Abstract
Plasmids pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4, isolated from Salmonella and Escherichia coli clinical strains from hospitals in Argentina, were completely sequenced. These plasmids include the qnrB19 gene and are 2,699, 3,082, 2,989, and 2,702 nucleotides long, respectively, and they share extensive homology among themselves and with other previously described small qnrB19-harboring plasmids. The genetic environment of qnrB19 in all four plasmids is identical to that in these other plasmids and in transposons such as Tn2012, Tn5387, and Tn5387-like. Nucleotide sequence comparisons among these and previously described plasmids showed a variable region characterized by being flanked by an oriT locus and a Xer recombination site. We propose that this arrangement could play a role in the evolution of plasmids and present a model for DNA swapping between plasmid molecules mediated by site-specific recombination events at oriT and a Xer target site.
INTRODUCTION
Qnrs are pentapeptide repeat proteins that mediate resistance to quinolones by protecting type II DNA topoisomerases (14, 28). They are known since 1998 when the first qnr gene was found in the multiresistance plasmid pMG252 harbored by a Klebsiella pneumoniae strain isolated from the urine of a patient at the University of Alabama (20). Since then five qnr families (qnrA, qnrB, qnrC, qnrD, and qnrS) have been found, usually hosted in large plasmids (31). The first qnrB gene (qnrB1) was identified in a plasmid from a K. pneumoniae strain isolated in South India (16), and 38 members of the family quickly followed (http://www.lahey.org/qnrStudies/) (13). The qnrB19 gene has been found in several genera of Enterobacteriaceae isolated from humans (healthy people and clinical isolates), animals, and food of animal origin in numerous geographical regions (6, 9, 12, 17, 21, 22, 27). An interesting characteristic of the qnrB19 allele is that it has been found within large plasmids, associated to ISEcp1C-based transposons (6, 9, 27), and in small plasmids (∼3 kbp) lacking ISEcp1C or any other insertion sequence (12, 17, 22) (Table 1). However, in spite of being located in such dissimilar elements, the qnrB19 genes share a conserved genetic environment (22).
Table 1.
qnrB19-harboring genetic elements
| Genetic element | Size (bp) | Country | Hostc | Origin | Source or reference (accession no.) |
|---|---|---|---|---|---|
| ISEcp1C-based transposonsa | |||||
| Tn2012 (pR4525, 40 kb) | 2,738 | Colombia | E. coli | Clinical isolate | 6 |
| Tn5387 (pLRM24, 80 kb) | 2,966 | USA | K. pneumoniae | Clinical isolate | 27 |
| Tn5387-like (p61/9, >21 kb) | 2,828 | Italy | S. enterica | Clinical isolate | 9 |
| Small plasmidsb | |||||
| pPAB19-1 | 2,699 | Argentina | Salmonella Infantis M7849* | Clinical isolate | This study (GQ412195) |
| pPAB19-2 | 3,082 | Argentina | E. coli M9996* | Clinical isolate | This study (JN979787) |
| pPAB19-3 | 2,989 | Argentina | E. coli M9888* | Clinical isolate | This study (JN985534) |
| pPAB19-4 | 2,702 | Argentina | Salmonella sp. strain M9397* | Clinical isolate | This study (JN995611) |
| pSGI15 | 2,699 | Netherlands | S. enterica | Clinical isolate | 12 |
| pECY6-7 | 2,699 | Bolivia, Peru | E. coli, E. fergusonii, E. hermanii, E. aerogenes, K. ascorbata, K. pneumoniae | Healthy people | 21, 22 |
| pECC14-9 | 3,071 | Bolivia, Peru | E. coli | Healthy people | 21, 22 |
| pMK100 | 2,699 | Colombia | S. enterica | Retail poultry | 17 |
| pMK102 | 2,750 | Colombia | S. enterica | Ground beef | 17 |
The plasmids hosting the transposons and their sizes are indicated in parentheses.
pPAB19-1 was found in 16 of the 45 E. coli and Salmonella isolates (8 E. coli, 1 Salmonella Infantis, and 7 Salmonella spp.); pPAB19-2 was found in 3 of the 45 E. coli and Salmonella isolates (2 E. coli and 1 Salmonella sp.); pPAB19-3 was found in 1 of the 45 E. coli and Salmonella isolates (E. coli); pPAB19-4 was found in 4 of the 45 E. coli and Salmonella isolates (Salmonella spp.).
