml) staining was performed with equal cell numbers from overnight cultures of the wild type and the sam37ΔΔ mutant. Staining was performed at room temperature for 5 min, followed by three washes in distilled water.
For transmission electron microscopy (TEM), mid-log-phase yeast cells grown in YPD were harvested by centrifugation and the pellets were fixed in 2.5% glutaraldehyde (vol/vol) in 0.1 M sodium cacodylate buffer (pH 7.4). Secondary fixation was performed in 1% osmium tetroxide in double-distilled water (ddH2O), and cells were then treated with 2% uranyl acetate en bloc stain. After dehydration with series of ethanol (50%, 75%, 95%, 100%) and propylene oxide, cells were embedded in Procure 812 resin before ultrathin sections were cut on a Leica UC6 ultramicrotome. Samples were then stained with uranyl acetate and lead citrate and examined with a Hitachi H7500 TEM. Cell wall thickness was measured manually for at least 10 individual yeast cells with E-ruler 1.1.
Candida-macrophage interaction experiments.
A C. albicans-macrophage interaction assay was carried out as described by Fernandez-Arenas et al. (20). Murine macrophages (2 × 105) (RAW 264.7) prepared in RPMI-1640 culture medium (containing 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 2.5 mM HEPES and supplemented with 10% fetal bovine serum) were plated onto sterile 18-mm coverslips placed in 24-well plates for 24 h at 37°C in a 5% CO2 incubator. Overnight cultures of C. albicans were grown in YPD, washed with PBS, and resuspended in RPMI-1640 to a cell density of 5 × 105 cells/ml. Macrophages were then allowed to interact with 1 ml of the C. albicans suspension for 30 min, 90 min, and 180 min at 37°C. At each time point, the coverslips were removed and washed three times with ice-cold PBS and then stained with 2.5 μM calcofluor white for 10 min. The cells were then fixed with 4% paraformaldehyde in PBS for 30 min. Photographs were taken using DIC and fluorescence microscopy (Olympus IX81). Digital images were processed with the ImageJ software.
Measurements of mitochondrial membrane potential.
Mitochondria from wild-type C. albicans or the sam37ΔΔ mutant were isolated by differential centrifugation as described previously (13). The experiment measures the drop in fluorescence of the cationic dye TMRM (tetramethylrhodamine methyl ester; Invitrogen Molecular Probes) on the addition of the antimycin-valinomycin-oligomycin (AVO) cocktail to mitochondria as a proxy for mitochondrial membrane potential. The AVO solution was made at 100× in ethanol and used at final concentrations as follows: antimycin (8 μM), valinomycin (1 μM), and oligomycin (20 μM). The measurements were performed at 25°C using a BMG Fluostar Optima spectrophotometer in a 96-well Lab Systems fluoro plate using 540 nm excitation and 590 nm emission. Mitochondria were diluted to 1.67 mg/ml in SEM (250 mM sucrose, 1 mM EDTA, 10 mM MOPS [morpholinepropanesulfonic acid], 10 mM succinate [pH 7], 10 mM malate [pH 7]), and 25 μg of mitochondria was added to 300 μl of potential buffer (0.6 M sorbitol, 10 mM MgCl2, 0.5 mM EDTA, 0.1% [wt/vol] bovine serum albumin [BSA], 20 mM KPi [pH 7.2]) containing TMRM. The results are averages (with standard deviations [SDs]) for three separate mitochondrial preparations.
Animal models.
Filamentation assays in the worm infection model were performed as described previously (55). Young adult nematodes were allowed to feed for 4 h on lawns of C. albicans grown on solid brain heart infusion (BHI) medium (Difco) containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), and streptomycin (200 μg/ml). Worms were washed with M9 medium and transferred into wells of a six-well microtiter dish with 2 ml of 80% M9 and 20% BHI medium, at 60 to 80 worms per well. The plates were incubated at 25°C. Worms were assessed at 24-h intervals for penetrative C. albicans filamentation using a DIC microscope. Photographs were taken using an Olympus IX81 microscope with the Olympus cell^M software. To determine whether C. albicans was ingested by the worms, yeast-infected worms were stained with calcofluor white (1 μg/ml) for 1 min and then examined by microscopy. The percentage of worms with penetrative filamentation was determined at day 3, from three independent experiments performed with three independent sam37ΔΔ mutant deletion clones. Means and the standard error were calculated, and the P value was determined using the Student t test.
The virulence assays in the mouse systemic candidiasis model were performed essentially as described previously (16). Briefly, cells from the wild type and mutant strains were grown for 20 h in YPD, and mice were infected via the tail vein with 7 × 105 cells/100 μl of PBS. Mice were euthanized at 20% weight loss or before if they showed debilitating clinical signs. An estimation of differences in survival (log rank test) using the Kaplan-Meier method was performed, and the survival curves were plotted with the SPSS version 16 statistical software. A P value of <0.05 was considered statistically significant. One-way analysis of variance (ANOVA) was used to compare means between groups. For histopathology, kidneys were fixed in 10% neutral buffered formalin (NBF) and selected tissue blocks were processed overnight using a routine overnight cycle in a tissue processor. The tissue blocks were then embedded in wax and serially sliced into 5-μm sections, and slides were stained with periodic acid-Schiff (PAS) stain.
qPCR analysis.
As a quantitative measure of mtDNA loss, quantitative PCR (qPCR) was used to determine the levels of genes on the mitochondrial genome (COX2 and ATP6) compared to ACT1 in the nucleus. qPCR was performed as described in reference 16 from total DNA isolated by the smash and grab method. The levels of COX2 and ATP6 were normalized to ACT1 to determine the relative mtDNA copy number. Three independent cultures were analyzed for each of the strains, and the average and the standard error are presented in Fig. 1C.
Bioinformatic analysis.
A curated set of fungal Sam35 and Sam37 sequences, as well as animal metaxins (1 and 2), was used for the bioinformatic analysis (see File S1 in the supplemental material). Sequences were clustered based on pairwise similarity scores using CLANS (21) with default settings. Hidden Markov models (HMM) for Sam37 were constructed as described by Likic et al. (43), using a set of yeast sequences. Pairwise identities shown in Table 1 were calculated from global pairwise alignment using the EMBOSS program “needle” (59).
Table 1.
