Abstract
The ampA gene plays a role in Dictyostelium discoideum cell migration. Loss of ampA function results in reduced ability of growing cells to migrate to folic acid and causes small plaques on bacterial lawns, while overexpression of AmpA results in a rapid-migration phenotype and correspondingly larger plaques than seen with wild-type cells. To help understand how the ampA gene functions, second-site suppressors were created by restriction enzyme-mediated integration (REMI) mutagenesis. These mutants were selected for their ability to reduce the large plaque size of the AmpA overexpresser strain. The lmbd2B gene was identified as a suppressor of an AmpA-overexpressing strain. The lmbd2B gene product belongs to the evolutionarily conserved LMBR1 protein family, some of whose known members are endocytic receptors associated with human diseases, such as anemia. In order to understand lmbd2B function, mRFP fusion proteins were created and lmbd2B knockout cell lines were established. Our findings indicate that the LMBD2B protein is found associated with endocytic cups. It colocalizes with proteins that play key roles in endocytic events and is localized to ruffles on the dorsal surfaces of growing cells. Vegetative lmbd2B-null cells display defects in cell migration. These cells have difficulty sensing the chemoattractant folic acid, as indicated by a decrease in their chemotactic index. lmbd2B-null cells also appear to have difficulty establishing a front/back orientation to facilitate migration. A role for lmbd2B in development is also suggested. Our research gives insight into the function of a previously uncharacterized branch of the LMBR1 family of proteins. We provide evidence of an LMBR1 family plasma membrane protein that associates with endocytic cups and plays a role in chemotaxis.
INTRODUCTION
Dictyostelium discoideum exists as a population of unicellular amoebas that feed on bacteria on the forest floor (4). When depleted of nutrients, they aggregate and begin multicellular differentiation (28). They develop into a fruiting body comprised of 3 cell types: spore cells, found in the head, or sorocarp; stalk cells that hold the sorocarp aloft; and anterior-like cells (ALCs). The ALCs are a specialized group of cells that undergo rapid movements during fruiting body formation. In the final fruiting body, they form the upper and lower cups and basal-disc support structures. Cell migration plays a major role in Dictyostelium growth and development. Growing cells migrate to find food, chemotaxing to folic acid produced by bacteria (44). Starving cells migrate by chemotaxis to secreted cyclic AMP (cAMP), forming multicellular aggregates. The developing cells then differentiate and move to their destined locations in the final fruiting body, movements reminiscent of embryo gastrulation (28). Investigating mechanisms of cell migration in Dictyostelium is simplified because of the ease of molecular manipulations due its haploid genome (7, 39, 40). Cell migration in Dictyostelium shares many features with higher systems and thus has implications for understanding cell migration in development and disease (14, 20, 26, 45, 53, 62).
A novel gene that plays a role in Dictyostelium migration is the ampA gene. During development, ampA plays a role in the migration of the ALCs to the upper cup of the fruiting body (57, 58). During growth, cells uniformly express the ampA gene as they reach high density (6). In vegetative cells, the loss of ampA activity by gene disruption results in the formation of much smaller plaques on bacterial lawns than are seen with wild-type (WT) cells (3; E. Noratel, C. Petty, Y. Zhang, and D. Blumberg, unpublished data). Extensive cell clumping and increased cell substrate adhesion is also detected in ampA-null cells (3; Noratel, Petty, Zhang, and Blumberg, unpublished). Overexpression of the ampA gene in growing cells has exactly the opposite phenotype. Cells make much larger plaques on lawns of bacteria than wild-type cells, and they show reduced cell-cell and cell-substrate adhesion. Recent work also indicates that ampA influences the level of actin polymerization, reducing it in knockout cells and increasing it above wild-type levels in overexpressing cells (Noratel, Petty, Zhang, and Blumberg, unpublished). Together, these defects result in significant effects on cell migration in response to the growth phase chemoattractant folic acid.
To gain understanding of the molecular mechanisms by which ampA influences cell migration, second-site suppressor screens were undertaken. Three suppressors of the increased cell migration observed in ampA overexpressers were identified. One of these disrupted the lmbd2B gene, whose product is a member of the LMBR1 protein family.
LMBR1-like proteins are a family (pfam PF04791) of integral membrane proteins, with an average size of 500 amino acid residues. They were originally named LMBR proteins, which stands for limb receptor proteins, because mutations in the lmbr1 gene resulted in mouse and human polydactyly, a limb defect (8). It was later determined that the disruption of lmbr1 is not responsible for the phenotype, but rather, the mutation in the lmbr1 gene actually disrupted a cis-acting regulator of Shh (32). Members of the LMBR1-like family of proteins in mammals include limb region 1 (LMBR1), lipocalin 1 receptor (LIMR), and two LMBR1-like proteins (LMBD1 and LMBD2) (19). There is also an LMBR2-like protein class, whose only known member is a RING finger protein (56).
Lipocalins are highly diverse but structurally similar small, soluble proteins (17, 27). They bind and transport hydrophobic molecules through cell membranes via their lipocalin receptors. Lipocalins are involved in an array of processes, such as apoptosis, metabolism, and animal behaviors (65). While some research has been devoted to the role of lipocalins, there is exceedingly little information and research on lipocalin receptors. One identified lipocalin receptor (the lipocalin 1 receptor, also referred to as LIMR) is a member of the LMBR1 family (64, 65). Lipocalin 1 (Lcn-1) is thought to act as a scavenger, responsible for the endocytosis of potentially harmful lipophilic compounds. It has been shown to bind fatty acids, cholesterol, retinol, glycolipids, and phospholipids (19, 65). In addition to Lcn-1, LIMR (the lipocalin 1 receptor) has been shown to bind other molecules similar to Lcn-1, such as β-lactoglobulin and uteroglobin. The binding of these molecules to LIMR results in antichemotatic effects in macrophages and anti-invasiveness in tumor cells (18, 33, 66). Because lipocalins and their receptors specialize in the transport of lipophilic molecules, they have huge potential in research on drug delivery and regulation of toxic compounds. Implications for cancer cell migration make the receptor proteins even more interesting.
Another group of members of the LMBR1-like family is the LMBD1 proteins. These proteins are putative cobalamin (vitamin B12) transporters (19, 48). Mammalian lmbd1 was found to encode a nine-span transmembrane protein that colocalizes with lysosomal membrane markers. Mutations in lmbd1 result in defects in cobalamin metabolism, with an increased accumulation of cobalamin apparent in lysosomes (19). These results suggest a role of LMBD1 as a lysosomal receptor responsible for the binding and shuttling of cobalamin. Understanding LMBD1 function is valuable, as defects in the cobalamin transporter can lead to a number of disorders, such as anemia and neurological and cognitive impairment (19, 48, 49).
The LMBD2 proteins, which include the suppressor of the ampA overexpression increased-migration phenotype, which we describe here, have not been characterized. Our data suggest the Dictyostelium lmbd2B gene encodes a plasma membrane protein that associates with endocytic cups. It appears to be associated, at least in part, with macropinocytosis events. It also plays a role in cell migration. lmbdB2 knockouts have defects in migration toward a chemoattractant and difficulty with proper orientation and pseudopod extension, explaining its role as a suppressor of the ampA overexpresser increased-migration phenotype. Our characterization of lmbd2B agrees with a role in endocytosis for LMBR1 domain-containing proteins and presents additional evidence of an LMBR1 domain-containing membrane protein affecting cell migration.
MATERIALS AND METHODS
Dictyostelium cell growth and development.
WT (AX3), ampAOE (G418-resistant), and ampA− cell lines were described by Varney et al. and Sussman and grown under standard conditions (54, 57, 58). The ampA− cell line was generated by a blastcidin resistance (bsr) cassette insertion into the ampA gene (57, 58). This insertion removed 500 bp of the ampA coding sequence. The blasticidin cassette was subsequently removed via a cre-loxP system, leaving the 500-bp deletion of ampA coding sequence followed by 3 in-frame termination codons (29). The transformed cell lines were grown in either 9.6 μg/ml G418 for G418/geneticin selection or 10 μg/ml blasticidin S for blasticidin resistance selection. For growth over bacteria, a concentration of Dictyostelium cells was plated with Escherichia coli B/r on LP agar plates (43). Dictyostelium cells were starved on filters to induce their multicellular development (15).
REMI mutagenesis.
Restriction enzyme-mediated integration (REMI) mutagenesis was adapted from Shaulsky et al. and Kuspa (31, 51). The plasmid used for integration was a pGEM3 plasmid (Promega) that has a 1,488-bp bsr with a Dictyostelium actin 15 promoter and an actin 8 terminator sequence. The cassette was added at an SmaI site in pGEM3. The original bsr gene cassette was from the plasmid pbsr519 provided by Frantisek Puta, Charles University, Czech Republic (47). The plasmid was linearized by BamHI. For REMI mutagenesis, ampAOE Dictyostelium cells were grown to a density of 2 × 106 to 4 × 106 cells/ml; cells were harvested and resuspended at a concentration of 1 × 107 cells/ml in cold electroporation (EP) buffer (10 mM NaPO4, 10 mM sucrose, pH 6.1), and 1 to 10 μg of linearized pGEM3-bsr plasmid was added to the cells, along with 10 units/ml of the restriction enzyme DpnII (New England BioLabs). The cells were electroporated at 1.0 kV, 3μF, and 200 Ω for 2 pulses with a Bio-Rad electroporator. The cells were then placed into HL5 medium (54), and after a 24-hour recovery period, 10 μg/ml blasticidin was added. Dead cells were removed daily with medium changes until colonies were visible (between 3 and 5 days of selection). Blasticidin-resistant colonies were plated with E. coli B/r on LP plates to screen for plaque sizes and to obtain individual clones (43).
To sequence and identify the disrupted gene, 1 μg of DNA from the REMI mutant was digested with ClaI (a restriction enzyme that cuts outside the REMI plasmid and in flanking Dictyostelium DNA). The enzyme was then deactivated, and the digested DNA product was ligated together. The ligation products were precipitated with 100% ethanol and dissolved in distilled water (dH2O). Approximately 2 to 6 μl was used for the subsequent Sure cell transformation (Stratagene; 200238). To determine the location of the REMI plasmid insert, internal pGEM3 primers were used to sequence out into the flanking genomic DNA. The primers for sequencing flanking Dictyostelium DNA were Sp6 primer, 5′ AGATGATAGGGTCTGCTTCAGTAAG 3′, and Bsr primer, 5′ TTCAAATAATAATTAACCAACCCAAG 3′ (see Fig. S1A in the supplemental material).