*, Strains from which the individual sequenced plasmid was isolated.
We have recently analyzed a collection of clinical enterobacterial isolates with decreased quinolone susceptibility, and we found four small plasmids harboring qnrB19 (2). We describe here their molecular features and characterize their relationships with other qnrB19-harboring genetic platforms. Furthermore, we propose possible pathways of evolution of the qnrB19 environment as well as a site-specific recombination-based model for DNA modifications at a variable region found in these plasmids.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmids pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4 analyzed in the present study were isolated from Salmonella enterica serovar Infantis M7849, Escherichia coli M9996, E. coli M9888, and Salmonella sp. strain M9397, respectively (Table 1). Salmonella Infantis M7849 was isolated at the Hospital Castro Rendón, Province of Neuquén (March 2006). E. coli strains M9888 and M9996 were isolated at the Policlínico Central de San Luis, Province of San Luis (May 2007 and August 2008, respectively), and Salmonella sp. strain M9397 was isolated at the Hospital de Niños Alassia, Province of Santa Fe (July 2007). These isolates are included in a group of 20 Salmonella and 25 E. coli strains that is part of a collection of 105 strains with decreased susceptibility to quinolones obtained from 31 hospitals in the city of Buenos Aires and 11 provinces of Argentina (2). A thorough characterization of these strains will be published elsewhere. Recombinant plasmids pPBR1 and pPBR2 were generated by ligating HindIII-digested pPAB19-1 or pPAB19-2 to HindIII-digested pUC19 (38). Dimers of plasmids pES and pKS492 were used as controls in dimer resolution experiments (4). E. coli DS941 (AB1157 recF143 lacIq lacZ; possesses wild-type xerC, xerD, argR, and pepA) (33), E. coli DS9028 (DS941 xerD3::fol) (30), and the hyper-recombinogenic E. coli JC8679 (DS945 recBC sbcA) (32) were used to carry out the Xer recombination experiments.
DNA sequencing and analysis.
Plasmids pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4 were screened by PCR using the divergent primers qnrB-Fout (5′-GACGTTCAGTGGTTCAGATCTCTC) and qnrB-Rout (5′-GACTAAAATTGCACCCTTTCTGACT) that bind to the qnrB19 gene, leading to amplifications of its surrounding plasmid sequences. The amplicons were sequenced using BigDye terminator methodology and sequence-based primers (DNA walking), with an ABI 3130xl genetic analyzer (Applied Biosystems/Perkin-Elmer, Foster City, CA). Nucleotide sequence editing and analyses were performed using ClustalX2 or ClustalW2 (v2.0.9; ftp://ftp.ebi.ac.uk/pub/software/clustalw2) (18), BioEdit (v7.0.9; http://www.mbio.ncsu.edu/bioedit/bioedit.html) (10), and the basic local alignment search tool (BLAST; http://www.ncbi.nlm.nih.gov/BLAST/) (1).
Xer recombination assays.
Xer recombination assays were carried out as described previously (36). Briefly, dimers of recombinant plasmids to be tested were prepared by transforming E. coli JC8679, culturing the transformed strains and extracting plasmid DNA, which was electrophoresed in 0.7% agarose gels. The DNA corresponding to the dimer was purified from the agarose gels using a QIAquick gel extraction kit (Qiagen). Since dimers run very close to the position of open circular monomer DNA, the isolated samples were used to transform the XerD-deficient E. coli DS9028. Since this strain cannot resolve the dimers, some colonies carry the monomer, and some others carry the dimers, allowing the isolation of transformants that have a plasmid dimer. Purified plasmid dimers were introduced by chemical transformation into E. coli DS941, the transformants were cultured overnight at 37°C in Lennox L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or medium containing the same concentrations of tryptone and yeast extract with no added NaCl, and the plasmid content was analyzed by agarose gel electrophoresis.