Percentage sequence similarity between fungal Sam37 and Sam35 proteins and the animal metaxins
| Comparison | % Identity for global pairwise sequence alignment |
|---|---|
| Sam37 C. albicans vs metaxin 1 human | 16.6 |
| Sam37 C. albicans vs metaxin 2 human | 10.6 |
| Sam35 C. albicans vs metaxin 1 human | 12.1 |
| Sam35 C. albicans vs metaxin 2 human | 7.5 |
RESULTS
Roles of SAM37 in fitness, mitochondrial DNA stability, and organelle morphology in C. albicans.
We previously observed that inactivation of SAM37 in C. albicans causes a substantial fitness defect (16). Here we sought to understand the basis of this phenotype, as the defect seen in C. albicans is much more pronounced than what is observed with the sam37Δ mutant in S. cerevisiae (Fig. 1A) (8, 17). We directly compared the sam37 mutants in the two yeasts during growth in rich (YPD) medium. In S. cerevisiae, the mean doubling time ± standard deviation for the sam37Δ mutant was 1.96 ± 0.07 h, compared to 1.89 ± 0.05 h for the wild type, while in C. albicans the mutant had a doubling time of 2.5 ± 0.2 h, compared to 1.4 ± 0.03 h for the wild type. The same growth defects were observed when synthetic medium was used. In C. albicans, the wild type had a doubling time of 1.98 ± 0.05 h compared to the 4.01 ± 0.5 h for the mutant, while in S. cerevisiae the doubling times were similar—3.93 ± 0.38 for the wild type and 3.88 ± 0.44 for the mutant (the growth curve for synthetic medium is shown in Fig. S1A in the supplemental material). The difference in the fitness defect between the sam37 mutants in C. albicans and S. cerevisiae was also evident in spot dilution assays on YPD or synthetic medium plates (see Fig. S1B in the supplemental material). This large fitness defect was observed in three independently constructed C. albicans sam37ΔΔ homozygous mutants.
Unlike S. cerevisiae, C. albicans is a petite-negative yeast, in which loss of the mitochondrial genome is expected to have a large effect on fitness. While Sam37 is not known to be required for mitochondrial DNA (mtDNA) stability in S. cerevisiae (48) and our experiments confirm these previous observations (see Fig. S1C in the supplemental material), we reasoned that Sam37 function might be somewhat different in C. albicans. Indeed, staining with DAPI revealed that 76.6% ± 3.4% of the cells in mutant cultures were devoid of mtDNA (Fig. 1B). Moreover, a few cells in mutant cultures displayed brighter, irregular mtDNA staining similar to what is observed in cells with partial loss of mtDNA (so-called rho− cells) (19), which is unlike the punctuate staining for mtDNA observed in the wild type (Fig. 1B). mtDNA loss was further confirmed by quantitative PCR, measuring the ratio of nuclear (ACT1) versus mtDNA genes (COX1 and ATP6) (Fig. 1C). The sam37ΔΔ mutant cells that were positive for mtDNA staining appeared to have retained wild-type mitochondrial function, as shown by the ability to grow on the respiratory carbon source glycerol (Fig. 1D). Staining mitochondria with Mitotracker Red revealed that Sam37 also affected the morphology of the organelle, with cells from the sam37ΔΔ mutant showing aggregated, clumped mitochondria, unlike the tubular structures observed in the wild type and reconstituted strain (Fig. 1E). We also assessed mitochondrial membrane potential by measuring the drop in fluorescence of the cationic dye TMRM (tetramethylrhodamine methyl ester) on addition of an inhibitor cocktail (valinomycin, oligomycin, and antimycin) to mitochondria isolated from the wild type or the sam37ΔΔ mutant. While the C. albicans sam37ΔΔ mutant was losing mitochondrial DNA, the mitochondrial membrane potential appeared unchanged: the drop in TMRM fluorescence was 9.0% ± 2.7% (standard deviation) for the wild type and 7.5% ± 1.3% (standard deviation) for the mutant (see Materials and Methods for more details on the experiment). We suspect that the normal mitochondrial membrane potential in the sam37ΔΔ mutant is due to the activation of compensatory mechanisms to maintain this essential cellular function in the petite-negative C. albicans. In S. cerevisiae, the deletion of SAM37 leads to reduced levels of another SAM complex subunit, Sam35, with Sam35 being essential for yeast viability (13, 49). When we overexpressed SAM35 by placing it under a strong constitutive promoter, we could partially rescue the growth defect of the C. albicans sam37ΔΔ mutant (Fig. 1F), suggesting that, in addition to mtDNA loss, lower levels and/or activity of Sam35 contribute to reduced fitness seen upon deletion of SAM37 in C. albicans.
SAM37 and cell wall integrity in C. albicans.
To start delineating the effect of Sam37 on the cell wall in C. albicans, we observed the structure of the cell wall in wild-type and mutant cultures by transmission electron microscopy (TEM) (Fig. 2A and B). The sam37ΔΔ mutant presented with a thicker cell wall compared to the wild type and the reconstituted strain (Fig. 2A and B). Specifically, the structure of the central, electron-translucent layer of the cell wall, which represents the cell wall glucan and chitin, was affected in the mutant (Fig. 2A, central panel). Changes in cell wall thickness are consistent with a cell wall defect, as they are often observed in bona fide cell wall mutants (for example, see references 44 and 58). We next isolated the cell walls from the wild type and the sam37ΔΔ mutant to analyze the carbohydrate composition by gas chromatography-mass spectrometry (GC-MS) of permethylated alditol acetates. This method measures the relative carbohydrate composition of the cell wall. As shown in Fig. 2C, the relative levels of mannan, 1,3-β-glucan, and 1,6-β-glucan were not affected in the sam37ΔΔ mutant compared to the wild type. In other words, the mannan:glucan ratio in the cell wall remained the same in the presence or absence of Sam37. Chitin levels were also determined by GC-MS and were found to be somewhat higher in the mutant (mole percentage in the wild type was 0.8 ± 0.1 [mean ± SD] and in the mutant it was 1.3 ± 0.4 [mean ± SD]; P = 0.048). This method, however, underestimates chitin, and therefore, for an independent measure of chitin content, we stained the wild-type and mutant cultures with calcofluor white and observed the cells with fluorescence microscopy. A proportion of cells from mutant cultures displayed more pronounced chitin staining (Fig. 2D). We noted that in particular mutant cells with a slightly more elongated cell morphology than the round yeast cells observed in the wild type were prone to stain more brightly with calcofluor white. Collectively, these data suggest that the levels of chitin are elevated in the cell wall of sam37ΔΔ cells, which represents a further indication of a cell wall defect.