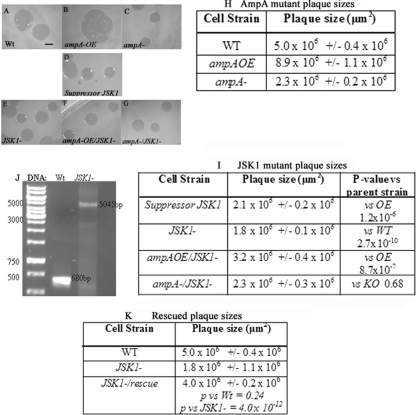
To verify insertion of the REMI plasmid into the DDB_G0281669/lmbd2B gene, the primers 5′ CAGGGATCCAATGTCAAGTAATACTACAACACC 3′ and 5′ CAGGAGCTCACACTTAGAATCACAATGATACCA 3′ were used in PCRs with genomic DNA. These primers flank the expected place of insertion in the gene. They produced a 680-bp fragment in wild-type cells and a 5,045-bp fragment when the pGEM3-BLAST REMI plasmid insert was present (Fig. 1J).
Fig 1.
Plaque sizes of potential suppressors of the ampA overexpresser phenotype. Cells were spread on LP plates with E. coli B/r and incubated at 22°C for 4 days. (A to I) Plaques were photographed on a dissecting scope (A to G), and the areas of the plaques were estimated using Metamorph (H and I). The cell types are indicated, as well as P values with the cell types tested against. The scale bar represents 1,000 μm. n > 17 plaques from at least 3 platings. (J) PCR results showing integration of the plasmid into the gene in JSK1− cells. Primers hybridized to sites flanking the REMI insert in the JSK1 gene. WT cells produce a 680-bp fragment. JSK1− cells produce a 5,045-bp fragment (4,365-bp pGEM3 plus 680-bp sequence). (K) Plaque sizes of the rescued JSK1− cells (by expressing an extrachromosomal copy of the gene in JSK1− cells).
Dictyostelium DNA preparation.
The Dictyostelium DNA preparation procedure was done according to the method of Nellen et al. (38).
Plasmid construction.
The original REMI plasmid with flanking lmbd2B genomic DNA linearized with BamHI was used to create knockout strains in WT, ampAOE, and ampA− backgrounds.
The LMBD2B-mRFP plasmid (see Fig. S3A in the supplemental material) contains 1,572 bp of the 3′ end of the LMBD2B coding region and 1,156 bp of 3′ LMBD2B noncoding region sequence immediately downstream of the LMBD2B termination site for promoting homologous recombination. It was designed with a proline-alanine linker (32 bp) directly following the last amino acid in the coding region of the LMBD2B gene. The linker is followed in frame by the mRFPmars coding sequence at the carboxy terminus of the protein (37). Following the mRFPmars coding region is a 270-bp terminator sequence. The plasmid also contains the floxed bsr cassette for selection (29). For 3′ coding region amplification and linker addition, the PCR primers used were 5′ CAGGGGCCCGCAGGTCAATACGACCCATTTAGAAT 3′ and 5′ CAGGTCGACCCTGCACCTGCACCTGCACCTGCACCTGCACCTTTTTATTTTTACGGCCAGTTCC 3′. The primers produce a 1,604-bp product containing the 3′ coding region plus the linker.
For the 3′ noncoding region, the primers used were 5′ CAGGCGGCCGCTCACCATTTAAGGAGATCAAAAGTGG 3′ and 5′ CAGCCGCGGAAGGTAATTTCAAGGTTGCCATTG 3′. The primers produce a 1,156-bp fragment. The plasmid was linearized with ApaI and SacII prior to electroporation. Electroporation into cells was done as described above for REMI mutagenesis. The primers used to verify insertion into the lmbd2B gene were 5′ AAAGGGTGCAGGTGCAGGTG 3′ and 5′ ACACTTACCAACAACCAGTACC 3′. The primers produce a 2,700-bp product in WT DNA and a 5,100-bp product with the lmbd2B-mRFP construct insertion.
An LMBD2B rescue plasmid was constructed to include the entire coding region of the lmbd2B gene. It contains the full-length lmbd2B cDNA inserted into pTIKL-MyD at XbaI-SacI restriction sites (13, 16). The insertion places the lmbd2B cDNA fused to the actin15 promoter and confers G418 resistance. The primers used were 5′ CAGGAGCTCATGTCAAGTAATACTACAACACC 3′ and 5′ CAGTCTAGATTACTTTTTATTTTTACGGCCAGTTC 3′.
Immunodetection.
For Western analysis, protein from 1 × 106 cells was run on 6% or 12% SDS-PAGE and transferred to nitrocellulose. A 1:500 dilution of red fluorescent protein (RFP) tag antibody (Ab) (Chromotek; 5F8 anti-Rat; 090428) and a 1:3,000 dilution of anti-rat alkaline phosphatase-conjugated antibody (S383A; Promega) were used as primary and secondary antibodies, respectively.
For indirect immunofluorescence, cells were fixed in 4% formaldehyde in 20 mM NaPO4 and permeabilized in methanol with 1% formaldehyde. Primary antibodies were diluted in 1% bovine serum albumin (BSA) at 1:300 for anti-RFP, 1:300 for anti-clathrin heavy chain (rabbit Ab P1663; Cell Signaling), and 1:50 for anti-coronin (mouse Ab 176-2-5; Hybridoma Bank [11]). For staining with DiI (Vybrant CM-DiI; Molecular Probes), cells were incubated in a 1:500 dilution for 5 min prior to fixation. In the case of dextran endocytosis, cells were incubated in HL5 plus fluorescein isothiocyanate (FITC)-dextran (Sigma; molecular weight, 20,000) at 2 mg/ml for 1 h prior to fixation (5). The secondary antibodies used were 1:200 dilutions of goat anti-rat, donkey anti-rabbit, and goat anti-mouse, 488 or 594 Alexa Fluor conjugated (Molecular Probes). It was necessary to use indirect immunofluorescence microscopy with fixed cells to detect the LMBD2B-mRFP fusion protein in Dictyostelium cells because the mRFP fluorescence was too weak to detect with laser power that did not kill the cells.
For actin staining, cells were fixed in 0.3% glutaraldehyde and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS), pH 7.4. Where utilized, application of primary antibodies was followed by incubation with secondary antibodies. Alexa Fluor 488-phalloidin (Molecular Probes) at a 1:500 dilution was included in the secondary incubation to label F-actin. If applicable, Alexa Fluor 594-conjugated DNase I (Molecular Probes) was added along with the phalloidin to label unpolymerized G-actin.
Confocal microscopy.
A Leica SP5 scanning confocal light microscope was utilized for imaging. For FITC- and Alexa Fluor 488-conjugated antibodies, the argon laser line 488 nm was used for excitation (20%) at an emission bandwidth of 500 to 550 nm. For 594 Alexa-conjugated antibody imaging, excitation was done with the DPSS laser line 561 nm (15%) and HeNe laser line 594 nm (30%), and emission was detected in a range of 600 nm to 767 nm. In DiI staining, the DPSS laser line 561 nm (30%) was used for excitation, and an emission range of 604 to 767 nm was used. For DAPI (4′,6-diamidino-2-phenylindole) staining and visualization of nuclei, the stain was excited with the diode laser line 405 nm (8%) and emission was recorded at 430 to 477 nm. Volocity software 5.5 (Perkin Elmer) was used to assemble images and calculate Pearson's coefficient of correlation (1).
Video microscopy (for cell migration and plaque sizes).
Agar plates were viewed under an Olympus dissecting scope (093467), and images were obtained using a DC330 video camera (DAGE-MTI, Inc., Michigan City, IN). Images used for calculating plaque sizes were processed and analyzed using the Metamorph Imaging series version 7.0r1 (Universal Imaging, West Chester, PA; Molecular Devices Corporation). For time lapse experiments, cells were imaged on the Leica SP5 confocal microscope with a low laser setting (5% argon laser line 488 nm) using bright-field images with either a 40× 0.55 numerical aperture (NA) long working objective or a 63× 1.4 NA oil objective. Slices of cells were captured at 20-s intervals for 5 min, and the images were processed with Volocity or Metamorph software.
Cell fractionation.
Fractionation was done using the Thermo Scientific subcellular protein fractionation kit according to the manufacturer's instructions. This involved pelleting 5 × 106 cells in the logarithmic growth phase and resuspending them in a cytoplasmic extraction buffer. The cells were then centrifuged at 500 × g, and the supernatant (cytoplasmic extract) was retrieved. The remaining pellet was then resuspended in the membrane extraction buffer, vortexed, and spun at 3,000 × g. The supernatant (membrane extract) was removed, and nuclear extraction buffer was added to the pellet. After vortexing and a 30-min incubation, the solution was centrifuged at 5,000 × g, and the supernatant (soluble nuclear extract) was retrieved. A chromatin-bound extraction buffer with micrococcal nuclease was added to the pellet and centrifuged at 16,000 × g. The supernatant (chromatin-bound nuclear extract) was separated, and the pellet was resuspended in the pellet extraction buffer. After centrifugation at 16,000 × g, the supernatant containing the cytoskeletal extract was removed. All buffers contained protease inhibitors. Fractions were run on SDS-PAGE, transferred, and immunoblotted as described above. The RFP antibody signal was used to indicate LMBD2B localization. The controls used were a strain containing N-golvesin green fluorescent protein (GFP) (16, 50), the fimbrin antibody 210-183-1 (Hybridoma Bank [46]), and the histone location as detected by Coomassie staining.
Chemotaxis.