Nucleotide sequence accession numbers.
The nucleotide sequences of plasmids pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4 have been deposited in GenBank under accession numbers GQ412195, JN979787, JN985534, and JN995611, respectively.
RESULTS AND DISCUSSION
qnrB19 genetic environment.
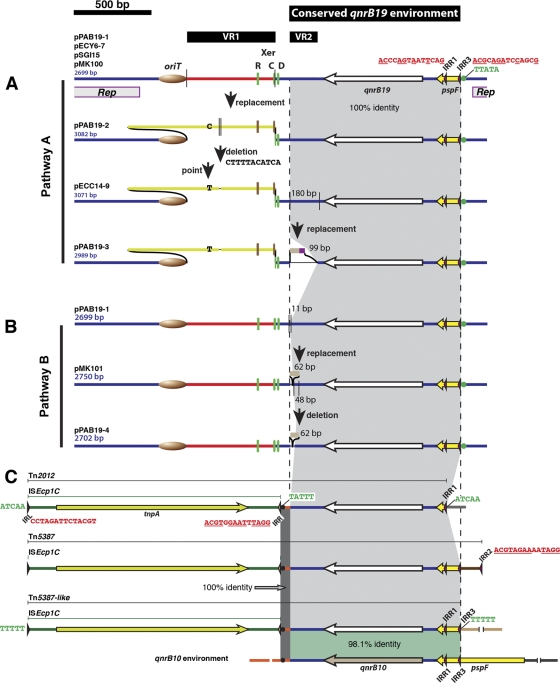
Four qnrB19-containing plasmids, pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4, isolated from clinical Salmonella spp. or E. coli isolates were sequenced and comparison analyses showed that they are identical or highly related to other known small qnrB19-carrying plasmids isolated from enterobacteria (Fig. 1A and B, also see Table 1 for information about these other plasmids). Plasmid pPAB19-1 is identical to a plasmid that has been isolated from E. coli, E. fergusonii, E. hermanii, Enterobacter aerogenes, K. pneumoniae, S. enterica, and Kluyvera ascorbata strains from the Netherlands, Bolivia, Peru, and Colombia by different research groups and has received several names (pSGI15, pECY6-7, and pMK100) (12, 17, 21, 22). Plasmids pPAB19-2, pPAB19-3, and pPAB19-4 are highly related to pPAB19-1 and other plasmids such as pECC14-9 (Bolivia and Peru) and pMK101 (Colombia) isolated from E. coli and S. enterica, respectively (17, 21, 22). A ColE1-type replication region locus is within the fragment that is common to all plasmids and share 93% identity with other non-qnrB-carrying plasmids from enterobacteria such as pJHCMW1 (29).
Fig 1.
Comparative diagram of qnrB19-harboring elements. Colors indicate identical nucleotide sequences. For the sake of clarity, in two regions across the different elements a gray shading was added to indicate the portions with identical sequences. In the case of the qnrB10 environment, a green shading was added to show the 98.1 identity region. Variable regions VR1 and VR2, as well as the conserved qnrB19 environment, are indicated by black bars at the top of the genetic maps. The thin vertical lines in the plasmid maps indicate the edges of the DNA fragments replaced or deleted in each rearrangement. The Xer recombination sites components are indicated as follows: R, ARG box (Arg-binding region); C, XerC-binding site; D, XerD-binding site. The different colors of XerC, XerD, and ArgR binding sites indicate that they have different sequences. The oriT is indicated as a brown oval. IRL, IRR, IRR1, IRR2, and IRR3 are indicated by slender arrowheads, and their sequences are shown in red (underlined nucleotides correspond to a perfect reverse complement of the IRL sequence). The target site duplications of ISEcp1C or ISEcp1C-based transposons are shown in green (the TTATA sequence after IRR3 in the different plasmids and the TATTT after IRR in the transposons are emphasized by green and black dots, respectively). The location of the replication region (Rep) of all plasmids is shown below the pPAB19-1 genetic map.