A previous report in the model yeast S. cerevisiae regarding a mitochondrial mutant deleted for the phosphatidylglycerol synthase Pgs1 indicated a link between mitochondrial function and activation of the protein kinase C (PKC)-dependent cell wall integrity (CWI) pathway (72). In C. albicans, dysfunctional mitochondria in the sam37ΔΔ mutant did not inhibit activation of the CWI pathway, as judged by the wild-type kinetics of appearance of the phosphorylated form of the downstream kinase Mkc1 upon treatment of cells with caspofungin (Fig. 2E). Quantification of phospho-Mkc1 levels over total Mkc1 showed that the levels of phospho-Mkc1 tend to stay higher in the sam37ΔΔ mutant than in the wild type at the late time points in the time course (see the 120-min time point in Fig. 2F; the difference was small but was reproducible in several independent experiments). This suggests a delay in adaptation by the mutant to cell wall stress inflicted by caspofungin treatment, which is consistent with the mutant exhibiting a cell wall defect.
Sensitivity of sam37ΔΔ mutants to antifungal compounds.
Specific mitochondrial mutations in C. albicans, Candida glabrata, and S. cerevisiae lead to either resistance or hypersensitivity to antifungal drugs that target cell membranes, such as the azoles and the polyenes (9, 10, 15, 16, 23, 24, 36, 62–64, 67). In particular, loss of mtDNA is known to lead to drug resistance, including resistance to the azole class of antifungal drugs (15, 26, 27, 36, 62, 63, 66; reviewed in reference 64). To address how deletion of SAM37 and the associated mitochondrial phenotypes affected drug tolerance in C. albicans, we tested the sensitivity of the C. albicans sam37ΔΔ mutant to the azole drug fluconazole, as well as to the polyene amphotericin B, by determining the MIC and the minimum fungicidal concentration (MFC) (Table 2). We also included cell wall-targeting agents—the echinocandin caspofungin, calcofluor white, and Congo red. The MIC and MFC of the sam37ΔΔ mutant in response to cell wall integrity inhibitors calcofluor white and Congo red was significantly reduced, consistent with a defective cell wall in the absence of Sam37 (Table 2). To our surprise, the caspofungin MIC was not altered, while the MFC was only 2-fold lower for the mutant compared to the wild type. This is in contrast to our previous observation that the sam37ΔΔ mutant was unable to grow on caspofungin in plate assays (16). We reasoned that this disparity could be due to differences in growth conditions (for example, the plate assays were done in YPD, while the MIC determination was done by using standardized approved protocols for which RPMI medium is used). Therefore, we next assayed caspofungin MIC and MFC in YPD and synthetic complete medium (Table 2). When YPD or synthetic medium was used, both the MIC and the MFC for caspofungin were significantly lower for the mutant than for the wild type, supporting our original observation that the sam37ΔΔ mutant is sensitive to caspofungin (16). Of note is that the MIC and MFC for caspofungin in YPD for the wild type were much lower than in RPMI medium (Table 2), which is in line with previous reports (35) and supports the idea that different growth conditions affect the susceptibility of C. albicans to this echinocandin drug. In terms of cell membrane-targeting antifungal drugs, no significant differences for MICs/MFCs were observed between the sam37ΔΔ mutant and the wild type (Table 2). The mutant displayed a 2-fold-lower MIC for fluconazole and nystatin but no changes to the MFC (Table 2). The MIC for amphotericin B was not changed, while the MFC was 2-fold higher for the mutant (Table 2). Collectively, these results show that while Sam37 has a profound effect on the cell's ability to deal with cell wall stress, it has at best a very mild defect in terms of tolerance of drugs that target the cell membrane.
Table 2.
Antifungal susceptibility testing for the C. albicans sam37ΔΔ mutant
| Antifungal drugb | MIC (μg/ml) |
MFC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| WT strain | sam37ΔΔ strain | sam37ΔΔ+SAM37 strain | Fold changea | WT | sam37ΔΔ strain | sam37ΔΔ+SAM37 strain | Fold changea | |
| Caspofungin | 0.25 | 0.25 | 0.25 | 1 | 0.5 | 0.25 | 0.5 | 2 |
| Congo red | 2 | 0.25 | 2 | 8 | 32 | 4 | 32 | 8 |
| Calcofluor white | 128 | 16 | 128 | 8 | >128 | 32 | >128 | >4 |
| Amphotericin B | 0.25 | 0.25 | 0.25 | 1 | 0.5 | 1 | 0.5 | 2 |
| Nystatin | 4 | 2 | 4 | 2 | 4 | 4 | 8 | 1 |
| Fluconazole | 0.5 | 0.25 | 0.5 | 2 | >64 | >64 | >64 | NA |
| Caspofungin (YPD) | 0.008 | 0.001 | 0.008 | 8 | 0.015 | 0.004 | 0.015 | 4 |
| Caspofungin (SC) | 0.25 | 0.06 | 0.25 | 4 | >8 | 1 | >8 | >8 |
Fold change: WT/sam37ΔΔ strain.Boldface indicates a significant change. NA, not available.
Unless otherwise stated, measurements were done using RPMI medium according to CLSI guidelines. SC, synthetic complete medium.
SAM37 and the yeast-to-hypha morphogenetic switch.