The chemotaxis procedure was adapted from Hadwiger and Srinivasan (23). Briefly, cells were washed and resuspended to 2 × 107 cells/ml in starvation buffer (20 mM NaKPO4). For cAMP chemotaxis, cells were starved for about 6 h at 22°C on filters; for folic acid chemotaxis, the cells could be used immediately (63). Agar (0.8%) was thinly spread in a chambered cover glass. A small drop of folic acid (1 mM) or cAMP (1 mM) was placed on the agar, and a drop of cells was placed less than 3 mm away. After 3 h, the chemotaxis was imaged. Cell pictures were taken at 20-s intervals for 5 min as described for video microscopy above.
Cell movement and shape were analyzed using DIAS (61) and ImageJ (W. S. Rasband, U.S. National Institutes of Health, Bethesda, Maryland, 1997 to 2011 [http://imagej.nih.gov/ij/]) software. Velocity and directionality values were obtained using the manual tracking (F. Cordelieres, Institute Curie, Orsay, France, 2004 [http://rsb.info.nih.gov/ij/plugins/track/]) and chemotaxis (G. Trapp and E. Horn, Munich, Germany [http://www.ibidi.de/applications/ap_chemo.html]) tools on ImageJ. The chemotactic index (CI) is equal to the cosine of the angle formed between the line of movement of the cell and the line representing direct movement to the chemoattractant (25). The slope of individual cell movements (from ImageJ) was used to help determine the angle and CI value. Using the slope, the angle from the origin could be calculated by taking the antitangent of the value. By subtracting that angle from the angle of placement of the chemoattractant, the angle difference between the two paths is established. Taking the cosine of the angle of difference gives the chemotactic index.
RNA extraction and RT-PCR.
RNA from 1 × 107 cells was isolated according to the TRIzol Reagent protocol (Invitrogen; 15596-018). cDNA was created from the RNA using the reverse transcription (RT)-PCR procedure and a first-strand cDNA synthesis kit (Fermentas). PCR amplification off the first-strand cDNA was done using a standard Taq (Fermentas) reaction procedure and primers designed for the specific amplification. The lmbd2B primers used in RT-PCR were the 3′ coding primers used in mRFP plasmid construction, 5′ GCAGGTCAATACGACCCATTTAGAAT 3′ and 5′ TTTTTATTTTTACGGCCAGTTCC 3′. As a control, Ig7 primers were used: 5′ TTACATTTATTAGACCCGAAACCAAGCG 3′ and 5′ TTCCCTTTAGACCTATGGACCTTAGCG 3′, which yield a 370-bp fragment (24). A Gel Doc digital imaging system (Alpha Innotech Corporation; IS-100) was used to quantify the band intensity under conditions where intensity was linearly dependent on loading. Unequal protein loading or unequal RNA levels were taken into account with the Coomassie and Ig7 controls. Then, the intensities of the bands relative to one another were calculated, with the most intense band equal to 100%.
Statistical analysis.
P values were calculated using the paired two-sample test on Excel. A P value of 0.05 or less was deemed significant.
RESULTS
Isolation of mutants that are candidates for suppression of an AmpA-overexpressing increased-migration phenotype.
REMI mutagenesis of a bacterial plasmid carrying a blasticidin resistance cassette was used to generate insertional mutations in an ampAOE strain (31). Overexpression of AmpA results in larger than normal plaques when individual Dictyostelium amoebas are clonally plated on a lawn of bacteria (compare Fig. 1B, ampAOE, to Fig. 1A, WT, as quantified in Fig. 1H). These large plaques are the result of an increase in the migration rate by cells that overexpress AmpA protein (E. Noratel, C. Petty, J. Kelsey, and D. Blumberg, unpublished data). The mutagenized cells were clonally plated on bacterial lawns and compared with WT, AmpAOE, and AmpA− strains. Out of 6,500 insertional (blasticidin-resistant) mutants, 6 were found to reproducibly reduce the size of the ampAOE strain plaques. The mutant plaques ranged in size from the smaller WT to the very small ampA− plaques. For three of these (JSK1, -2, and -3), we were able to recover the inserted plasmid and flanking Dictyostelium DNA. This report focuses on only one (JSK1) of these potential suppressors of the ampAOE phenotype. The original JSK1 mutant produced by the suppressor screen inhibited the large ampAOE plaques (Fig. 1D and I). Sequencing of the Dictyostelium DNA flanking the inserted plasmid indicated that the plasmid had inserted into a DpnII site at the start of the predicted second exon of a previously uncharacterized gene (DDB_G0281669). The insertion disrupted the coding sequence of the predicted protein between the 94th and 95th amino acids (see Fig. S1B in the supplemental material).
In order to confirm that the disruption of this particular gene was responsible for suppressing the large-plaque phenotype of the ampAOE strain, the recovered plasmid and flanking DNA were used to generate new knockout mutants in the ampAOE strain and in the WT and ampA− strains. Not only does a disruption of DDB_G0281669 in ampAOE cells reproduce a small-plaque phenotype (Fig. 1F), its disruption in WT cells also causes small plaques (Fig. 1E, JSK1−).
Disrupting DDB_G0281669 in an ampA− background to generate a double-knockout mutant (ampA− JSK1−) resulted in plaques that were no smaller than those generated by either single mutation alone in a WT background. This suggests that there is no additive effect of the two null mutants together (Fig. 1C and G, ampA− and ampA− JSK1). If the mutations were in different pathways affecting plaque size, one might expect the double mutant to produce a smaller plaque than either single mutant. This was not the case, which suggests the possibility that the JSK1 mutant may indeed disrupt a gene in the ampA pathway.
Disruption of the DDB_G0281669 gene by the REMI plasmid was confirmed by PCR indicating that the plasmid was inserted into the gene coding region (Fig. 1J). Also, RT-PCR results confirmed disruption, as the knockout strain does not produce lmbd2B gene transcripts (see Fig. S1C in the supplemental material). To further ensure the phenotypes observed in lmbd2B− cells are a result of a knockout in LMBD2B function, a rescue plasmid was expressed in the lmbd2B− cells. The expression of functional LMBD2B should be able to rescue the lmbd2B− phenotypes. Rescue constructs containing the lmbd2B coding region were constructed (see Fig. S7 in the supplemental material). Expression of the full-length lmbd2B cDNA in lmbd2B− cells rescued the small-plaque phenotype (Fig. 1K).
The JSK1 suppressor protein was identified as a member of the LMBR1 family of proteins.
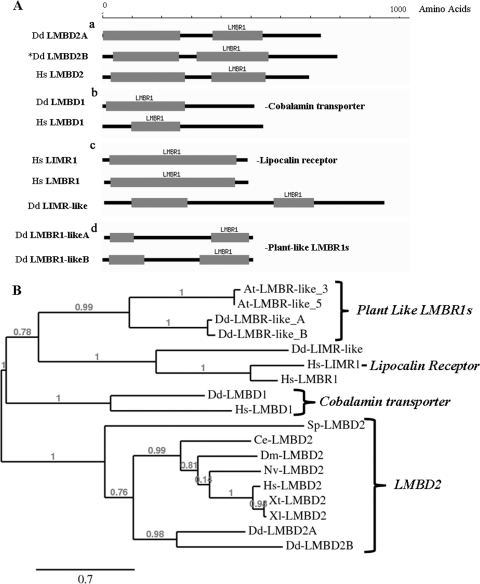
The sequence of the DDB_G0281669 gene product indicates that it is an uncharacterized member of the LMBR1 family of nine-span membrane receptor proteins (34, 35, 36). The proteins in the LMBR1 family are comprised of 4 general types grouped according to the number, length, and organization of their LMBR1 domains, as well as homology and function (Fig. 2A). A search of the D. discoideum genome revealed 6 genes that encode various members of the LMBR1 family of nine-span membrane proteins. The organization of the LMBR1 domains of these 6 proteins is depicted in Fig. 2A and is compared to the Homo sapiens LMBR1 proteins.
Fig 2.
Domain structure and phylogenetic analysis of LMBR1 domain-containing proteins. (A) LMBR1 domain-containing proteins are displayed, with their LMBR1 domains represented by gray blocks. The scale shows the length in amino acids. (a) LMBR1-containing type 2 proteins (LMBD2): Dd-LMBD2A (sp|Q54Q92 D. discoideum), Dd-LMBD2B (sp|Q54TM2 D. discoideum), and Hs-LMBD2 (sp|Q68DH5 H. sapiens). (b) LMBR1-containing type 1 proteins that are probable cobalamin transporters: Dd-LMBD1 (sp|Q54KD1 D. discoideum) and Hs-LMBD1 (sp|Q9NUN5 H. sapiens). (c) The lipocalin 1 receptor: Hs-LIMR1 (sp|Q6UX01 H. sapiens) and the limb region 1 protein homologue, Hs-LMBR1 (sp|Q8WVP7 H. sapiens), as well as Dd-LIMR-like (XP_645368.1 D. discoideum). (d) The two plant-like LMBR1 proteins: Dd-LMBR1-likeA (sp|Q54BI3 D. discoideum) and Dd-LMBR1-likeB (sp|Q54QP7 D. discoideum) (34, 35, 36). (B) Phylogenetic analysis of LMBR1 domain-containing proteins across species using Muscle v3.7 on the phylogeny.fr platform. The phylogenetic tree was constructed using the maximum- likelihood method implemented in the PhyML program v3.0 aLRT and was drawn using TreeDyn v198.3 (12). Sequences utilized included the Dictyostelium and human protein sequences from panel A, as well as the protein sequences for Xt-LMBD2 (NP_001123692 X. tropicalis), Xl-LMBD2 (NP_001080584 X. laevis), Nv-LMBD2 (XP_001637236 N. vectensis), At-LMBR1-like 3 (sp|Q9SR93 Arabidopsis thaliana), At-LMBR1-like 5 (sp|Q9M028 A. thaliana), Dm-LMBD2 (gb|AAF54372 D. melanogaster), Ce-LMBD2 (NP_496413 C. elegans), and Sp-LMBD2 (NP_588183 S. pombe).
The LMBD2 proteins.