The genetic variations observed in this group of plasmids can be fitted into two pathways of plasmid evolution starting from the most common plasmid pPAB19-1/pSGI15/pECY6-7/pMK100. In pathway A, the plasmids pPAB19-2 and pECC14-9 could have been generated by a first event consisting of a replacement of a DNA region that we named variable region 1 (VR1), flanked by the oriT locus and a Xer site-specific recombination site, followed by a second event consisting of a point mutation and a deletion of one 11-bp copy from a region that includes 12 11-bp imperfect repeats (Fig. 1A). We cannot discard the possibility that pECC14-9 is the direct result of a DNA replacement in pPAB19-1 and in that case pPAB19-2 was generated by a point mutation and the addition of a copy of the 11-bp imperfect repeat (Fig. 1A). Plasmids pMK101 and pPAB19-4 could have resulted from a replacement and a deletion in a separate region that we called variable region 2 (VR2) (Fig. 1B). Plasmid pPAB19-3 has undergone modifications in both VR1 and VR2 (Fig. 1A).
The genetic environment of qnrB19 in these plasmids, as well as in the ISEcp1C-based transposons Tn2012, Tn5387, and Tn5387-like, which have been proposed as important players in qnrB19 mobilization (6, 9, 27), are very well conserved (shaded area in Fig. 1). ISEcp1 has 14-bp inverted repeats (IRs) and preferentially uses 5-bp AT-rich sites as the insertion targets. ISEcp1 is characterized by its ability to mobilize DNA fragments located adjacent to the IR right (IRR) of the element most probably by using alternative IRR-like sequences (named as IRR1, IRR2, IRR3, and so on) that have partial identity with IRR and might be recognized as such by the ISEcp1 transposase (24). Our analyses also showed that the common qnrB19 genetic environment is highly homologous (98.1% identity) to that of the qnrB10 allele (Fig. 1C), which was recently described in a study on clinical enterobacteria from Argentina (25), where it was found to be associated to ISCR1. In that study it was also proposed that several ISCR1-associated qnrB alleles could have been originally located in similar chromosomal contexts, i.e., downstream of pspF, the transcriptional activator of the stress-inducible psp operon (25). This hypothesis is supported by recent results indicating that the chromosome of Citrobacter spp. is the likely source of plasmid-mediated qnrB (15).
On the basis of the findings described above, we hypothesize that the original environment of qnrB19 could have been similar to that of qnrB10, i.e., it was located downstream of pspF. Then, ISEcp1C could have transposed to the TATTT site downstream of qnrB19 (see Fig. 1C) and captured this gene through subsequent transposition events by using the alternative IRR-like sequences IRR1 or IRR3, giving rise to Tn2012 or Tn5387/Tn5387-like, respectively. In turn, Tn5387-like could have transposed to an ancestor of the small ColE1-type plasmids analyzed here. As recently suggested by Palecchi et al. (22), the presence of a TTATA site at the edge of the conserved qnrB19 environment in all plasmids (Fig. 1A and B) supports this assumption. However, these plasmids lack ISEcp1C. It has been proposed that this insertion sequence could have been lost by excision using another IRR-like site located a few base pairs beyond IRR (22). However, we think that this is just one of many possibilities. One point of excision is at the edge of a region with high variability (VR2, Fig. 1), which could indicate other mechanisms of excision at this end of the insertion sequence. Furthermore, in Tn5387 and Tn5387-like we did not find any putative 14-bp sequence at the left edge of the conserved qnrB19 environment that matched the signatures of an IRR-like site (≥3 nucleotide identities with IRR, including a GG or CG at its 3′ end) (24). Therefore, other rearrangements, including the possibility of another sequence playing a role similar to the IRRs but that has the potential to induce further modifications, may have occurred during excision of ISEcp1C.