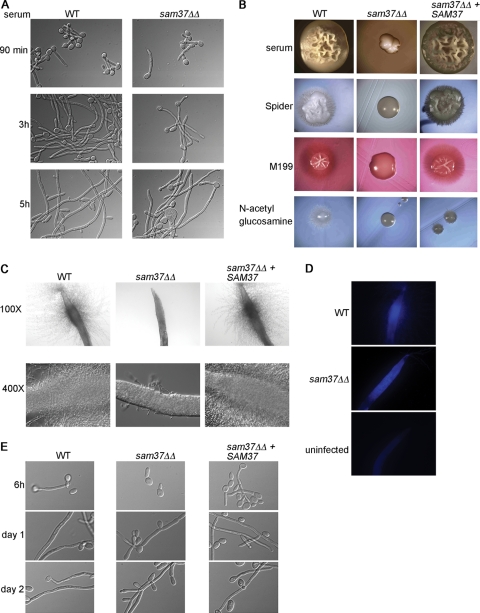
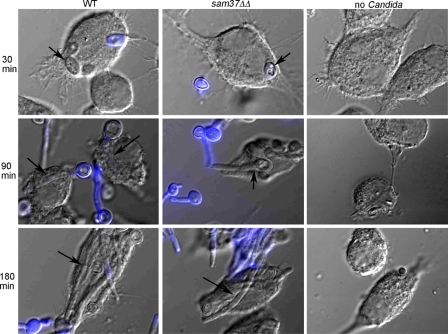
Mitochondrial function plays a role in the ability of C. albicans to undergo the switch to hyphal growth (2, 37, 45, 50, 68). The sam37ΔΔ mutant was able to form filaments in vitro in YPD plus 10% serum medium (Fig. 3A), and also in M199 and in medium supplemented with N-acetylglucosamine (see Fig. S2 in the supplemental material) (in M199 medium, filamentation by the mutant was somewhat delayed, perhaps due to slower growth). The mutant was able to form a substantial number of long hyphal cells, but we also noticed that a proportion of the filaments were somewhat shorter and wider than in the wild type. In liquid Spider medium, the sam37ΔΔmutant displayed a filamentous growth defect, and even after 5 h a large proportion of cells was in yeast form, although filaments could be observed after 8 h (see Fig. S2). On plates, the sam37ΔΔ mutant was unable to form filaments in any of the conditions tested even after prolonged incubation (Fig. 3B). We also tested the ability of the mutant to filament in an in vivo host system, using the Caenorhabditis elegans-C. albicans infection model (7, 55). In the host, filamentation was severely crippled, with the mutant only producing very few short filaments, and this was true even after prolonged incubation of up to 7 days (Fig. 3C and data not shown). The percentages of worms with penetrative filamentation for wild-type C. albicans and the reconstituted strain were 50.67% ± 7.5% (mean ± SD) and 53.67% ± 11.0%, and this was reduced to 11% ± 1.0% for worms infected with the sam37ΔΔ mutant. Staining with calcofluor white indicated that the mutant cells were ingested by the worms (Fig. 3D). As a control, we tested growth and filamentation in vitro in the medium used for the worm assays (20% BHI in M9; see Materials and Methods) at 25°C, which is the temperature at which the worm assay is performed, and also at 37°C. The sam37ΔΔ mutant was able to filament in 20% BHI/M9 medium at 37°C, but with somewhat slower kinetics. At the 6-h time point, filamentation was seen for the wild type and the reconstituted strain, while the mutant was still in yeast form, but at day 1 all strains, including the mutant, were filamentous (Fig. 3E) (the full time course is shown in Fig. S3 in the supplemental material). At 25°C all strains, including the wild type, stayed in yeast form (see Fig. S3B in the supplemental material). The sam37ΔΔ mutant grew slower in the 20% BHI/M9 medium in vitro, but even the wild type grew very slowly in this low-nutrient medium (Fig. S3A in the supplemental material). We cannot rule out that the filamentation defect in the worm is due to the fitness defect of the sam37ΔΔ mutant; however, the fact that the mutant was able to filament substantially under similar conditions in vitro, despite the fitness defect, suggests that Sam37 might be required for filamentous growth in the worm host. In contrast to what we observed in the worm, the sam37ΔΔ mutant could filament upon phagocytosis by macrophages. Internalization by macrophages of both wild-type and mutant C. albicans cells could be observed after 30 min, and at 90 min filaments piercing through the macrophages were clearly seen for both wild-type and mutant strains (Fig. 4). We noticed that a proportion of the filaments formed by the sam37ΔΔ mutant were shorter than those produced by the wild type. In conclusion, the requirement for Sam37 for filamentous growth of C. albicans varies between different hyphal conditions and is most pronounced on solid medium and in the worm host.
Fig 3.
Filamentous growth phenotypes of the C. albicans sam37ΔΔ mutant. (A) Filamentous growth in vitro was tested in 10% serum medium at 37°C. (B) Filamentation on solid medium was assessed by streaking out the wild type, sam37ΔΔ mutant or the complemented strain on the indicated plates and incubating at 37°C. Colonies from the wild type and the complemented strain on all media and the sam37ΔΔ mutant on serum plates were photographed at 20× magnification. The mutant colonies on Spider, M199, and N-acetylglucosamine-containing media were photographed at 60X magnification due to a much smaller size of the colony. (C) The worm C. elegans was infected with wild type C. albicans, the sam37ΔΔ mutant or the complemented strain and penetrative filamentation was monitored over time. The mutant only ever formed short and very few filaments. (D) Calcofluor white staining performed at day 5 of the infection showed that mutant cells were ingested by the worm. (E) Filamentation was assessed in vitro by microscopy, using growth medium equivalent to the worm assay (20% BHI in M9 buffer), at 37°C. In this medium, filaments were not visible in the wild type at 90 min or 3 h, and appeared only at the 6-h time point. The full time course is shown in Fig. S3B in the supplemental material.
Fig 4.
Filamentation by the sam37ΔΔ mutant upon phagocytosis by macrophages. Figures are overlaps of DIC images and fluorescent images and were constructed using ImageJ. Calcofluor white staining was used to differentiate intracellular from external C. albicans (the yeast cells inside the macrophage do not stain with calcofluor white). Internalized C. albicans cells are marked by the arrows.
SAM37 is required for virulence in the mouse systemic candidiasis model.