The JSK1 protein represents the LMBR1 domain 2 (LMBD2) proteins. These proteins are larger than the other LMBR1 members, contain 2 LMBR1 domains, and have largely unknown functions (Fig. 2A, a). They appear to form a separate branch of the phylogenic tree in Fig. 2B. The JSK1 suppressor protein (DDB_G0281669 gene product), which we have named LMBD2B, shares 49% similarity with the human LMBD2 protein and 53% similarity with a second Dictyostelium homologue of the human LMBD2 protein (LMBD2A). In the phylogenetic tree (Fig. 2B), Dd-LMBD2A and -B cluster on a branch of the tree with the other LMBD2 domain proteins from H. sapiens, Xenopus tropicalis and Xenopus laevis, Nematostella vectensis, Drosophila melanogaster, Caenorhabditis elegans, and Schizosaccharomyces pombe (an alignment is shown in Fig. S2 in the supplemental material).
The LMBD1 proteins.
The LMBR1 domain proteins that contain only 1 LMBR1 domain fall into two groups. One group includes the lysosomal cobalamin transporters, which are named the LMBD1 proteins (Fig. 2A, b). Dictyostelium contains one LMBD1 protein with significant homology to the human cobalamin transporter.
The LIMR/LMBR proteins.
The other group with only one LMBR1 domain is named for its major member, the lipocalin 1 receptor (LIMR, for lipocalin-interacting membrane receptor). It contains 1 long LMBR1 domain (Fig. 2A, c). The human LMBR1, the original defining member of the LMBR1 family of proteins (which was originally identified as being involved in limb defects), is very closely related to LIMR. Dictyostelium has a highly diverged homologue of the LIMR group. It contains 2 LMBR1 domains and in the phylogenetic tree is located in the cluster with the LIMR/LMBR1 proteins, so we refer to it as LIMR-like.
The LMBR1-like proteins.
There is another group of LMBR1 proteins that contain 2 LMBR1 domains but are smaller than the other LMBD2 proteins (Fig. 2A, d). They appear to have split off from the LMBD1 and LIMR proteins and appear to be more closely related to the LIMR/LMBR1 proteins than to the LMBD2 proteins (Fig. 2B). These LMBR1 proteins are found mostly in the genomes of plants and ciliates, and we refer to them as LMBR1-like. Interestingly, Dictyostelium contains 2 LMBR1 proteins that most closely resemble the plant and ciliate LMBR1-like proteins. We have called these proteins LMBR1-likeA and -B.
Thus, Dictyostelium appears to have representatives of the 4 distinct classes of LMBR1 proteins. The gene responsible for the JSK1 mutation (LMBD2B) is clearly a member of the LMBD2 class. Since the LMBD2 members of the LMBR1 protein family have largely unknown functions, the fact that LMBD2B has a distinct phenotype when knocked out suggests that it may prove useful in elucidating some of the functions of this family of putative nine-span membrane receptors.
The LMBD2B protein in Dictyostelium is located in the plasma membrane and is associated with sites of endocytosis.
To visualize the localization of the LMBD2B protein, an lmbd2B-mRFP fusion construct was generated and knocked into WT cells to generate a gene replacement (see Fig. S3A in the supplemental material). Genomic PCR analysis confirmed that the WT lmbd2B gene had been replaced with the larger lmbd2B-mRFP fusion and that the expected 116-kDa fusion protein was produced (an 89-kDa LMBD2B plus a 27-kDa mRFP) (see Fig. S3B and C in the supplemental material). The LMBD2B-mRFP fusion protein gene replacement strain showed near-wild-type function. It made larger plaques than the lmbd2B− strain that were similar in size to the WT plaques, and it displayed normal development where the lmbd2B− strain shows a significant developmental delay (see below and Fig. S3D and E in the supplemental material).
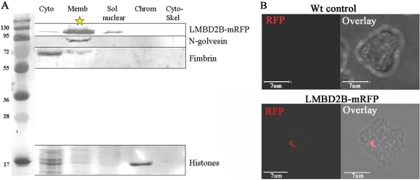
Cell fractionation using the lmbd2B-mRFP gene replacement strain indicates that LMBD2B is largely localized to the membrane fraction (Fig. 3A). In nonpermeabilized cells, where the antibody cannot penetrate the cell membrane, the LMBD2B-mRFP fusion protein is accessible to the antibody and gives a strong localized signal (Fig. 3B). This argues that the protein is localized to the plasma membrane. Since the mRFP protein is at the carboxy terminus, it also suggests that the carboxy terminus is extracellular. This suggestion is borne out by the predicted topography of the LMBD2B protein (see Fig. S6 in the supplemental material). The Dictyostelium LMBD2B protein is predicted to have its C terminus on the extracellular side of the plasma membrane (9, 59).
Fig 3.
The LMBD2B protein is localized to the plasma membrane. (A) Cell fractionation using the Thermo Scientific subcellular protein fractionation kit. The fractions were run on SDS-PAGE, transferred, and probed with an appropriate antibody. A molecular mass ladder (kDa) is in the far left lane, followed by fractions: cytosolic (Cyto), membrane (Memb), soluble (Sol) nuclear, chromatin bound (Chrom), and cytoskeletal (Cyto-Skel). An RFP antibody was used to detect LMBD2B-mRFP. To detect Golgi and Golgi-derived vesicle locations, GFP–N-golvesin (91 kDa) and a GFP antibody were used. Localization of the soluble actin binding protein fimbrin is shown, as well (fimbrin antibody, 67 kDa). The Coomassie staining showed histone localization to the chromatin fraction (∼17 kDa). The 116-kDa LMBD2B-mRFP protein was mostly localized to the membrane fraction. (B) Indirect immunofluorescence using the RFP antibody is shown for both WT control cells and LMBD2B-mRFP cells. The WT control is a 3D flattened image displaying little or no RFP fluorescence in the cell. Below is shown immunofluorescent LMBD2B localization in a nonpermeabilized cell. Growing cells were placed on a coverslip for 2 h, fixed, and stained (as described in Materials and Methods, with the methanol step of permeabilization omitted). A dorsally shifted Z-axis slice (a slice through the top of the cell) is displayed. Overlays of fluorescence on transmitted images are also shown. LMBD2B fluorescence is indicated by the red signal. LMBD2B was detected on the dorsal surface of the cell.
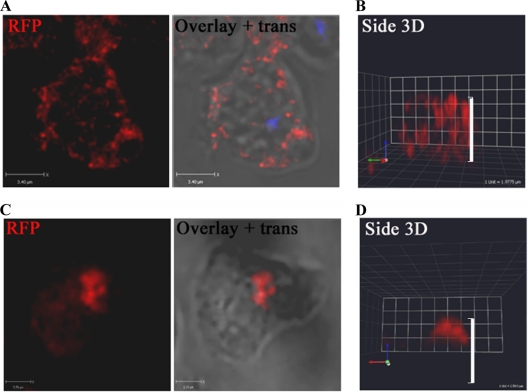
In fixed and permeabilized vegetative cells, LMBD2B-mRFP staining is seen largely at the cell periphery. The longer a cell was in contact with a substrate before fixing, the more the protein appeared to localize to patches on the dorsal side, or top, of the cell (Fig. 4). The dorsal side, or top, of the cell is considered to be the side opposite the side in contact with the substratum. Vegetative Dictyostelium cells were grown in suspension shaking culture and then placed on coverslips. After 15 min or less of substrate contact, LMBD2B was seen throughout the cell periphery, and a slice through the center of the cell displayed punctate red spots of LMBD2B around the edge of the cell (Fig. 4A). In the side view of the cell sitting for 15 min (Fig. 4B), the red signal is spread from the base of the cell to the top. This dispersed expression of LMBD2B changed with prolonged contact with a substrate. Figure 4C shows a slice toward the top of a cell that had been sitting for 2 h. A more concentrated location of LMBD2B was detected. The concentrated LMBD2B is often seen located toward the dorsal side of the cell. This is depicted in Fig. 4D, which shows a side view of a three-dimensional (3D) reconstruction of a cell that had prolonged substrate contact. A concentrated signal toward the top of the cell is observed.
Fig 4.
LMBD2B is localized in punctate regions on the cell periphery but clusters with prolonged substrate contact. Growing cells were placed on a coverslip for 15 min (A and B) or >2 h (C and D), fixed, and stained. Images of LMBD2B-mRFP indirect immunofluorescence are shown. Overlays of fluorescence over transmitted (trans) images are also shown. LMBD2B fluorescence is indicated by the red signal. (A and C) Z-axis slices through a cell (blue signal indicates nuclei stained with DAPI). (B and D) Side views of 3D-reconstructed cells; the white side bars indicate cell height.