The VR1 region: an oriT/Xer recombination-based mechanism of plasmid evolution?
VR1 regions are flanked by an oriT locus and a Xer site-specific recombination-like site (Fig. 1). The oriT locus of the plasmids described here shares extensive homology with others that can utilize the ColE1 mob functions such as that in the plasmid pJHCMW1 (Fig. 2). The pJHCMW1 oriT was shown to be functional in mating experiments using as donor strain E. coli DH5α harboring a recombinant plasmid that includes oriT and a helper plasmid carrying the ColE1 mob genes and the RK2 tra genes (8).
Fig 2.
Comparison of oriT regions. Comparison was carried out using CLUSTAL W2. The oriT sites from pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4 are identical.
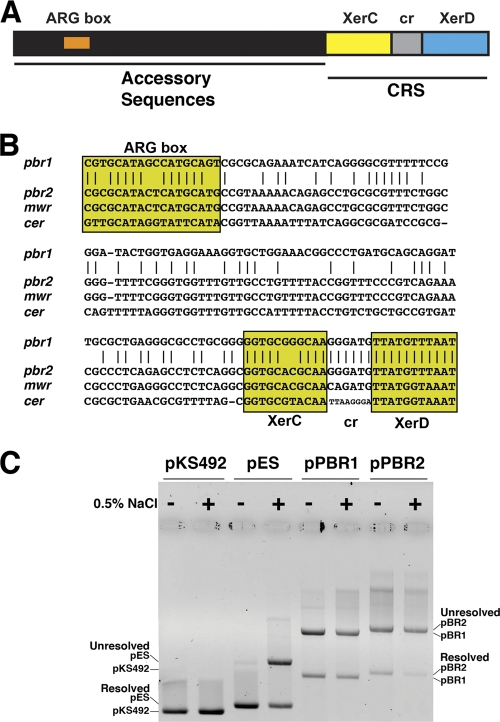
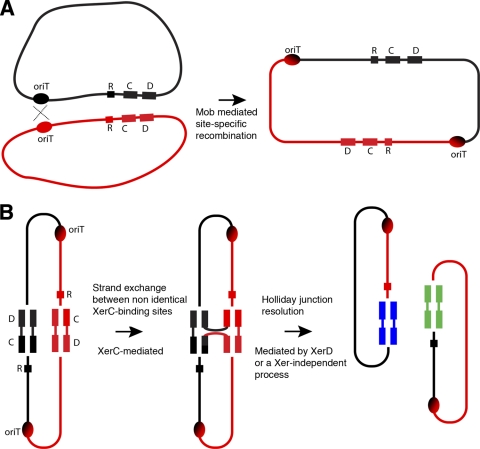
A diagram of the general structure of plasmids' Xer recombination sites is shown in Fig. 3A. They usually consist of a core recombination site were the strand exchanges occur and a stretch of ∼180 bp known as accessory sequences that possess binding sites for the architectural proteins PepA and ArgR (Fig. 3B). The core recombination site includes two 11-nucleotide binding sites for the tyrosine recombinases XerC and XerD, separated by a 6- to 8-nucleotide central region. Binding of the architectural proteins to the accessory sequences facilitates formation of a synaptic complex (26), where XerC is activated through interaction with XerD and catalyzes the exchange of the first pair of strands, which results in the formation of a Holliday junction (11) that, in the case of cer (ColE1) or mwr (pJHCMW1), is resolved by Xer-independent processes (3, 34). Two different versions of Xer recombination sites were found at one end of the VR1 in plasmids pPAB19-1, pPAB19-2, pPAB19-3, and pPAB19-4 (schematically shown in Fig. 1). The nucleotide sequence of these sites, from here on called pbr1 (pPAB19-1 Xer recombination site present in pPAB19-1 and pPAB19-4) and pbr2 (pPAB19–2 Xer recombination site present in pPAB19-2 and pPAB19-3), as well as a comparison among themselves and to the nucleotide sequences of other well-known Xer recombination sites are shown in Fig. 3B. The pbr1 and pbr2 sites include identical central regions and XerD-binding sites but seem to derive all or part of the XerC-binding site and the accessory sequences from a different source (Fig. 3B). To test whether they are functional, we generated dimers of recombinant clones pPBR1 and pPBR2, which include pbr1 or pbr2, respectively, and carried out resolution assays. Dimers of recombinant clones including mwr or cer, the Xer recombination sites from pJHCMW1 and ColE1, were used as controls. We have shown before that the concentration of NaCl in the medium where E. coli harboring the dimers is cultured affects the efficiency of resolution for some Xer recombination sites. Therefore, we carried out these assays using culture medium containing 0 or 0.5% added NaCl. It is known that dimers containing cer are efficiently resolved regardless of the NaCl concentration in the culture medium and dimers containing mwr increase the resolution efficiency as the NaCl in the medium decreases (23, 36). Figure 3C shows that