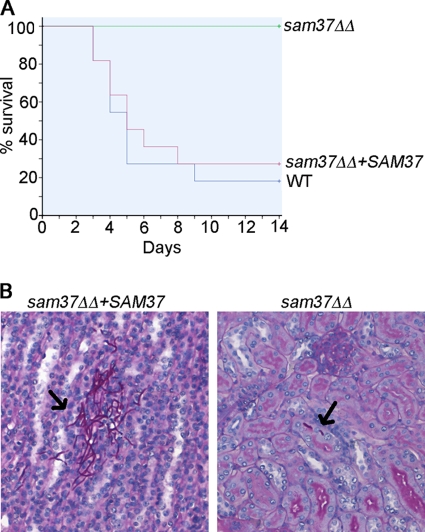
Genes required for fitness of C. albicans are promising drug targets. The significant reduction in fitness of the sam37ΔΔ mutant in vitro predicts a defect in fitness within the host environment. However, it is important to test this prediction directly, as, for example, in the screen of C. albicans genes essential for growth in vitro Becker et al. (3) demonstrated a spectrum of outcomes in vivo. These ranged from being essential for virulence to more moderate defects (“attenuated virulence”). To test the role of Sam37 in C. albicans virulence, we used the tail vein injection model of systemic candidasis. As shown in Fig. 5A, mice infected with the sam37ΔΔ mutant did not succumb to infection, unlike those infected with the wild type or the reconstituted strain. Because of the observation that SAM37 is required for filamentation in a host-pathogen context in the worm (Fig. 3), we performed histopathology analysis of the infected kidneys. Very few infection foci were visible in kidneys from animals infected with the mutant, and the fungal cells that were present lacked the extensive filamentation that is seen in the robust infection foci formed by the reconstituted strain (sam37ΔΔ+SAM37 strain) (Fig. 5B). To obtain a semiquantitative measure of the ability of the sam37ΔΔ mutant to infect the kidney, we counted the infection foci in kidney sections from animals infected with the mutant or the reconstituted strain. In 12 sections, we counted 38 cells in 18 infection foci for the mutant and 383 cells in 33 infection foci for the reconstituted strain. Only 5.5% of infection foci in the mutant, compared to 82% in the reconstituted strain, had four or more cells, indicating that the mutant is compromised in the ability to establish robust infection foci. We conclude that SAM37 cripples fitness of C. albicans in vivo and is essential for virulence.
Fig 5.
Sam37 is required for virulence of C. albicans in the mouse systemic candidiasis model. (A) Mice were infected with the C. albicans strains via the tail vein, and disease progression was monitored as described in Materials and Methods. Differences in survival were assessed by the Kaplan-Meier method (P < 0.05). (B) Kidney histopathology was performed at day 1 of the infection, and C. albicans cells were visualized by PAS staining. Arrows indicate C. albicans cells found in the infected kidneys.
The fungal Sam37 proteins are distant from their functional counterparts in animals.
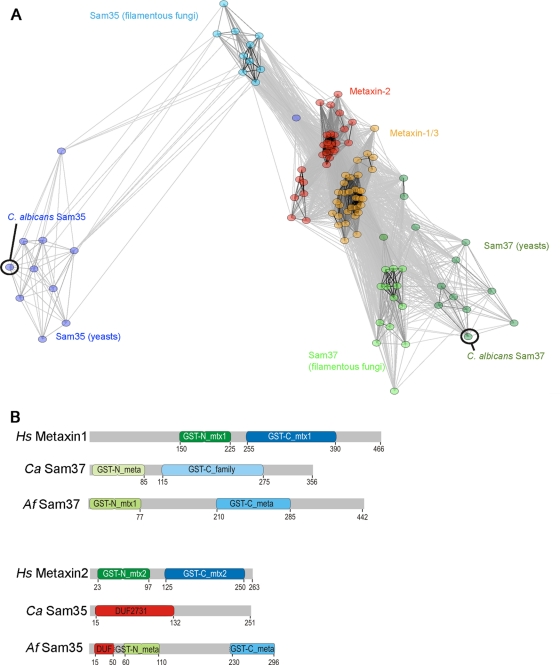
The close relationship of fungi to humans can lead to host toxicity of antifungal compounds, and is therefore an issue worth considering when thinking of potentially suitable targets for antifungal drug development. While the metaxin proteins (metaxin 1 and metaxin 2) from humans have been suggested as functional orthologs of the Sam37 and Sam35 subunits of the fungal SAM complex (1, 40), sequence analysis demonstrates a significant divergence between the fungal SAM proteins and the metaxins (a ClustalW alignment is shown in Fig. S4 in the supplemental material). To further address how different/similar the metaxins are to their fungal counterparts, we performed all-against-all BLAST similarity searches using a curated set of fungal Sam35 and Sam37 sequences and the animal metaxins 1 and 2 (see File S1 in the supplemental material) and visualized the results using CLANS (21). A CLANS plot of these sequences (Fig. 6A) shows the distinct clusters representing the metaxin 1 (orange) and metaxin 2 (red) groups. These two groups sit close together because of the relatively high BLAST scores between the individual metaxin sequences. The Sam37 sequences from fungi (green) spread across two poles with the yeast sequences tending away from the sequences from filamentous fungi. This variation is reflected in the Pfam domain predictions for the sequences (Fig. 6B): the sequences from C. albicans (CaSam37) and Aspergillus fumigatus (AfSam37) both were predicted to have glutathione S-transferase (GST)-N and GST-C domains that would give them a GST-like protein fold, but neither conforms to the strict sequence composition of the metaxin (i.e., GST-N_mtx1 and GST-C_mtx1). We included Sam35 in addition to Sam37 in these analyses because the SAM35 gene is essential for viability in S. cerevisiae (33, 49, 69) and could thus be considered as potentially useful in the context of drug development. The Sam35 sequences in fungi show even more drift than the Sam37 sequences: this is particularly true in the Sam35 sequences from the yeasts, which are distanced from the metaxin 2 sequence cluster (Fig. 6A, purple). Sensitive sequence similarity searches against proteins of known structure using HHpred (65) suggest that each of these proteins adopts an overall GST-like fold (A. Perry, unpublished data); however, the sequence identity between the human and C. albicans proteins is very low, well below the threshold that would indicate clear structural similarity, suggesting the detailed three-dimensional (3D) structures of Sam35 and Sam37 proteins are likely to be significantly different from those of the human metaxins (Table 1). As an independent measure of similarity, we also used a set of yeast Sam37 sequences to construct a hidden Markov model (HMM) (43), which was used to search the UniProtKB/TrEMBL data set. The C. albicans Sam37 protein has an HMM score of 6.9 ×10−105, while the human metaxin 1 HMM score was 6.4 × 10−6, demonstrating the significantly closer relationship of the C. albicans protein to the fungal sequences and the high divergence of the metaxins.
Fig 6.