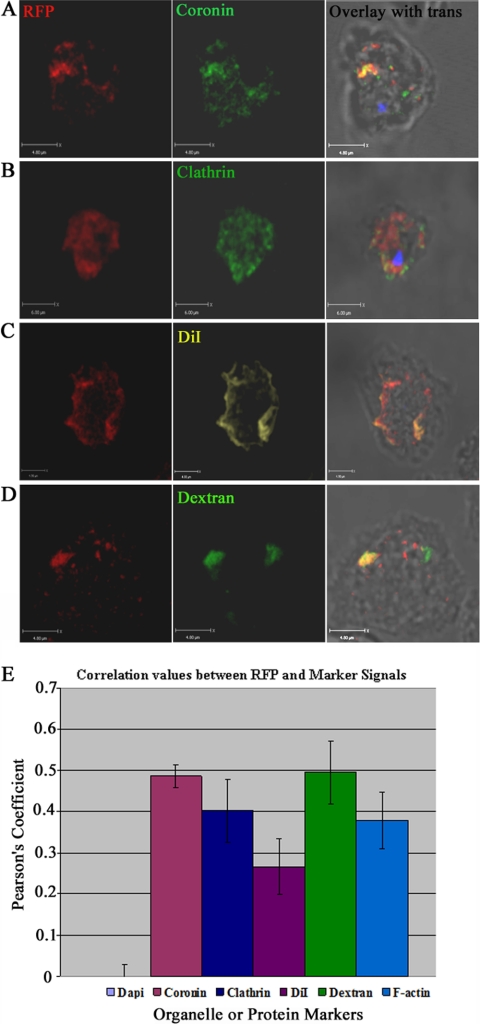
Based on the functions of known LMBR1 domain-containing proteins as endocytic receptors, it seemed possible that LMBD2B was involved in endocytosis. Accordingly, protein localization of LMBD2B with endocytic components was investigated. Coronin is an actin binding protein involved in endocytosis and vesicle trafficking (55). Definite overlap was detected, as displayed in Fig. 5A, with a positive correlation coefficient, indicating that the colocalization was significant (Fig. 5E). Another protein involved in endocytosis is clathrin. Clathrin is essential for substrate internalization and vesicle formation (41). A portion of LMBD2B punctate spots colocalized with clathrin vesicles (Fig. 5B), and a positive correlation was also detected (Fig. 5E). DiI is a membrane marker and in cells at room temperature is often endocytosed (15). The image in Fig. 5C shows an upper slice through a cell (notice there is no nuclear staining), where any membrane labeled with DiI likely represents the plasma membrane. There is a large amount of colocalization, and a positive correlation between LMBD2B and DiI was detected (Fig. 5E). Another interesting thing about the expression of LMBD2B and DiI was the shape it takes on the cell. Membrane ruffles are formed in the plasma membrane. These membrane ruffles are labeled by membrane staining DiI, but surprisingly, LMBD2B signal is present on the ruffles, as well (Fig. 5C). Dictyostelium cells take up dextran through endocytosis, and therefore, it is used as a general indicator of endocytosis (9, 15). If LMBD2B is associated with endocytic processes, colocalization of LMBD2B with regions of uptake of FITC-dextran should be detected. This was indeed the case, as seen in Fig. 5D, where an overlap of LMBD2B and dextran is apparent. A positive correlation was detected between LMBD2B and dextran (Fig. 5E). The positive correlation with membrane and endocytic markers provides evidence of association of LMBD2B with sites of endocytosis. There is not complete colocalization with any of the markers, which is not surprising, as LMBD2B might be expected to be associated with only a subset of endocytic events.
Fig 5.
Colocalization of LMBD2B with endocytic and membrane markers. Growing cells were placed on a coverslip, fixed, and stained. Images of LMBD2B-mRFP indirect immunofluorescence are displayed on the left, other protein markers in the center, and overlays with transmitted light images on the right. (A) Coronin Ab. (B) Clathrin Ab. (C) DiI. (D) FITC-dextran. LMBD2B fluorescence is indicated by the red signal. All images are Z-axis slices through a cell. Blue signal indicates nuclei stained with DAPI. Yellow/orange signal indicates colocalization. (E) Values of colocalization. A positive correlation is indicated by a positive Pearson's coefficient. Colocalization of LMBD2B with DAPI was used as a negative control, since there should be no colocalization, so the value would represent 0 correlation. n > 30 cells from at least 2 rounds of fixations. The error bars indicate standard errors.
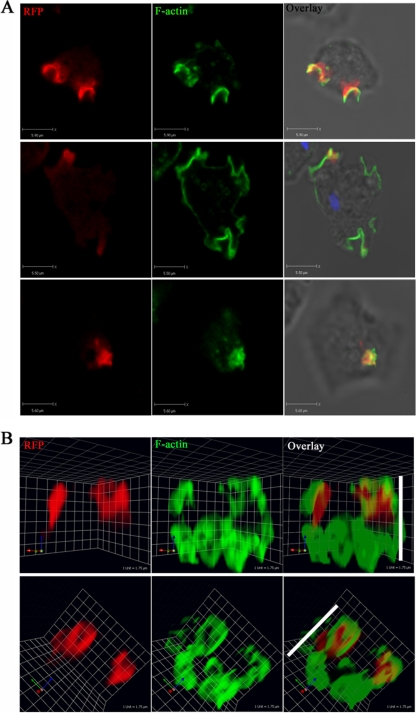
More striking evidence of LMBD2B association with endocytic events was seen when LMBD2B localization with F-actin was investigated. Actin polymerization is necessary for a number of cellular processes, such as cellular extension. Figure 6 displays images of the actin organization in growing cells. Regions of F-actin polymerization are shown in green, and in growing cells, it is largely distributed to cortical actin and to endocytic cups. A number of endocytic cups can be seen in Fig. 6. The LMBD2B protein was detected surrounding and overlapping some of these actin-rich cups. The 3D images displayed in Fig. 6B help illustrate the often dorsal localization of the actin-rich/LMBD2B-rich endocytic cups. The shape of distribution of the LMBD2B protein was similar to that of the F-actin endocytic cups, and a positive correlation was determined for colocalization (Fig. 5E). This would be expected if LMBD2B played a role in endocytosis, like other members of the LMBR family of nine-span membrane receptors. Complete colocalization was not seen between LMBD2B and all of the actin-rich cups, but only a subset. The colocalization of LMBD2B with endocytosis-specific markers and with endocytic cups provides compelling evidence that LMBD2B is localized to regions of endocytosis and that it may function in the endocytosis process. Interestingly, when lmbd2B is knocked out, there is actually a slight increase in the rate of dextran endocytosis (see Fig. S4A in the supplemental material). This may be because other endocytic events are increased and there is more efficient uptake of dextran in the absence of LMBD2B-associated endocytosis.
Fig 6.
LMBD2B localization to actin-rich endocytic processes. Growing cells were fixed and stained. Images of LMBD2B-mRFP indirect immunofluorescence are displayed on the left, 488-phalloidin-stained F-actin is displayed in the center, and overlays with transmitted-light images are on the right. (A) Z-axis slices through a cell. Blue signal indicates nuclear staining. (B) Side views of 3D-reconstructed cells. The white side bars indicate cell height. An overlap in signal is yellow. A positive correlation between LMBD2B-mRFP staining and F actin was determined for 21 cells from at least 3 different phalloidin stainings (Fig. 5E).
Identification of a role for LMBD2B in cell motility.
Null mutations of the LMBD2B protein result in a small-plaque phenotype in both WT and ampAOE backgrounds. A small-plaque phenotype can mean that cellular functions affecting cell growth, phagocytosis, or cell migration have been disrupted. Comparison of lmbd2B-null cells with WT parent strains show no difference in growth rates (see Fig. S4C in the supplemental material). A slight increase in phagocytosis was seen in lmbd2B− cells (see Fig. S4B in the supplemental material). This, if anything, would likely cause larger plaques. The lack of a significant phagocytosis defect, or any indications of a growth defect in LMBD2B-null cells, suggested the possibility of a cell migration defect. Since ampAOE cells induce significantly more F-actin than the WT and migrate more rapidly than the WT (Noratel, Petty, Zhang, and Blumberg, unpublished), the possibility of a suppressor of the ampAOE phenotype harboring a cell migration defect is not unrealistic.
The ability of a cell to migrate is based on a number of pathways working together to permit proper motility. Some of those pathways include proper sensing of a chemoattractant, actin polymerization, extension of a dominant pseudopod toward the chemoattractant, and a change in cell-substrate adhesion. Each of these components was examined as a possible defect in lmbd2B− cells. There did not appear to be any significant change in the cell's ability to adhere to the substrate (see Fig. S5A in the supplemental material). Also, there was no change in the levels of polymerized F-actin in lmbd2B− compared to WT cells (see Fig. S5B in the supplemental material).
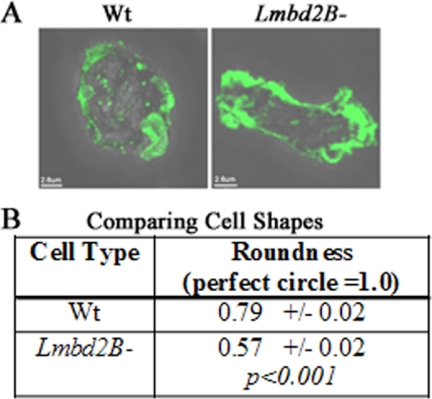
When comparing actin organization, a change in cell shape was noticed in LMBD2B-null cells. When lmbd2B− cells were placed on a coverslip to adhere overnight, the cells acquired a more elongated shape than WT cells (Fig. 7). The degree of roundness of WT cells was calculated to be about 0.79, closely resembling a perfect circle, which would give a value of 1.0. lmdb2B− cells were significantly more elongated, with a roundness value of only 0.57. The cells were sitting in medium, which should cause them to have a rounder shape, as their environment was readily providing nutrients, so the cells had no need to move in search of food. In spite of that, lmbd2B− cells started to elongate.
Fig 7.
Vegetative LMBD2B-null cells show an elongated shape. Growing cells were placed on coverslips and allowed to sit overnight. The cells were stained with Alexa Fluor 488-phalloidin to label F-actin. (A) Images of the different cell types (an overlay of 3D flattened fluorescence and transmitted-light images). Polymerized actin is green. (B) Comparison of the roundness of WT and lmbd2B− cells. ImageJ was used to calculate the roundness of the cells as follows; roundness = 4 × area/(π × major axis2). A perfect circle would equal a roundness of 1.0. lmbd2B− cells were significantly less round (or more elongated) than WT cells. n = 25 cells from at least 3 different phalloidin stainings.
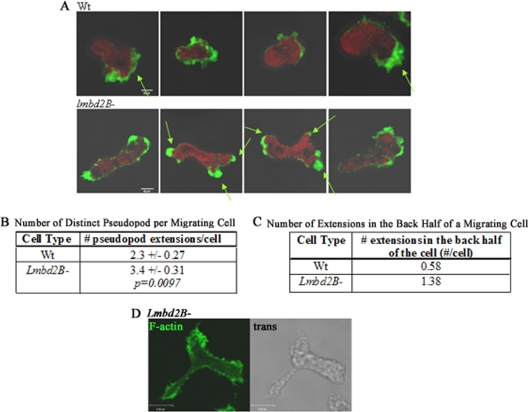
Simple elongation of lmbd2B knockout cells would not account for a small-plaque-size defect. Differences in migration of these cells were more closely examined. Cell motility in growing cells was promoted by placing a drop of a chemoattractant, folic acid, near a drop of cells on a coverslip. The cells were then fixed and stained with phalloidin to identify F-actin organization in the migrating cells. lmbd2B− cells showed more pseudopodia than WT cells (Fig. 8) and also extended pseudopodia in regions other than the front of the chemotaxing cell. Often, F-actin-rich pseudopodia were detected in the front and back simultaneously in a moving cell (Fig. 8A, bottom). Determining the number of pseudopods per migrating cell indicated that lmbd2B− cells had on average 1 more pseudopod than WT cells and had more pseudopods forming in the back half of the cell than WT cells (Fig. 8B and C). This F-actin organization suggests a lack of polarization and difficulty in producing a dominant pseudopod.