the level of resolution of dimers containing pbr1 or pbr2 is considerably lower than those of the controls containing cer or mwr. We have shown before that levels of resolution as low as that one exhibited by mwr-harboring dimers in cells grown in L broth containing 0.5% NaCl are not enough to confer stability by multimer resolution (34, 35). As shown in Fig. 3C, the levels of resolution of dimers containing pbr1 or pbr2 are not significantly modified when the cells are cultured in the absence of added NaCl, and in both cases the efficiency is significantly lower than those of mwr, suggesting that they are unable to stabilize the host plasmid by multimer resolution under the conditions commonly tested in the laboratory. It is possible that pbr1 and pbr2, as well as other Xer recombination sites that present low efficiency, may not have as a main role plasmid stabilization, but they contribute to plasmid evolution by working in concert with the nearby oriT locus. Based on their comparison of the pECY6-7 and pECC14-9 nucleotide sequences, Pallechi et al. recently suggested that the Xer recombination sites found in these two plasmids could play a role in plasmid evolution (22). We postulate here a mechanism for swapping of DNA fragments flanked by an oriT locus and a Xer recombination site (Fig. 4). In this model the first event involves site-specific recombination at oriT, a process that has been described before for ColE1 and other plasmids' oriT loci and depends exclusively on the presence of the oriT site and the nickase activity (19, 39). Site-specific recombination at the oriT sites of two plasmids, represented as red and black in Fig. 4, leads to integration (Fig. 4A). The cointegrate includes two directly positioned nonidentical Xer recombination sites that may serve as substrate for a second site-specific recombination event mediated by Xer (Fig. 4B). Although Xer site-specific recombination usually occurs between directly repeated identical Xer sites, recombination events between nonidentical Xer sites has been described before (7, 37). It is not possible for us at this time to determine whether the accessory sequences play a role in positioning the XerC and XerD binding sites to form a synaptic complex. In the second recombination event the recombinase XerC mediates the strand exchange of one pair of strands to form a Holliday junction. In Fig. 4B the point of action of XerC has been drawn at the position it usually happens, but we do not know whether the nick and religation in this particular reaction can occur at another position. The Holliday junction is then resolved by XerD-catalyzed strand exchange of the second pair of strands or by a Xer-independent process such as replication (Fig. 4B). Since we do not know the site of action of XerC or the mechanism of resolution of the Holliday junction, we cannot predict the final nucleotide sequence of the newly formed Xer recombination sites. However, if the XerC binding sites and/or the central regions are not identical, we can predict that the newly formed Xer recombination sites will have a modified structure with respect to the original ones (represented by the different colors of the newly formed sites in Fig. 4B).
Fig 3.