Bioinformatic analysis of the fungal Sam37 and Sam35 proteins and the metaxins. (A) Sequence similarity between Sam37, Sam35, and metaxins, visualized using CLANS (18). Sequences are clustered based on the pairwise P value between two sequences in an all-against-all BLAST search. Circles represent individual sequences, while lines are drawn between any two sequences that have P values of <10−6. (B) Domain structure of the metaxins and the fungal Sam35 and Sam37 proteins from C. albicans and Aspergillus fumigatus. The indicated domains are from the conserved domain database, in which one of the major groupings is the glutathione S-transferase (GST) family, to which the metaxins and the SAMs belong. The presence of a conserved N-terminal domain with a thioredoxin fold casts a protein in the GST-N_family, while the presence of a conserved C-terminal alpha helical domain casts it in a GST-C_family. GST-N_meta(xin) and GST-C_meta(xin) are conserved subgroups of the GST-N and GST-C families, respectively, which include many of the metaxin/SAM subunits. Within the “meta” families, the proteins most conserved with metaxin 1 from animals are designated “mtx1” (e.g., GST_N_mtx1), and those most conserved with metaxin 2 are called “mtx2.” CaSam35 does not conform to the GST family but, like many other Sam35 homologs, it has conserved domain features classified as DUF2731 (DUF is domain of unknown function). DUF2731 must share some sequence-based features with the GST-N_family, given that AfSam35 partly conforms to the DUF2731 group and partly to the GST-N_meta(xin) group of sequences. We attempted homology modeling of Sam35, Sam37, and the metaxins; however, no reliable models could be made based on template proteins with a GST-like fold that we used.
DISCUSSION
In this study, we analyzed the cellular roles of the mitochondrial outer membrane protein Sam37 in mitochondrial physiology and cell wall integrity, as well as in the fitness and pathogenicity of C. albicans. The drop in fitness upon inactivation of SAM37 in C. albicans was unexpected, as the same does not occur in S. cerevisiae (Fig. 1A). We suggest that this substantial fitness defect of the C. albicans sam37ΔΔ mutant is in part due to the instability of the mitochondrial genome and in part due to loss of the essential SAM complex subunit Sam35. mtDNA loss is expected to have a profound effect on fitness in a petite-negative yeast such as C. albicans. To our knowledge, so far only one other factor has been identified with roles in mtDNA maintenance in C. albicans, the mitochondrial DNA helicase Hmi1 (34). Our sam37ΔΔ mutant resembles the hmi1ΔΔ mutant in several respects. Both mutants are slow growing but viable because a proportion of cells retain mtDNA, both mutants can grow on respiratory carbon sources, indicating that the cells which retained mtDNA are respiratory proficient, and in both cases mtDNA loss and the fitness defect are rescued by complementation with a wild-type copy of the SAM37 or HMI1 genes, respectively (Fig. 1) (34). The latter result suggests that the cells that retain wild-type mtDNA can restore the population to a wild-type phenotype after complementation. In the context of the growth defect, it is worth noting that inactivation of SAM37 has a greater effect on fitness than inactivation of HMI1, and we suggest that the loss of Sam35 activity contributes to the larger effect of Sam37 on cell growth. Sam37 is not required for mtDNA stability in S. cerevisiae (see Fig. S1C in the supplemental material) (48), and it was thus surprising that in C. albicans inactivation of SAM37 led to mtDNA loss. How exactly Sam37 affects mtDNA stability in C. albicans is not clear at this stage. From studies in S. cerevisiae, it is known that a large number of mitochondrial proteins/processes are necessary for mtDNA maintenance, but only for some (mostly those affecting mtDNA metabolism directly) is the molecular basis for this function understood (48). In S. cerevisiae, the SAM complex has been functionally linked to mitochondrial membrane complexes which have roles in mtDNA stability, such as ERMES (ER-Mitochondria Encounter Structure), which spans the ER and mitochondrial outer membranes (6, 11, 16, 28, 31, 38, 39, 46, 47), and the MICOS (Mitochondrial Contact Site) complex that links the mitochondrial outer and inner membranes (29). The exact biochemical role of these membrane-spanning complexes in mtDNA stability is not fully understood, but it is possible that the requirement for Sam37 in mtDNA maintenance stems from functional interactions with ERMES and/or MICOS. The fact that Sam37 has a role in mtDNA stability in C. albicans but not in S. cerevisiae could suggest that the function of Sam37 within the SAM complex is somewhat different in the two yeasts and/or that the functional links between the SAM and the ERMES/MICOS complexes is different. In this context, although in S. cerevisiae the essential protein Sam35 is destabilized in the sam37 mutant (13), this does not lead to a large reduction in fitness, while our data suggest that in C. albicans loss of Sam35 activity contributes to slow growth of cells lacking Sam37. These data support the notion that the structural/functional organization of the SAM complex differs between S. cerevisiae and C. albicans. Of note is that comprehensive sequence analysis using hidden Markov models to recognize protein sequences related to Sam35 and Sam37 have been used previously to interrogate the yeast genome (18). No proteins of any similarity to Sam37 are found in S. cerevisiae and it is therefore unlikely that the explanation for the less pronounced defects of the sam37 mutation in S. cerevisiae than in C. albicans is the presence of a protein which functions in a redundant manner. A final consideration about the mtDNA loss phenotype of the C. albicans sam37ΔΔ strain is in regard to the susceptibility of the mutant to antifungal drugs, particularly the azole fluconazole. mtDNA loss is known to render yeast cells drug resistant; for example, the connection between mtDNA instability and azole resistance has been well documented in the petite-positive fungal pathogen Candida glabrata (reviewed in reference 64). However, the C. albicans sam37ΔΔ mutant was not less susceptible to fluconazole and even displayed mild hypersusceptibility, suggesting that the relationship between mtDNA instability and azole resistance might be different in petite-negative yeasts. It has to be noted, though, that lower azole susceptibility has been reported for a C. albicans mitochondrial mutant with uncoupled oxidative phosphorylation (15). Therefore, specific mitochondrial mutations can lead to different outcomes in terms of drug resistance in C. albicans, and further experiments are required to understand the molecular basis of the azole susceptibility phenotypes.