Fig 8.
Multiple pseudopodial extensions in LMBD2B-null cells. Growing cells were forced to undergo migration by spotting folic acid near a drop of cells on coverslips. The cells were stained with Alexa Fluor 488-phalloidin to label F-actin. (A) Z-axis slices of WT and lmbd2B− cells. Polymerized actin is green, and red represents G-actin. The arrows point to distinct areas of F-actin pseudopodial extensions. (B) The number of distinct pseudopods per cell was calculated. lmbd2B− cells have on average one extra pseudopod per migrating cell compared to WT cells. n > 8 cells from 3 separate phalloidin staining experiments. (C) Results of counting actin-rich extensions in the rear of the migrating cells. lmbd2B− cells have more pseudopods forming in the back half of a chemotaxing cell than WT cells. n > 12 cells from 3 separate phalloidin staining experiments. (D) Same as panel A but showing an actin-rich pseudopodial extension from the middle of an lmbd2B− cell; a Z-axis slice of an F-actin fluorescent staining and a transmitted image are displayed.
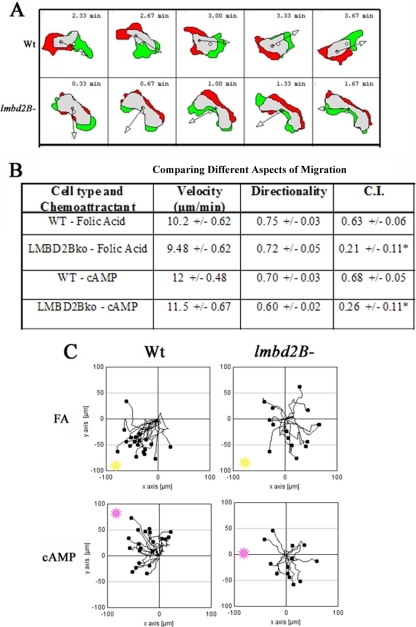
The motility of lmbd2B− cells was compared to that of WT cells. lmbd2B− cells often migrated sideways during chemotaxis. Figure 9A depicts this phenotype, and difference plots of a WT and lmbd2B− cell are displayed for comparison. Green represents extending areas, and red represents retracted areas; the small arrows point in the direction of movement. The LMBD2B-null cell appears to extend and retract pseudopods from the same side of each end of the cell. It is as if the cell cannot establish a dominant front and back at the cell poles.
Fig 9.
LMBD2B-null cells are defective in chemotaxis. Images were taken every 20 s for 5 min. (A) Difference plots of chemotaxing cells are shown (DIAS). Green represents areas of extension from the previous image, red indicates retraction regions from the previous image, and gray indicates areas that remain the same. Consistency is seen in the WT cell's amount of extension and retraction at the poles of the cell. This is not the case for lmbd2B− cells, which often travel sideways, extending from the broad side of a cell as opposed to one of the narrow poles. (B) Values for the velocity, directionality, and CI of WT and lmbd2B− cells migrating toward folic acid and cAMP. ∗, significant difference, P < 0.05, by two-tailed student's t test. (C) Chemotaxis plots. Cells were tracked on agar every 20 s for a total of 5 min. The black dots represent cells, and their individual paths are shown by lines. The direction and distance of each cell from its origin at 0 is shown. The location of the edge of the source of chemoattractant is shown in each plot as a pink or yellow spot. In the upper plots, folic acid (FA) (yellow) and in the bottom plots cAMP (pink) was used as the chemoattractant. WT cells travel more directly toward folic acid and cAMP than lmbd2B− cells. n > 30 cells from 3 or 4 separate chemotaxis experiments.
Migration rates and directionality were unaffected by the defect in lmbd2B− cells (velocity in Fig. 9B). The chemotaxis plots displayed in Fig. 9C illustrate a defect in the ability of the LMBD2B-null cells to properly move toward a chemoattractant. Folic acid was used as a chemoattractant to mimic the presence of bacteria. Most of the migrating cells in the WT chemotaxis plots are migrating toward the folic acid source (Fig. 9C, top left). The lmbd2B− chemotaxis plots show some cells moving in the correct direction, toward folic acid, but there are more cells moving in directions away from the chemoattractant (Fig. 9C, top right). To test if the chemotaxis defect was specific to folic acid, movement toward cAMP, the chemoattractant during development, was tested as well. Results similar to those seen in the folic acid chemotaxis were observed (Fig. 9C, bottom), with lmbd2B− cells having difficulty moving toward the cAMP source. The chemotactic index of lmbd2B− cells most clearly illustrates the lmbd2B− chemotaxis defect. Figure 9B gives the chemotactic indexes for WT and lmbd2B− cells. With both folic acid and cAMP chemoattractants, there is a drop in values to about one-third that of the WT, confirming the difficulty in the cell's ability to move in the correct direction to the chemoattractant.
A role for LMBD2B expression during development.
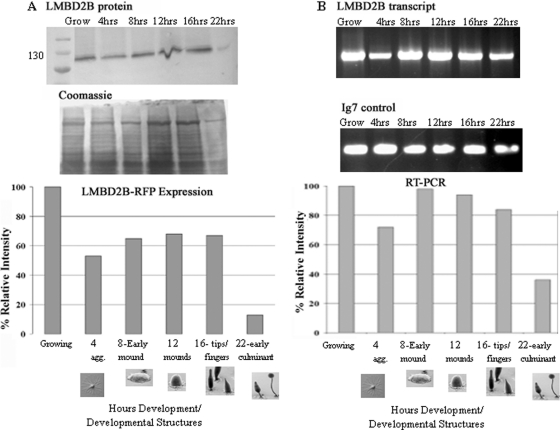
The mRFP-tagged LMBD2B fusion protein was used to determine protein expression levels throughout Dictyostelium growth and development, using an RFP antibody. RNA was isolated during growth and at different time points during development, and RT-PCR was used to determine the relative lmbd2B transcript levels. A fairly high level of LMBD2B-mRFP protein and RNA was detected throughout growth and development, with a peak in the growth phase and at the mound stage of development and a decrease at culmination. This suggests a role for LMBD2B during growth and in the mound stages of development (Fig. 10).
Fig 10.
LMBD2B protein and mRNA are expressed throughout growth and development. Analysis of LMBD2B-mRFP protein expression (A) and RT-PCR with relative levels of lmbd2B transcript (B) are shown. (A) During growth and after the indicated number of hours of development, cells were washed, harvested, and run on an SDS-6% PAGE gel. (Top) After transfer, nitrocellulose was probed with an RFP antibody. The LMBD2B-mRFP product is about 116 kDa. (Bottom) Coomassie stain was used to correct quantification for unequal loading. (B) At specific time points, the RNA from the developing structures was isolated. A cDNA copy of total RNA was used for PCR. lmbd2B-specific primers were used to amplify the transcript levels in the cells. (Top) Agarose gels of the RT-PCR product of LMBD2B transcripts and Ig7 control transcripts. (Bottom) Calculation of the relative intensity of each band. The developmental pictures are from http://dictybase.org (copyright, M. J. Grimson and R. L. Blanton, Biological Sciences Electron Microscopy Laboratory, Texas Tech University; reported with permission).
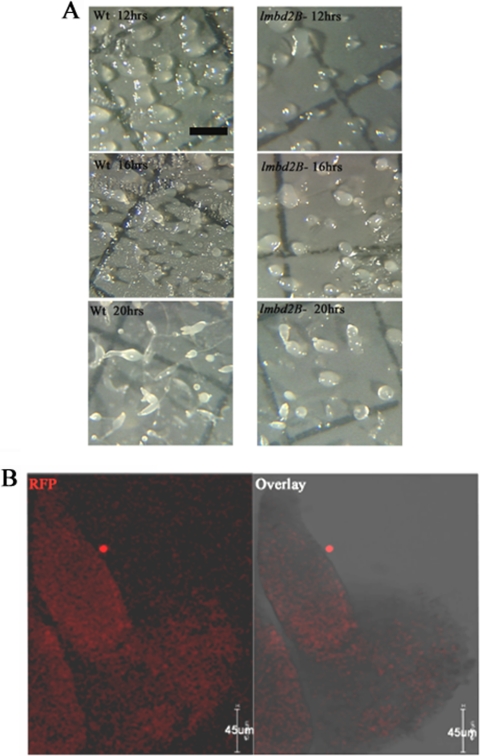
Consistent with a role in development, defects in Dictyostelium developmental structures were detected in lmbd2B− cells (Fig. 11A). A delay in development was seen in lmbd2B knockout cells. At 16 h of development, lmbd2B− cells were still in the mound stage while WT cells had progressed well into tipped mounds and fingers. The difference in structures suggests lmbd2B− cells have about a 4-h delay in development. Also, the mounds appeared sparse in lmbd2B− cells compared to WT cells, an indication that it takes longer for LMBD2B-null cells to aggregate to mound stage and also that fewer aggregates form and go on to culminate. Immunofluorescent staining of whole mounts indicated that cells in both the prespore and prestalk regions of the culminants express the LMBD2B-mRFP protein construct (Fig. 11B).
Fig 11.
LMBD2B has a role during development. (A) Cells were plated for development on nitrocellulose filters. At the indicated times (12, 16, and 20 h), images were taken using Metamorph software and a Dage-MTI video camera mounted on an Olympus dissecting microscope. The scale bar represents 1,000 μm. (B) Indirect immunofluorescence of LMBD2B-mRFP localization in whole mounts imaged on a Leica SP5 confocal microscope. Optical Z-axis slices through the developing structure are shown, with red signal indicating LMBD2B locations; shown are immunofluorescent images and overlays with transmitted images. The developing structure is a finger (∼16 h development).