(A) Schematic diagram of plasmid's Xer recombination sites. The sites contain a core recombination region that includes the 11-bp XerC and XerD binding sites and a central region (6 to 8 bp),and accessory sequences (180 bp) with which the architectural proteins ArgR and PepA interact. The diagram is not drawn to scale. (B) Comparison of the nucleotide sequences of pbr1 (present in pPAB19-1 and pPAB19-4), pbr2 (present in pPAB19-2 and pPAB19-3), mwr, and cer. The ArgR-binding site (ARG box) and different regions of the core recombination site (CRS) are shown. XerC, XerC-binding site; XerD, XerD-binding site; cr, central region. Identical nucleotides between pbr1 and pbr2 are indicated by vertical lines. (C) Dimer resolution assay. Dimers of plasmids pKS492 (cer), pES (mwr), pPBR1 (pbr1), and pPBR2 (pbr2) were introduced by transformation into E. coli DS941. The cells were cultured in medium containing 0 or 0.5% added NaCl in the presence of 100 μg of ampicillin per ml for 20 generations. Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The positions of dimers and monomers are indicated at the sides.
Fig 4.
Model of exchange of the DNA region flanked by oriT and Xer recombination sites. (A) Two plasmids (black and red) form a cointegrate through oriT site-specific recombination mediated by the nickase supplied by a resident plasmid that includes a mob gene. R, ARG box; C, XerC-binding site; D, XerD-binding site. (B) The cointegrate is resolved through Xer site-specific mediated recombination. Whether the accessory sequences are needed to facilitate formation of a synaptic complex is still unknown. Double-stranded DNA is shown only at the core recombination site to show the exchange of strands. XerC mediates the exchange of the first pair of strands at an undetermined nucleotide inside the XerC-binding site and forms a Holliday junction. Resolution of the Holliday junction may occur through XerD-mediated exchange of the second pair of strands or through a Xer-independent process such as replication. Since we do not know the mechanism of resolution of the Holliday junction we cannot predict the final nucleotide sequence of the newly formed Xer recombination sites. For this reason we indicate them in blue and green. Resolution of the Holliday junction results in two plasmids that have swapped the region limited by oriT and the Xer recombination site.
As a consequence of the two successive site-specific recombination reactions, the plasmids have exchanged the fragment that includes the whole region between oriT and the Xer recombination site, and the Xer recombination site in each plasmid has also been modified (see Fig. 4B). The extension of the modification of the Xer recombination site is dependent on the similarities of the recombination sites, the point of action of XerC, and the mechanism of resolution of the Holliday junction. Future studies will permit us to test the model and define it with more exactitude. The combination oriT-Xer recombination site could be considered one more element used by plasmids to evolve by swapping DNA regions. Unlike integrons that mediate acquisition or shedding of genes flanked by specific target sites (attI and attC) by site-specific recombination mediated by an integrase (5), this oriT-Xer element swaps DNA regions that do not necessarily include genes and do not present any requirements other than being located between an oriT and a Xer recombination site.
ACKNOWLEDGMENTS
This study was supported by a grant from ANPCYT (PICT 2007–01804), Buenos Aires, Argentina, to A.P.; a Public Health Service grant 2R15AI047115 (to M.E.T.) from the National Institutes of Health; and a grant from the CSU State Fund to M.E.T. A.S.-B. and E.A. were supported by fellowships from CONICET and ANPCYT, respectively. T.T. was supported by grant MHIRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institutes of Health.