The link between mitochondrial function and cell wall integrity in fungi has been made mostly based on altered sensitivities of mitochondrial mutants to cell wall-targeting drugs (12, 14, 16, 30). Our result that the C. albicans sam37ΔΔ mutant displays altered cell wall structure is the first direct demonstration that in C. albicans a mitochondrial factor is necessary for cell wall integrity. The C. albicans sam37ΔΔ mutant displayed thicker cell walls and higher chitin levels, while the mannan:glucan ratio in the cell wall was not affected. Upregulation of cell wall chitin is a well-known compensatory response to cell wall defects (42), and changes to cell wall thickness are also common in cell wall mutants (for example, see references 44 and 58). Therefore, the changes that we observed in the structure and composition of walls derived from the sam37ΔΔ mutant support a role for Sam37 in cell wall integrity in C. albicans. To our knowledge, only one other mitochondrial mutant in any fungal species has been characterized in terms of the cell wall defect, the S. cerevisiae mutant deleted for the PGS1 phosphatidylglycerol synthase (71, 72). The S. cerevisiae pgs1Δ mutant displays lower levels of β-1,3-glucan in the wall and defective CWI pathway activation (71, 72). We note that the cell wall defects resulting from inactivation of SAM37 in C. albicans are different from those observed in the S. cerevisiae pgs1Δ cells, as cell wall glucans and PKC pathway activation are not inhibited in the C. albicans sam37ΔΔ mutant. Our previously published result with the S. cerevisiae sam37Δ mutant supports the C. albicans data shown here, in that PKC activation in response to cell wall stress was not affected (16). These very different cell wall phenotypes arising from inactivation of PGS1 or SAM37 suggest that mitochondria have complex roles affecting multiple pathways important for cell wall construction and the maintenance of cell wall integrity. What the primary cell wall defect is in the absence of Sam37 is not clear at this stage. Our data that glucan levels are not affected in walls derived from sam37ΔΔ cells argue against gross changes in glucose availability for cell wall construction due to metabolic changes in response to mitochondrial dysfunction. Sam37 is required for phosphatidylethanolamine (PE) biosynthesis in mitochondria, with PE being required for the synthesis of the glycosylphosphatidylinositol (GPI) anchor and therefore the maturation of GPI-anchored cell wall proteins (4, 16, 32). The levels of mannan in sam37ΔΔ mutant walls were not lower than in the wild type, arguing against gross changes to cell wall mannoprotein levels. However, it is still possible that more subtle changes to GPI-anchored proteins in the absence of Sam37 are at the basis of the observed cell wall defects, and we are currently developing quantitative proteomics approaches based on SILAC (stable isotope labeling by amino acids in cell culture) to address this question.
A final point that we would like to discuss is the roles of mitochondrial proteins in fitness of C. albicans and the possibility to explore mitochondrial factors as drug targets. C. albicans is a petite-negative yeast, as are other important fungal pathogens, such as Cryptococcus neoformans. This predicts that genes required for mitochondrial genome stability will be essential in these pathogens and could potentially represent antifungal drug targets. The factors important for mtDNA stability in S. cerevisiae are relatively well defined, including using a whole-genome screen (48), but in C. albicans this is far from being the case. In fact, in addition to SAM37, there is only one other gene with a described role in mtDNA maintenance, HMI1 (34). Our result showing that Sam37 affects mtDNA maintenance in C. albicans but not in S. cerevisiae underscores the importance of studying mitochondrial factors directly in pathogenic species. With eukaryotic human pathogens and drug targets, it is important to consider the issue of conservation of protein structure and function between fungi and humans. In this context, it is worth noting that several mitochondrial proteins do not have close homologs in animals (53). Here we investigated this for Sam37, as well as for the Sam35 subunit of the SAM complex, which is essential in all fungal species tested so far, and it could therefore be useful in the context of drug targets. Based on initial BLAST analysis, metaxin 1 was suggested to be the ortholog of Sam37 and metaxin 2 to be the ortholog of Sam35 (1, 40). By cluster analysis of a large group of sequences, CLANS analysis supports this relationship. While these proteins may share a common evolutionary origin, functional analysis suggests the metaxins to be quite different to Sam37 and Sam35: (i) metaxin 1 cannot complement the S. cerevisiae sam37Δ mutant (1), and metaxin 2 cannot complement the S. cerevisiae sam35Δ mutant (K. Vascotto, A. Perry, and T. Lithgow, unpublished data); and (ii) Sam37 and Sam35 in S. cerevisiae and Neurospora crassa form a stable complex with the β-barrel channel protein Sam50 (33, 41, 49, 69), and we have evidence that in C. albicans the Sam37 protein also functions in the SAM complex (V. L. Hewitt, A. Traven, and T. Lithgow, unpublished data). However, the metaxins are not in the same complex with the animal Sam50 protein (40). Collectively, these observations suggest differences in structure and/or function between the animal and fungal proteins. Such differences are borne out also by Pfam-based characterization of the domain structures of Sam35, Sam37, and the metaxins (Fig. 5B). Moreover, the very low sequence conservation between Sam35 and Sam37 and the human metaxins (<20%) predicts that the 3D structures of these proteins will be significantly different. While it will be necessary to determine the structure of the fungal Sam37 and Sam35 proteins and of the metaxins to continue exploring the possibility of inhibiting the SAM complex as a strategy against fungal pathogens, our functional and bioinformatic analyses, as well as the virulence experiments in the mouse model, indicate that Sam37 might be a good candidate for inhibition by antifungal drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jesus Plà and Elvira Román for providing antibodies against Mkc1 and their advice on the Western blot experiments, and Georg Ramm (Monash Microimaging) for expert advice and help with transmission electron microscopy. We further thank Kip Gabriel for comments on the manuscript, expert advice on experiments, and many discussions on the fungal SAM complex. We acknowledge the Fungal Genetic Stock Center for providing the C. albicans kinase mutant library from which the mkc1 mutant strain was obtained.
The work in A.T.'s laboratory on mitochondrial proteins in C. albicans is supported by a project grant from the National Health and Medical Research Council of Australia (NH&MRC). Y.Q. is supported by a Super Science fellowship from the Australian Research Council (ARC). Work of B.J. in A.T.'s laboratory was made possible by an Australian Group of Eight European fellowship. T.H.B. and T.L. are supported by ARC fellowships. F.B. is supported by a postgraduate scholarship from the Saudi Arabian Government. A.P. is an NH&MRC biomedical fellow. A.Y.P. is supported by grants and a fellowship from the NH&MRC.