DISCUSSION
lmbd2B is a member of the LMBR1 family of proteins. Like other LMBD2 proteins in this family, it contains 2 LMBR1 domains. Topography predictions suggest LMBD2B is an eight (possibly nine)-span transmembrane protein, which is consistent with other LMBR1 family proteins (9, 59). LMBD2B is localized to membranes. The fact that it is present on the periphery of cells and that it colocalizes with DiI at the top of the cell suggests its localization in the plasma membrane. The protein was found localized in punctate spots around the periphery of growing cells. These spots appeared to be more dispersed in cells that were in suspension than in those that were attached to a substrate. After prolonged substrate contact, more dense patches of LMBD2B were found, often localized on the dorsal side/top of the cell. What would cause the protein to congregate? It is likely that when the cell utilizes ventral substrate attachments, LMBD2B, which is associated with endocytic cups, becomes restricted to other areas of the cell.
LMBD2B in receptor-mediated endocytosis.
As a member of a family of proteins involved in receptor-mediated endocytosis, it seems likely that LMBD2B would also be involved in endocytosis. The process of receptor-mediated endocytosis begins with cargo binding and activation of receptors. Subsequent recruitment of the effector molecules essential to vesicle formation then occurs. Clathrin is assembled at activated receptors, and clathrin-coated pits are formed at the plasma membrane and pinch off into vesicles (41). The pinching off of vesicles is dependent on actin polymerization. LMBD2B was seen colocalizing with proteins involved in receptor-mediated endocytosis (namely, clathrin and F-actin). Since LMBD2B may be associated only with a subset of endocytosis events occurring in the cell, it was not surprising to see that endocytic cups and proteins were also found without LMBD2B.
LMBD2B involvement in macropinocytosis.
LMBD2B colocalized with FITC-dextran and coronin, suggesting association with endocytosis events via fluid phase macropinocytosis. This large-scale form of endocytosis involves membrane ruffling, followed by the nonselective engulfment of fluid in the cells (30). This result was surprising, as plasma membrane receptors are expected to be involved in receptor-mediated clathrin-dependent endocytosis. The endocytic proteins coronin and actin are thought to be involved in macropinocytosis. Coronin is an actin binding protein that regulates Arp2/3, allowing actin polymerization, which is important for macropinocytosis (55). It is a little less clear whether clathrin participates in fluid phase uptake. Dictyostelium clathrin-null cells display an 80% reduction in macropinocytosis (22). Based on the knockout phenotype results, this suggests a role for clathrin in macropinocytosis, although no direct involvement has been shown.
The characteristic dorsal ruffling of the plasma membrane that occurs during macropinocytosis is similar to the pattern sometimes seen with LMBD2B localization. LMBD2B was often detected concentrated toward the top of the cell and in patterns similar to ruffling, or waves, as was most clearly seen in the LMBD2B DiI colocalization result. It is possible that LMBD2B is associated with events specific to macropinocytosis, but conflicting with that idea was the detection of punctate colocalization of LMBD2B with clathrin, suggesting association with clathrin-dependent receptor-mediated endocytosis. A more likely scenario is that which occurs with epidermal growth factor (EGF) and its receptor (EGFR), which has been shown to be involved in multiple endocytic processes (42). At low concentrations of EGF, the mode of endocytosis is almost entirely clathrin dependent. At high EGF concentrations, a large-scale clathrin-independent endocytic pathway emerges (52). Also, another switch in the mode of uptake of EGFR is seen in migrating endothelial and mesenchymal cells. EGFR internalization by wave-like ruffles on the dorsal surfaces of migrating cells was identified. At times, a single endocytic event could involve up to 50% of the surface EGFR (30). The authors suggested the large-scale endocytic uptake was a result of actin wave endocytosis rather than membrane-ruffling macropinocytosis. The processes are similar, and both involve the use of coronin and F-actin, but they differ in other specific proteins associated with actin structures (21). Determining whether LMBD2B is involved with actin waves or membrane-ruffling endocytosis would require further protein-specific markers. Nonetheless, it does appear that LMBD2B may be associated with two different routes of endocytosis.
A role for LMBD2B in migration.
lmbd2B has a role during cell migration, as evidenced by knockout phenotypes. lmbd2b− cells in contact with a substrate have a tendency to elongate, as if the cells are randomly remodeling the actin organization for pseudopod extensions, readying the cell for movement. In migrating lmbd2B− cells, more than one pseudopod is formed. There were often pseudopods in the front and back of elongated cells, suggesting these cells had difficulty establishing a dominant pseudopod. The defect in cell orientation and pseudopod projection did not affect cell migration rates. It did affect the ability of cells to move toward a chemoattractant. lmbd2B− cells are significantly less directed in their movement toward folic acid, and also toward cAMP. The fact that chemotaxis to two different attractants is defective suggests that LMBD2B is necessary for a basic component of motility and pseudopod extension, rather than an attractant-specific detection mechanism. The inability of the cells to properly “sense” or orient toward a chemoattractant likely plays a role in the small-plaque phenotype that is observed in the lmbd2B-null cells.
A model for LMBD2B.
How could a protein associated with endocytic events be affecting cell migration? There are numerous examples of endocytic processes affecting cell motility. In Dictyostelium, clathrin-null mutants display defects in lateral pseudopod extensions and chemotaxis (60). Even more applicable to lmbd2B, lipocalin 1 receptors have been shown to have effects on macrophage and cancer cell motility. Lipocalin 1 receptor (LIMR) substrate binding in cancer cells was shown to suppress extracellular matrix invasion while having no effect on adhesion and chemotaxis (33, 66). The LIMR-ligand receptor binding and endocytosis inhibited the invasiveness/responsiveness of tumor cells. While no effect on chemotaxis was determined in tumor cells, LIMR-substrate endocytosis did have anti-chemotactic effects in macrophages (33).
It is possible that the LMBD2B transmembrane protein acts as a receptor, binds a substrate, is endocytosed, and so signals the cell for continued growth and suppresses motility. In lmbd2B-null cells, without this signal, the cell undergoes actin remodeling and elongates. This lmbd2B endocytic signal may also be important during chemotaxis. LMBD2B receptor endocytosis could act as a sampling mechanism during movement. It could help orient the cell, permitting dominant pseudopod production toward a chemoattractant. If LMBD2B functions as a receptor, its substrate remains elusive. Presumably it would be something similar to those of other LMBR1 family substrates, such as a lipophilic molecule or a specific nutrient. It seems unlikely that LMBD2B would be the folic acid receptor, since the lmbd2B-null cells also fail to undergo directional migration toward the chemoattractant cAMP.
lmbd2B is expressed during D. discoideum growth and development. There appears to be a definite role for LMBD2B during development, as lmbd2B− cells display delayed development, and a defect is apparent in fruiting body structures. Possibly, the defect in motility seen in growing lmbd2B− cells occurs during aggregation and early development. The inability of the lmbd2B− cells to move together and continue development could account for the developmental delay and reduced number of developing structures that form.
It is unclear at this point what the relationship could be between the LMBD2B protein and the ampA pathway. LMBD2B does not appear to play any role in the location/secretion of the AmpA protein. What is clear is that selecting for suppressors of the large-plaque phenotype of the AmpA overexpresser strain has indeed resulted in the identification of a gene involved in cell motility whose loss reduces directional cell motility. This loss of directionality in the lmbd2B-null cells potentially compensates for the increased migration velocity of the AmpA overexpresser and could lead to the small-plaque phenotype of the double mutant.
Previous work has identified another class of nine-span transmembrane proteins in Dictyostelium that are involved in cellular adhesion, phagocytosis, growth, and development (2, 10). These proteins include the products of the Phg1a, -b, and -c genes. They appear to be members of the EMP7 family of endomembrane proteins but are unrelated to the LMBR1 domain proteins that we describe here. Thus, Dictyostelium contains multiple members of two different nine-span membrane protein families.
Further research on lmbd2B and other LMBR1-like receptors utilizing a simple system like Dictyostelium should prove to be interesting because of the ease of generating mutants and examining phenotypes. If other members have the ability to take in compounds similar to those taken up by the lipocalin receptors, the implications as possible carriers for drug delivery or toxic compounds are enormous. Also, with effects on cell motility, these receptors present another possible target of cancer therapy. Our results provide another angle of research into understanding and manipulating motility-regulating proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Annette Muller-Taubenberger and the Dictyostelium Stock Center for the mRFP plasmid and Alice Rutatangwa and Julie Wolf for help with plasmid construction. Also, the Dictyostelium Stock Center and Gunther Gerish supplied the GFP N-golvesin construct. The coronin and fimbrin antibodies developed by Gunther Gerish were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biology, University of Iowa, Iowa City, IA 52242. We are grateful to Cheré Petty and the UMBC Keith R. Porter Imaging Facility for help with microscopy and to Elizabeth F. Noratel for discussions.