Footnotes
Published ahead of print 30 January 2012
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Andres P, et al. 2010. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr C2-1471 [Google Scholar]
- 3. Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. 2000. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 299:391–403 [DOI] [PubMed] [Google Scholar]
- 4. Bui D, et al. 2006. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 188:2812–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu. Rev. Genet. 44:141–166 [DOI] [PubMed] [Google Scholar]
- 6. Cattoir V, Nordmann P, Silva-Sanchez J, Espinal P, Poirel L. 2008. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother. 52:2929–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Das B, Bischerour J, Barre FX. 2011. VGJphi integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc. Natl. Acad. Sci. U. S. A. 108:2516–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dery KJ, et al. 1997. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid 38:97–105 [DOI] [PubMed] [Google Scholar]
- 9. Dionisi AM, Lucarelli C, Owczarek S, Luzzi I, Villa L. 2009. Characterization of the plasmid-borne quinolone resistance gene qnrB19 in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53:4019–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 11. Hallet B, Arciszewska LK, Sherratt DJ. 1999. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell 4:949–959 [DOI] [PubMed] [Google Scholar]
- 12. Hammerl JA, et al. 2010. pSGI15, a small ColE-like qnrB19 plasmid of a Salmonella enterica serovar Typhimurium strain carrying Salmonella genomic island 1 (SGI1). J. Antimicrob. Chemother. 65:173–175 [DOI] [PubMed] [Google Scholar]
- 13. Jacoby G, et al. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jacoby GA, Gacharna N, Black TA, Miller GH, Hooper DC. 2009. Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob. Agents Chemother. 53:1665–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacoby GA, Griffin C, Hooper DC. 2011. Citrobacter spp. as a source of qnrB alleles. Antimicrob. Agents Chemother. 55:4979–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacoby GA, et al. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karczmarczyk M, et al. 2010. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 313:10–19 [DOI] [PubMed] [Google Scholar]
- 18. Larkin MA, et al. 2007. CLUSTAL W and CLUSTAL X, version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 19. Llosa M, Bolland S, Grandoso G, de la Cruz F. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J. Bacteriol. 176:3210–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez-Martinez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799 [DOI] [PubMed] [Google Scholar]
- 21. Pallecchi L, et al. 2011. Small qnrB-harboring ColE-like plasmids widespread in commensal enterobacteria from a remote Amazonas population not exposed to antibiotics. J. Antimicrob. Chemother. 66:1176–1178 [DOI] [PubMed] [Google Scholar]
- 22. Pallecchi L, et al. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob. Agents Chemother. 54:678–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pham H, Dery KJ, Sherratt DJ, Tolmasky ME. 2002. Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination. J. Bacteriol. 184:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quiroga MP, et al. 2007. Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob. Agents Chemother. 51:4466–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reijns M, Lu Y, Leach S, Colloms SD. 2005. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 57:927–941 [DOI] [PubMed] [Google Scholar]
- 27. Rice LB, et al. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3427–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodriguez-Martinez JM, Cano ME, Velasco C, Martinez-Martinez L, Pascual A. 2011. Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17:149–182 [DOI] [PubMed] [Google Scholar]
- 29. Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spiers AJ, Sherratt DJ. 1997. Relating primary structure to function in the Escherichia coli XerD site-specific recombinase. Mol. Microbiol. 24:1071–1082 [DOI] [PubMed] [Google Scholar]
- 31. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Summers DK, Sherratt DJ. 1984. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097–1103 [DOI] [PubMed] [Google Scholar]
- 33. Summers DK, Sherratt DJ. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tolmasky ME, Colloms S, Blakely G, Sherratt DJ. 2000. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146:581–589 [DOI] [PubMed] [Google Scholar]
- 35. Tran T, Sherratt DJ, Tolmasky ME. 2010. fpr, a deficient Xer recombination site from a Salmonella plasmid, fails to confer stability by dimer resolution: comparative studies with the pJHCMW1 mwr site. J. Bacteriol. 192:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trigueros S, et al. 2009. mwr Xer site-specific recombination is hypersensitive to DNA supercoiling. Nucleic Acids Res. 37:3580–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Val ME, et al. 2005. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19:559–566 [DOI] [PubMed] [Google Scholar]
- 38. Vieira J, Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268 [DOI] [PubMed] [Google Scholar]
- 39. Warren GJ, Clark AJ. 1980. Sequence-specific recombination of plasmid ColE1. Proc. Natl. Acad. Sci. U. S. A. 77:6724–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]