Footnotes
Published ahead of print 27 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P. 1997. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 272:6510–6518 [DOI] [PubMed] [Google Scholar]
- 2. Bambach A, et al. 2009. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot. Cell 8:1706–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Becker JM, et al. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. U. S. A. 107:22044–22049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birner R, Burgermeister M, Schneiter R, Daum G. 2001. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boldogh IR, et al. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell 14:4618–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breger J, et al. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breslow DK, et al. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brun S, et al. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brun S, et al. 2005. Biological consequences of petite mutations in Candida glabrata. J. Antimicrob. Chemother. 56:307–314 [DOI] [PubMed] [Google Scholar]
- 11. Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chamilos G, Lewis RE, Kontoyiannis DP. 2006. Inhibition of Candida parapsilosis mitochondrial respiratory pathways enhances susceptibility to caspofungin. Antimicrob. Agents Chemother. 50:744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan NC, Lithgow T. 2008. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 19:126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen YL, et al. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol. Microbiol. 75:1112–1132 [DOI] [PubMed] [Google Scholar]
- 15. Cheng S, Clancy CJ, Nguyen KT, Clapp W, Nguyen MH. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51:1855–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dagley MJ, et al. 2011. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol. Microbiol. 79:968–989 [DOI] [PubMed] [Google Scholar]
- 17. Deutschbauer AM, et al. 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169:1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dolezal P, Likic V, Tachezy J, Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318 [DOI] [PubMed] [Google Scholar]
- 19. Doudican NA, Song B, Shadel GS, Doetsch PW. 2005. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:5196–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez-Arenas E, et al. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell. Proteomics 6:460–478 [DOI] [PubMed] [Google Scholar]
- 21. Frickey T, Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704 [DOI] [PubMed] [Google Scholar]
- 22. Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geraghty P, Kavanagh K. 2003. Disruption of mitochondrial function in Candida albicans leads to reduced cellular ergosterol levels and elevated growth in the presence of amphotericin B. Arch. Microbiol. 179:295–300 [DOI] [PubMed] [Google Scholar]
- 24. Geraghty P, Kavanagh K. 2003. Erythromycin, an inhibitor of mitoribosomal protein biosynthesis, alters the amphotericin B susceptibility of Candida albicans. J. Pharm. Pharmacol. 55:179–184 [DOI] [PubMed] [Google Scholar]
- 25. Gratzer S, et al. 1995. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 129:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gulshan K, Schmidt JA, Shahi P, Moye-Rowley WS. 2008. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 28:5851–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hallstrom TC, Moye-Rowley WS. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347–37356 [DOI] [PubMed] [Google Scholar]
- 28. Hanekamp T, et al. 2002. Maintenance of mitochondrial morphology is linked to maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Genetics 162:1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harner M, et al. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hillenmeyer ME, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Imhof I, Canivenc-Gansel E, Meyer U, Conzelmann A. 2000. Phosphatidylethanolamine is the donor of the phosphorylethanolamine linked to the alpha1,4-linked mannose of yeast GPI structures. Glycobiology 10:1271–1275 [DOI] [PubMed] [Google Scholar]
- 33. Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. 2004. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J. Cell Biol. 166:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joers P, Gerhold JM, Sedman T, Kuusk S, Sedman J. 2007. The helicase CaHmi1p is required for wild-type mitochondrial DNA organization in Candida albicans. FEMS Yeast Res. 7:118–130 [DOI] [PubMed] [Google Scholar]
- 35. Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaur R, Castano I, Cormack BP. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim SY, Kim J. 2010. Roles of dihydrolipoamide dehydrogenase Lpd1 in Candida albicans filamentation. Fungal Genet. Biol. 47:782–788 [DOI] [PubMed] [Google Scholar]
- 38. Kornmann B, et al. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kornmann B, Walter P. 2010. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 123:1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozjak-Pavlovic V, et al. 2007. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 8:576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lackey SW, Wideman JG, Kennedy EK, Go NE, Nargang FE. 2011. The Neurospora crassa TOB complex: analysis of the topology and function of Tob38 and Tob37. PLoS One 6:e25650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenardon MD, Munro CA, Gow NA. 2010. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 13:416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Likic VA, Dolezal P, Celik N, Dagley M, Lithgow T. 2010. Using hidden markov models to discover new protein transport machines. Methods Mol. Biol. 619:271–284 [DOI] [PubMed] [Google Scholar]
- 44. Martinez-Lopez R, Park H, Myers CL, Gil C, Filler SG. 2006. Candida albicans Ecm33p is important for normal cell wall architecture and interactions with host cells. Eukaryot. Cell 5:140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McDonough JA, Bhattacherjee V, Sadlon T, Hostetter MK. 2002. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Genet. Biol. 36:117–127 [DOI] [PubMed] [Google Scholar]
- 46. Meeusen S, Nunnari J. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meisinger C, et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26:2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merz S, Westermann B. 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milenkovic D, et al. 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279:22781–22785 [DOI] [PubMed] [Google Scholar]
- 50. Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nobile CJ, et al. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamoto K, Shaw JM. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39:503–536 [DOI] [PubMed] [Google Scholar]
- 54. Ortega M, et al. 2010. Candida spp. bloodstream infection: influence of antifungal treatment on outcome. J. Antimicrob. Chemother. 65:562–568 [DOI] [PubMed] [Google Scholar]
- 55. Peleg AY, et al. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 105:14585–14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Plaine A, et al. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 60. Roemer T, et al. 2011. Confronting the challenges of natural product-based antifungal discovery. Chem. Biol. 18:148–164 [DOI] [PubMed] [Google Scholar]
- 61. Roman E, Cottier F, Ernst JF, Pla J. 2009. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanglard D, Ischer F, Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shahi P, Moye-Rowley WS. 2009. Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 1794:852–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shingu-Vazquez M, Traven A. 2011. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot. Cell 10:1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. 2001. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276:4020–4027 [DOI] [PubMed] [Google Scholar]
- 67. Vandeputte P, et al. 2009. Hypersusceptibility to azole antifungals in a clinical isolate of Candida glabrata with reduced aerobic growth. Antimicrob. Agents Chemother. 53:3034–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vellucci VF, Gygax SE, Hostetter MK. 2007. Involvement of Candida albicans pyruvate dehydrogenase complex protein X (Pdx1) in filamentation. Fungal Genet. Biol. 44:979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Waizenegger T, et al. 2004. Tob38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria. EMBO Rep. 5:704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wiedemann N, et al. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424:565–571 [DOI] [PubMed] [Google Scholar]
- 71. Zhong Q, Gvozdenovic-Jeremic J, Webster P, Zhou J, Greenberg ML. 2005. Loss of function of KRE5 suppresses temperature sensitivity of mutants lacking mitochondrial anionic lipids. Mol. Biol. Cell 16:665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhong Q, Li G, Gvozdenovic-Jeremic J, Greenberg ML. 2007. Up-regulation of the cell integrity pathway in Saccharomyces cerevisiae suppresses temperature sensitivity of the pgs1Delta mutant. J. Biol. Chem. 282:15946–15953 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.