This work was supported by NSF grants MCB-0444883 to D.D.B. and MRI-0722569 to D.D.B. and Theresa Good.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1. Barlow AL, Macleod A, Noppen S, Sanderson J, Guérin CJ. 2010. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson's correlation coefficient. Microsc. Microanal. 16:710–724 [DOI] [PubMed] [Google Scholar]
- 2. Benghezal M, et al. 2003. Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol. Biol. Cell 14:2890–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blumberg DD, Ho HN, Petty CL, Varney TR, Gandham S. 2002. AmpA, a modular protein containing disintegrin and ornatin domains, has multiple effects on cell adhesion and cell fate specification. J. Muscle Res. Cell Motil. 23:817–828 [DOI] [PubMed] [Google Scholar]
- 4. Bonner JT. 1967. The cellular slime molds. Princeton University Press, Princeton, NJ [Google Scholar]
- 5. Brazill DT, Meyer LR, Hatton RD, Brock DA, Gomer RH. 2001. ABC transporters required for endocytosis and endosomal pH regulation in Dictyostelium. J. Cell Sci. 114:3923–3932 [DOI] [PubMed] [Google Scholar]
- 6. Casademunt E, Varney T, Dolman J, Petty C, Blumberg D. 2002. A gene encoding a novel anti-adhesive protein is expressed in growing cells and restricted to anterior-like cells during development of Dictyostelium. Differentiation 70:23–35 [DOI] [PubMed] [Google Scholar]
- 7. Chisholm RL, et al. 2006. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 34:D423–D427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark RM, Marker PC, Kingsley DM. 2000. A novel candidate gene for mouse and human preaxial polydactyly with altered expression in limbs of Hemimelic extra-toes mutant mice. Genomics 67:19–27 [DOI] [PubMed] [Google Scholar]
- 9. Claros MG, von Heijne G. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685–686 [DOI] [PubMed] [Google Scholar]
- 10. Cornillon S, et al. 2000. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J. Biol. Chem. 275:34287–34292 [DOI] [PubMed] [Google Scholar]
- 11. de Hostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. 1991. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 10:4097–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egelhoff TT, Lee RJ, Spudich JA. 1993. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75:363–371 [DOI] [PubMed] [Google Scholar]
- 14. Eichinger L, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eichinger L, Rivero F. 2010. Dictyostelium discoideum protocols. Humana Press, Totowa, NJ [Google Scholar]
- 16. Fey P, Gaudet P, Pilcher KE, Franke J, Chisholm RL. 2006. dictyBase and the Dicty Stock Center. Methods Mol. Biol. 346:51–74 [DOI] [PubMed] [Google Scholar]
- 17. Flower DR. 2000. Beyond the superfamily: the lipocalin receptors. Biochim. Biophys. Acta 1482:327–336 [DOI] [PubMed] [Google Scholar]
- 18. Fluckinger M, Merschak P, Hermann M, Haertlé T, Redl B. 2008. Lipocalin-interacting-membrane-receptor (LIMR) mediates cellular internalization of beta-lactoglobulin. Biochim. Biophys. Acta 1778:342–347 [DOI] [PubMed] [Google Scholar]
- 19. Gailus S, et al. 2010. Insights into lysosomal cobalamin trafficking: lessons learned from cblF disease. J. Mol. Med. 88:459–466 [DOI] [PubMed] [Google Scholar]
- 20. Geiger J, Wessels D, Soll DR. 2003. Human polymorphonuclear leukocytes respond to waves of chemoattractant, like Dictyostelium. Cell Motil. Cytoskeleton 56:27–44 [DOI] [PubMed] [Google Scholar]
- 21. Gerisch G. 2010. Self-organizing actin waves that simulate phagocytic cup structures. PMC Biophys. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hacker U, Albrecht R, Maniak M. 1997. Fluid-phase uptake by macropinocytosis in Dictyostelium. J. Cell Sci. 110:105–112 [DOI] [PubMed] [Google Scholar]
- 23. Hadwiger JA, Srinivasan J. 1999. Folic acid stimulation of the Galpha4 G protein-mediated signal transduction pathway inhibits anterior prestalk cell development in Dictyostelium. Differentiation 64:195–204 [DOI] [PubMed] [Google Scholar]
- 24. Hopper NA, Harwood AJ, Bouzid S, Véron M, Williams JG. 1993. Activation of the prespore and spore cell pathway of Dictyostelium differentiation by cAMP-dependent protein kinase and evidence for its upstream regulation by ammonia. EMBO J. 12:2459–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iijima M, Devreotes P. 2002. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109:599–610 [DOI] [PubMed] [Google Scholar]
- 26. Janetopoulos C, Firtel RA. 2008. Directional sensing during chemotaxis. FEBS Lett. 582:2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kehrer JP. 2010. Lipocalin-2: pro- or anti-apoptotic? Cell Biol. Toxicol. 26:83–89 [DOI] [PubMed] [Google Scholar]
- 28. Kessin RH. 2001. Dictyostelium: evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 29. Kimmel AR, Faix J. 2006. Generation of multiple knockout mutants using the Cre-loxP system. Methods Mol. Biol. 346:187–199 [DOI] [PubMed] [Google Scholar]
- 30. Kumari S, Mg S, Mayor S. 2010. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 20:256–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuspa A. 2006. Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol. Biol. 346:201–209 [DOI] [PubMed] [Google Scholar]
- 32. Lettice LA, et al. 2002. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. U. S. A. 99:7548–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leyton J, et al. 1994. Recombinant human uteroglobin inhibits the in vitro invasiveness of human metastatic prostate tumor cells and the release of arachidonic acid stimulated by fibroblast-conditioned medium. Cancer Res. 54:3696–3699 [PubMed] [Google Scholar]
- 34. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Müller-Taubenberger A, et al. 2006. Monomeric red fluorescent protein variants used for imaging studies in different species. Eur. J. Cell Biol. 85:1119–1129 [DOI] [PubMed] [Google Scholar]
- 38. Nellen W, et al. 1987. Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 28:67–100 [DOI] [PubMed] [Google Scholar]
- 39. Noegel A, Gerisch G, Stadler J, Westphal M. 1986. Complete sequence and transcript regulation of a cell adhesion protein from aggregating Dictyostelium cells. EMBO J. 5:1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noegel AA, Schleicher M. 2000. The actin cytoskeleton of Dictyostelium: a story told by mutants. J. Cell Sci. 113:759–766 [DOI] [PubMed] [Google Scholar]
- 41. O'Halloran TJ, Anderson RG. 1992. Clathrin heavy chain is required for pinocytosis, the presence of large vacuoles, and development in Dictyostelium. J. Cell Biol. 118:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orth JD, Krueger EW, Weller SG, McNiven MA. 2006. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 66:3603–3610 [DOI] [PubMed] [Google Scholar]
- 43. Oyama M, Blumberg D. 1986. Changes during differentiation in requirements for cAMP for expression of cell-type-specific mRNAs in the cellular slime mold, Dictyostelium discoideum. Dev. Biol. 117:550–556 [DOI] [PubMed] [Google Scholar]
- 44. Pan P, Hall EM, Bonner JT. 1972. Folic acid as second chemotactic substance in the cellular slime moulds. Nat. New Biol. 237:181–182 [DOI] [PubMed] [Google Scholar]
- 45. Parent CA, Devreotes PN. 1999. A cell's sense of direction. Science 284:765–770 [DOI] [PubMed] [Google Scholar]
- 46. Prassler J, et al. 1997. Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Mol. Biol. Cell 8:83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pùta F, Zeng C. 1998. Blasticidin resistance cassette in symmetrical polylinkers for insertional inactivation of genes in Dictyostelium. Folia Biol. (Prague) 44:185–188 [PubMed] [Google Scholar]
- 48. Rutsch F, et al. 2009. Identification of a putative lysosomal cobalamin exporter altered in the cblF defect of vitamin B12 metabolism. Nat. Genet. 41:234–239 [DOI] [PubMed] [Google Scholar]
- 49. Rutsch F, Gailus S, Suormala T, Fowler B. 2011. LMBRD1: the gene for the cblF defect of vitamin B12 metabolism. J. Inherit. Metab. Dis. 34:121–126 [DOI] [PubMed] [Google Scholar]
- 50. Schneider N, et al. 2000. Golvesin-GFP fusions as distinct markers for Golgi and post-Golgi vesicles in Dictyostelium cells. Biol. Cell 92:495–511 [DOI] [PubMed] [Google Scholar]
- 51. Shaulsky G, Escalante R, Loomis WF. 1996. Developmental signal transduction pathways uncovered by genetic suppressors. Proc. Natl. Acad. Sci. U. S. A. 93:15260–15265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sigismund S, et al. 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. U. S. A. 102:2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soll D. 2003. Dictyostelium may model diseases involving defects in cellular chemotaxis. ASM News 69:246 [Google Scholar]
- 54. Sussman M. 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28:9–29 [DOI] [PubMed] [Google Scholar]
- 55. Uetrecht AC, Bear JE. 2006. Coronins: the return of the crown. Trends Cell Biol. 16:421–426 [DOI] [PubMed] [Google Scholar]
- 56. van Baren MJ, et al. 2002. A double RING-H2 domain in RNF32, a gene expressed during sperm formation. Biochem. Biophys. Res. Commun. 292:58–65 [DOI] [PubMed] [Google Scholar]
- 57. Varney T, et al. 2002. A novel Dictyostelium gene encoding multiple repeats of adhesion inhibitor-like domains has effects on cell-cell and cell-substrate adhesion. Dev. Biol. 243:226–248 [DOI] [PubMed] [Google Scholar]
- 58. Varney T, Ho H, Petty C, Blumberg D. 2002. A novel disintegrin domain protein affects early cell type specification and pattern formation in Dictyostelium. Development 129:2381–2389 [DOI] [PubMed] [Google Scholar]
- 59. von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 60. Wessels D, Kuhl S, Soll DR. 2006. Application of 2D and 3D DIAS to motion analysis of live cells in transmission and confocal microscopy imaging. Methods Mol. Biol. 346:261–279 [DOI] [PubMed] [Google Scholar]
- 61. Wessels D, et al. 2000. Clathrin plays a novel role in the regulation of cell polarity, pseudopod formation, uropod stability and motility in Dictyostelium. J. Cell Sci. 113:21–36 [DOI] [PubMed] [Google Scholar]
- 62. Williams RS, et al. 2006. Towards a molecular understanding of human diseases using Dictyostelium discoideum. Trends Mol. Med. 12:415–424 [DOI] [PubMed] [Google Scholar]
- 63. Witke W, Schleicher M, Noegel AA. 1992. Redundancy in the microfilament system: abnormal development of Dictyostelium cells lacking two F-actin cross-linking proteins. Cell 68:53–62 [DOI] [PubMed] [Google Scholar]
- 64. Wojnar P, Lechner M, Merschak P, Redl B. 2001. Molecular cloning of a novel lipocalin-1 interacting human cell membrane receptor using phage display. J. Biol. Chem. 276:20206–20212 [DOI] [PubMed] [Google Scholar]
- 65. Wojnar P, Lechner M, Redl B. 2003. Antisense down-regulation of lipocalin-interacting membrane receptor expression inhibits cellular internalization of lipocalin-1 in human NT2 cells. J. Biol. Chem. 278:16209–16215 [DOI] [PubMed] [Google Scholar]
- 66. Zhang Z, et al. 2006. Interaction of uteroglobin with lipocalin-1 receptor suppresses cancer cell motility and invasion. Gene 369:66–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.